Abstract
In the title compound, C16H14N2O3, the essentially planar 1,3,4-oxadiazole ring [maximum deviation = 0.0021 (11) Å] is inclined at dihedral angles of 8.06 (6) and 11.21 (6)° with respect to the two benzene rings; the dihedral angle between the latter rings is 11.66 (5)°. In the crystal, short intermolecular C⋯O interactions [2.9968 (15) Å] connect adjacent molecules into chains propagating in [203]. The crystal structure is further stabilized by weak intermolecular C—H⋯π interactions.
Related literature
For general background to and applications of the title compound, see: Andersen et al. (1994 ▶); Clitherow et al. (1996 ▶); Hegde et al. (2008 ▶); Rai et al. (2008 ▶); Showell et al. (1991 ▶). For closely related 2,5-diphenyl-1,3,4-oxadiazole structures, see: Reck et al. (2003a
▶,b
▶); Franco et al. (2003 ▶). For the stability of the temperature controller used in the data collection, see: Cosier & Glazer (1986 ▶).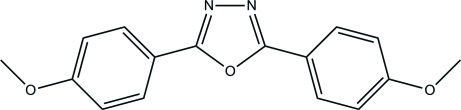
Experimental
Crystal data
C16H14N2O3
M r = 282.29
Monoclinic,

a = 10.7525 (2) Å
b = 11.8973 (2) Å
c = 11.6340 (2) Å
β = 115.434 (1)°
V = 1344.04 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.53 × 0.49 × 0.09 mm
Data collection
Bruker SMART APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.950, T max = 0.992
17061 measured reflections
4744 independent reflections
3744 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.137
S = 1.03
4744 reflections
246 parameters
All H-atom parameters refined
Δρmax = 0.38 e Å−3
Δρmin = −0.30 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810044405/hb5714sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810044405/hb5714Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of C9–C14 and C1–C6 benzene rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C15—H15A⋯Cg1i | 1.031 (14) | 2.563 (16) | 3.4903 (14) | 149.4 (10) |
| C15—H15B⋯Cg2ii | 0.992 (14) | 2.994 (16) | 3.8804 (14) | 149.4 (11) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors thank Universiti Sains Malaysia (USM) for the Research University Grant (No. 1001/PFIZIK/811160).
supplementary crystallographic information
Comment
Heterocyclic compounds are becoming increasingly important in recent years due to their pharmacological activities (Rai et al., 2008). Nitrogen- and oxygen-containing five/six-membered heterocyclic compounds are of enormous significance in the field of medicinal chemistry (Hegde et al., 2008). Oxadiazoles play a very vital role in the preparation of various biologically active drugs with anti-inflammatory (Andersen et al., 1994) and anti-cancer (Showell et al., 1991) activities. The results of biological studies showed that oxadiazole derivatives also possess significant anti-inflammatory, analgesic and minimum ulcerogenic and lipid per-oxidation (Clitherow et al., 1996) properties.
In the title oxadiazole compound (Fig. 1), the 1,3,4-oxadiazole ring (C7/C8/N1/N2/O1) is essentially planar, with a maximum deviation of -0.0021 (11) at atom N1. The C1-C6 and C9-C14 phenyl rings are inclined at dihedral angles of 11.21 (6) and 8.06 (6)°, respectively, with the 1,3,4-oxadiazole ring. The geometric parameters agree well with those observed in closely related 2,5-diphenyl-1,3,4-oxadiazole structures (Reck et al., 2003a,b; Franco et al., 2003).
In the crystal, no significant intermolecular hydrogen bond is observed. The interesting feature of the crystal structure is the intermolecular short C15···O3 interactions [2.9968 (15) Å, symmetry code: x+1, -y+1/2, z+3/2], which is significantly shorter than the sum of the Van der Waals radii of the relevant atoms (3.22 Å), interconnecting adjacent molecules into one-dimensional chains propagating along the [203] direction (Fig. 2). Further stabilization of the crystal structure is provided by weak intermolecular C15—H15A···Cg1 and C15—H15B···Cg2 interactions (Table 1) where Cg1 and Cg2 are the centroids of C9-C14 and C1-C6 phenyl rings, respectively.
Experimental
An equimolar mixture of 4-methoxybenzoic acid (0.01 mol) and 4-methoxybenzhydrazide was dissolved in POCl3 (25–30 ml) by gentle warming. The solution is refluxed for about 12 h. Excess of POCl3 is distilled off, and then the reaction mixture is poured into crushed ice. The separated solid was filtered, washed with water and dried. It was then recrystallized from ethanol. Yellow blocks of (I) were obtained from ethanol by slow evaporation.
