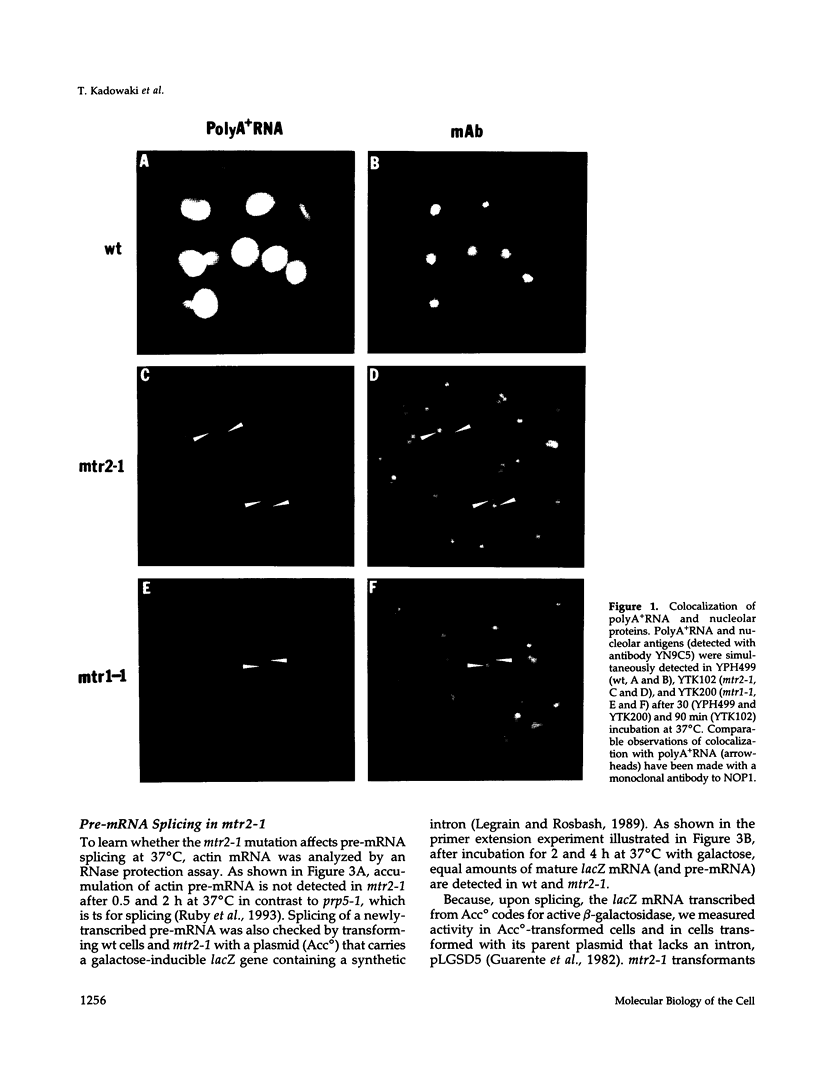
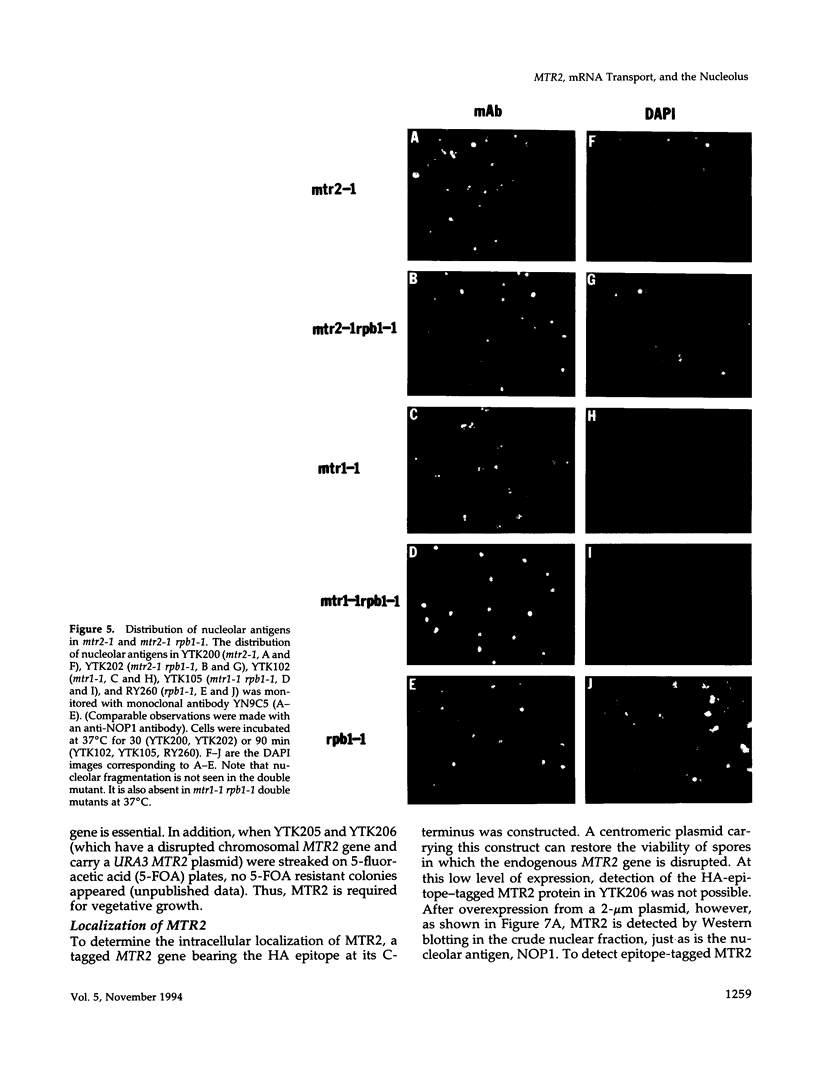