Refinement
All H atoms were located from difference Fourier map and allowed to refine freely with range of C—H = 0.916 (14) – 1.031 (14) Å.
Figures
Fig. 1.
The asymmetric unit of the title compound, showing 50 % probability displacement ellipsoids for non-H atoms.
Fig. 2.

The crystal structure of the title compound, viewed along the a axis, showing molecules being interconnected into one-dimensional chains. H atoms have been omitted for clarity and intermolecular short interactions are shown as dashed lines.
Crystal data
| C16H14N2O3 | F(000) = 592 |
| Mr = 282.29 | Dx = 1.395 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5014 reflections |
| a = 10.7525 (2) Å | θ = 2.6–32.2° |
| b = 11.8973 (2) Å | µ = 0.10 mm−1 |
| c = 11.6340 (2) Å | T = 100 K |
| β = 115.434 (1)° | Block, yellow |
| V = 1344.04 (4) Å3 | 0.53 × 0.49 × 0.09 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD diffractometer | 4744 independent reflections |
| Radiation source: fine-focus sealed tube | 3744 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| φ and ω scans | θmax = 32.3°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −16→15 |
| Tmin = 0.950, Tmax = 0.992 | k = −17→17 |
| 17061 measured reflections | l = −14→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.137 | All H-atom parameters refined |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.079P)2 + 0.1649P] where P = (Fo2 + 2Fc2)/3 |
| 4744 reflections | (Δ/σ)max = 0.001 |
| 246 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1)K. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.89032 (6) | 0.25303 (6) | 0.21170 (6) | 0.01974 (15) | |
| O2 | 0.49848 (8) | 0.12006 (7) | −0.35883 (8) | 0.0333 (2) | |
| O3 | 1.26608 (8) | 0.16634 (6) | 0.80451 (7) | 0.02542 (17) | |
| N1 | 0.84586 (9) | 0.42831 (7) | 0.14199 (9) | 0.02542 (19) | |
| N2 | 0.93713 (9) | 0.43051 (7) | 0.27255 (8) | 0.02513 (19) | |
| C1 | 0.64742 (10) | 0.33907 (9) | −0.11316 (10) | 0.0243 (2) | |
| C2 | 0.56651 (10) | 0.29145 (9) | −0.23102 (10) | 0.0257 (2) | |
| C3 | 0.57391 (10) | 0.17639 (9) | −0.24904 (10) | 0.0247 (2) | |
| C4 | 0.66395 (10) | 0.10958 (9) | −0.14829 (10) | 0.0264 (2) | |
| C5 | 0.74483 (10) | 0.15763 (8) | −0.03171 (10) | 0.0234 (2) | |
| C6 | 0.73715 (9) | 0.27300 (8) | −0.01231 (9) | 0.02039 (18) | |
| C7 | 0.82192 (9) | 0.32333 (8) | 0.11073 (9) | 0.02031 (18) | |
| C8 | 0.95953 (9) | 0.32608 (7) | 0.30883 (9) | 0.01932 (18) | |
| C9 | 1.04150 (9) | 0.28123 (7) | 0.43534 (9) | 0.01863 (17) | |
| C10 | 1.06169 (9) | 0.16598 (8) | 0.45607 (9) | 0.01989 (18) | |
| C11 | 1.13636 (9) | 0.12351 (8) | 0.57833 (9) | 0.02060 (18) | |
| C12 | 1.19097 (9) | 0.19768 (8) | 0.68094 (9) | 0.01974 (18) | |
| C13 | 1.17027 (10) | 0.31372 (8) | 0.66105 (10) | 0.0232 (2) | |
| C14 | 1.09745 (10) | 0.35469 (8) | 0.53959 (10) | 0.02276 (19) | |
| C15 | 0.42071 (11) | 0.18563 (12) | −0.47047 (11) | 0.0320 (2) | |
| C16 | 1.29532 (13) | 0.05018 (10) | 0.82938 (11) | 0.0312 (2) | |
| H1A | 0.6395 (13) | 0.4157 (12) | −0.1004 (13) | 0.031 (3)* | |
| H2A | 0.5040 (16) | 0.3408 (13) | −0.2994 (15) | 0.044 (4)* | |
| H4A | 0.6699 (14) | 0.0323 (13) | −0.1637 (14) | 0.034 (4)* | |
| H5A | 0.8051 (14) | 0.1104 (12) | 0.0334 (13) | 0.033 (4)* | |
| H10A | 1.0288 (13) | 0.1131 (11) | 0.3879 (13) | 0.028 (3)* | |
| H11A | 1.1508 (14) | 0.0475 (12) | 0.5874 (14) | 0.033 (3)* | |
| H13A | 1.2011 (15) | 0.3619 (13) | 0.7306 (14) | 0.039 (4)* | |
| H14A | 1.0822 (13) | 0.4342 (12) | 0.5266 (13) | 0.031 (3)* | |
| H15A | 0.3446 (14) | 0.2322 (11) | −0.4612 (13) | 0.031 (3)* | |
| H15B | 0.4821 (14) | 0.2357 (12) | −0.4910 (14) | 0.030 (3)* | |
| H15C | 0.3802 (16) | 0.1299 (14) | −0.5399 (15) | 0.043 (4)* | |
| H16A | 1.2087 (16) | 0.0060 (13) | 0.8025 (15) | 0.044 (4)* | |
| H16B | 1.3518 (15) | 0.0229 (13) | 0.7875 (14) | 0.040 (4)* | |
| H16C | 1.3490 (16) | 0.0457 (14) | 0.9238 (16) | 0.046 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0201 (3) | 0.0161 (3) | 0.0210 (3) | 0.0006 (2) | 0.0069 (2) | 0.0022 (2) |
| O2 | 0.0298 (4) | 0.0347 (4) | 0.0238 (4) | −0.0009 (3) | 0.0003 (3) | 0.0011 (3) |
| O3 | 0.0325 (4) | 0.0234 (4) | 0.0183 (3) | 0.0021 (3) | 0.0089 (3) | 0.0011 (3) |
| N1 | 0.0316 (4) | 0.0193 (4) | 0.0244 (4) | 0.0024 (3) | 0.0112 (3) | 0.0035 (3) |
| N2 | 0.0324 (4) | 0.0184 (4) | 0.0236 (4) | 0.0022 (3) | 0.0111 (3) | 0.0020 (3) |
| C1 | 0.0247 (4) | 0.0219 (5) | 0.0262 (5) | 0.0025 (3) | 0.0108 (4) | 0.0062 (4) |
| C2 | 0.0220 (4) | 0.0283 (5) | 0.0246 (5) | 0.0024 (3) | 0.0080 (4) | 0.0086 (4) |
| C3 | 0.0197 (4) | 0.0282 (5) | 0.0224 (4) | −0.0015 (3) | 0.0054 (3) | 0.0031 (4) |
| C4 | 0.0243 (4) | 0.0222 (5) | 0.0265 (5) | 0.0002 (3) | 0.0050 (4) | 0.0023 (4) |
| C5 | 0.0210 (4) | 0.0217 (4) | 0.0237 (4) | 0.0009 (3) | 0.0058 (3) | 0.0050 (3) |
| C6 | 0.0183 (4) | 0.0213 (4) | 0.0219 (4) | 0.0005 (3) | 0.0090 (3) | 0.0045 (3) |
| C7 | 0.0204 (4) | 0.0192 (4) | 0.0223 (4) | 0.0031 (3) | 0.0101 (3) | 0.0054 (3) |
| C8 | 0.0210 (4) | 0.0165 (4) | 0.0217 (4) | 0.0007 (3) | 0.0103 (3) | −0.0001 (3) |
| C9 | 0.0192 (4) | 0.0161 (4) | 0.0213 (4) | 0.0001 (3) | 0.0092 (3) | −0.0003 (3) |
| C10 | 0.0201 (4) | 0.0161 (4) | 0.0220 (4) | −0.0010 (3) | 0.0076 (3) | −0.0020 (3) |
| C11 | 0.0222 (4) | 0.0156 (4) | 0.0228 (4) | −0.0009 (3) | 0.0086 (3) | −0.0005 (3) |
| C12 | 0.0212 (4) | 0.0206 (4) | 0.0185 (4) | 0.0006 (3) | 0.0096 (3) | 0.0004 (3) |
| C13 | 0.0284 (4) | 0.0195 (4) | 0.0223 (4) | 0.0010 (3) | 0.0113 (4) | −0.0039 (3) |
| C14 | 0.0275 (4) | 0.0163 (4) | 0.0246 (4) | 0.0017 (3) | 0.0112 (4) | −0.0015 (3) |
| C15 | 0.0237 (4) | 0.0470 (7) | 0.0213 (5) | 0.0004 (4) | 0.0059 (4) | 0.0067 (4) |
| C16 | 0.0390 (6) | 0.0239 (5) | 0.0242 (5) | 0.0018 (4) | 0.0074 (4) | 0.0053 (4) |
Geometric parameters (Å, °)
| O1—C8 | 1.3652 (11) | C6—C7 | 1.4552 (13) |
| O1—C7 | 1.3704 (11) | C8—C9 | 1.4537 (13) |
| O2—C3 | 1.3597 (13) | C9—C10 | 1.3930 (12) |
| O2—C15 | 1.4359 (13) | C9—C14 | 1.4036 (13) |
| O3—C12 | 1.3641 (11) | C10—C11 | 1.3937 (13) |
| O3—C16 | 1.4193 (13) | C10—H10A | 0.954 (14) |
| N1—C7 | 1.2954 (13) | C11—C12 | 1.3952 (13) |
| N1—N2 | 1.4110 (12) | C11—H11A | 0.916 (14) |
| N2—C8 | 1.3010 (12) | C12—C13 | 1.4017 (14) |
| C1—C2 | 1.3905 (15) | C13—C14 | 1.3774 (14) |
| C1—C6 | 1.3975 (13) | C13—H13A | 0.929 (16) |
| C1—H1A | 0.934 (14) | C14—H14A | 0.960 (14) |
| C2—C3 | 1.3923 (15) | C15—H15A | 1.031 (14) |
| C2—H2A | 0.985 (16) | C15—H15B | 0.992 (14) |
| C3—C4 | 1.4029 (14) | C15—H15C | 0.990 (16) |
| C4—C5 | 1.3819 (14) | C16—H16A | 0.996 (16) |
| C4—H4A | 0.944 (15) | C16—H16B | 0.982 (16) |
| C5—C6 | 1.3992 (14) | C16—H16C | 0.999 (17) |
| C5—H5A | 0.943 (14) | ||
| C8—O1—C7 | 102.83 (7) | C10—C9—C8 | 121.21 (8) |
| C3—O2—C15 | 117.56 (9) | C14—C9—C8 | 119.66 (8) |
| C12—O3—C16 | 117.39 (8) | C9—C10—C11 | 120.82 (8) |
| C7—N1—N2 | 106.41 (8) | C9—C10—H10A | 122.1 (8) |
| C8—N2—N1 | 106.13 (8) | C11—C10—H10A | 117.1 (8) |
| C2—C1—C6 | 120.85 (9) | C10—C11—C12 | 119.35 (9) |
| C2—C1—H1A | 119.5 (9) | C10—C11—H11A | 118.0 (9) |
| C6—C1—H1A | 119.6 (9) | C12—C11—H11A | 122.5 (9) |
| C1—C2—C3 | 119.80 (9) | O3—C12—C11 | 124.75 (9) |
| C1—C2—H2A | 118.4 (9) | O3—C12—C13 | 115.05 (8) |
| C3—C2—H2A | 121.8 (9) | C11—C12—C13 | 120.20 (9) |
| O2—C3—C2 | 125.20 (9) | C14—C13—C12 | 119.87 (9) |
| O2—C3—C4 | 115.14 (9) | C14—C13—H13A | 120.6 (10) |
| C2—C3—C4 | 119.66 (9) | C12—C13—H13A | 119.5 (10) |
| C5—C4—C3 | 120.21 (10) | C13—C14—C9 | 120.66 (9) |
| C5—C4—H4A | 121.6 (9) | C13—C14—H14A | 119.5 (8) |
| C3—C4—H4A | 118.2 (9) | C9—C14—H14A | 119.8 (8) |
| C4—C5—C6 | 120.55 (9) | O2—C15—H15A | 112.1 (8) |
| C4—C5—H5A | 117.9 (9) | O2—C15—H15B | 110.8 (8) |
| C6—C5—H5A | 121.6 (9) | H15A—C15—H15B | 110.0 (11) |
| C1—C6—C5 | 118.92 (9) | O2—C15—H15C | 104.8 (10) |
| C1—C6—C7 | 120.62 (9) | H15A—C15—H15C | 110.8 (12) |
| C5—C6—C7 | 120.46 (8) | H15B—C15—H15C | 108.1 (12) |
| N1—C7—O1 | 112.27 (9) | O3—C16—H16A | 110.8 (9) |
| N1—C7—C6 | 129.63 (9) | O3—C16—H16B | 110.5 (9) |
| O1—C7—C6 | 118.09 (8) | H16A—C16—H16B | 111.3 (13) |
| N2—C8—O1 | 112.35 (8) | O3—C16—H16C | 104.4 (10) |
| N2—C8—C9 | 128.77 (9) | H16A—C16—H16C | 109.8 (13) |
| O1—C8—C9 | 118.84 (8) | H16B—C16—H16C | 109.8 (13) |
| C10—C9—C14 | 119.09 (9) | ||
| C7—N1—N2—C8 | −0.41 (11) | N1—N2—C8—O1 | 0.33 (11) |
| C6—C1—C2—C3 | 0.44 (16) | N1—N2—C8—C9 | −177.28 (9) |
| C15—O2—C3—C2 | 9.99 (16) | C7—O1—C8—N2 | −0.13 (10) |
| C15—O2—C3—C4 | −170.60 (10) | C7—O1—C8—C9 | 177.74 (8) |
| C1—C2—C3—O2 | 178.82 (10) | N2—C8—C9—C10 | −175.90 (10) |
| C1—C2—C3—C4 | −0.55 (16) | O1—C8—C9—C10 | 6.62 (13) |
| O2—C3—C4—C5 | −179.38 (10) | N2—C8—C9—C14 | 6.42 (16) |
| C2—C3—C4—C5 | 0.05 (16) | O1—C8—C9—C14 | −171.05 (8) |
| C3—C4—C5—C6 | 0.57 (16) | C14—C9—C10—C11 | −0.04 (14) |
| C2—C1—C6—C5 | 0.18 (15) | C8—C9—C10—C11 | −177.72 (9) |
| C2—C1—C6—C7 | 179.80 (9) | C9—C10—C11—C12 | 0.20 (14) |
| C4—C5—C6—C1 | −0.69 (15) | C16—O3—C12—C11 | 2.71 (14) |
| C4—C5—C6—C7 | 179.69 (9) | C16—O3—C12—C13 | −177.29 (9) |
| N2—N1—C7—O1 | 0.35 (11) | C10—C11—C12—O3 | −179.73 (8) |
| N2—N1—C7—C6 | −178.82 (9) | C10—C11—C12—C13 | 0.27 (14) |
| C8—O1—C7—N1 | −0.15 (10) | O3—C12—C13—C14 | 179.08 (9) |
| C8—O1—C7—C6 | 179.12 (8) | C11—C12—C13—C14 | −0.92 (15) |
| C1—C6—C7—N1 | −11.61 (16) | C12—C13—C14—C9 | 1.10 (15) |
| C5—C6—C7—N1 | 168.01 (10) | C10—C9—C14—C13 | −0.62 (14) |
| C1—C6—C7—O1 | 169.27 (9) | C8—C9—C14—C13 | 177.10 (9) |
| C5—C6—C7—O1 | −11.12 (13) |
Hydrogen-bond geometry (Å, °)
| Cg1 and Cg2 are the centroids of C9–C14 and C1–C6 benzene rings, respectively. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C15—H15A···Cg1i | 1.031 (14) | 2.563 (16) | 3.4903 (14) | 149.4 (10) |
| C15—H15B···Cg2ii | 0.992 (14) | 2.994 (16) | 3.8804 (14) | 149.4 (11) |
Symmetry codes: (i) x−1, y, z−1; (ii) x, −y−1/2, z−3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5714).
References
- Andersen, K. E., Jørgensen, A. S. & Braestrup, C. (1994). Eur. J. Med. Chem.29, 393–399.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Clitherow, J. W., Beswick, P., Irving, W. J., Scopes, D. I. C., Barnes, J. C., Clapham, J., Brown, J. D., Evans, D. J. & Hayes, A. G. (1996). Bioorg. Med. Chem. Lett.6, 833–838.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst.19, 105–107.
- Franco, O., Reck, G., Orgzall, I., Schulz, B. W. & Schulz, B. (2003). J. Mol. Struct.649, 219–230.
- Hegde, J. C., Girisha, K. S., Adhikari, A. & Kalluraya, B. (2008). Eur. J. Med. Chem.43, 2831–2834. [DOI] [PubMed]
- Rai, N. S., Kalluraya, B., Lingappa, B., Shenoy, S. & Puranic, V. G. (2008). Eur. J. Med. Chem.43, 1715–1720. [DOI] [PubMed]
- Reck, G., Orgzall, I. & Schulz, B. (2003a). Acta Cryst. E59, o1135–o1136. [DOI] [PubMed]
- Reck, G., Orgzall, I., Schulz, B. & Dietzel, B. (2003b). Acta Cryst. C59, o634–o635. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Showell, G. A., Gibbons, T. L., Kneen, C. O., MacLeod, A. M., Merchant, K., Suanders, J., Freedman, S. B., Patel, S. B. & Baker, R. (1991). J. Med. Chem.34, 1086–1094. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810044405/hb5714sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810044405/hb5714Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