Abstract
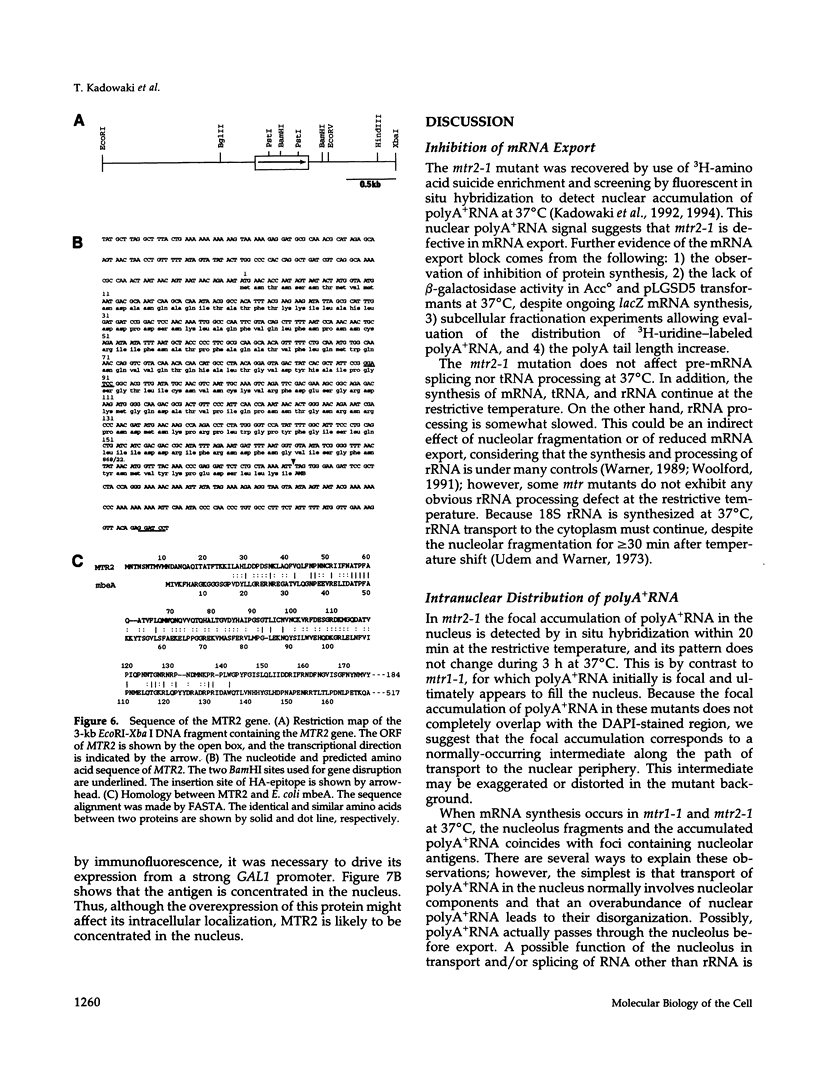
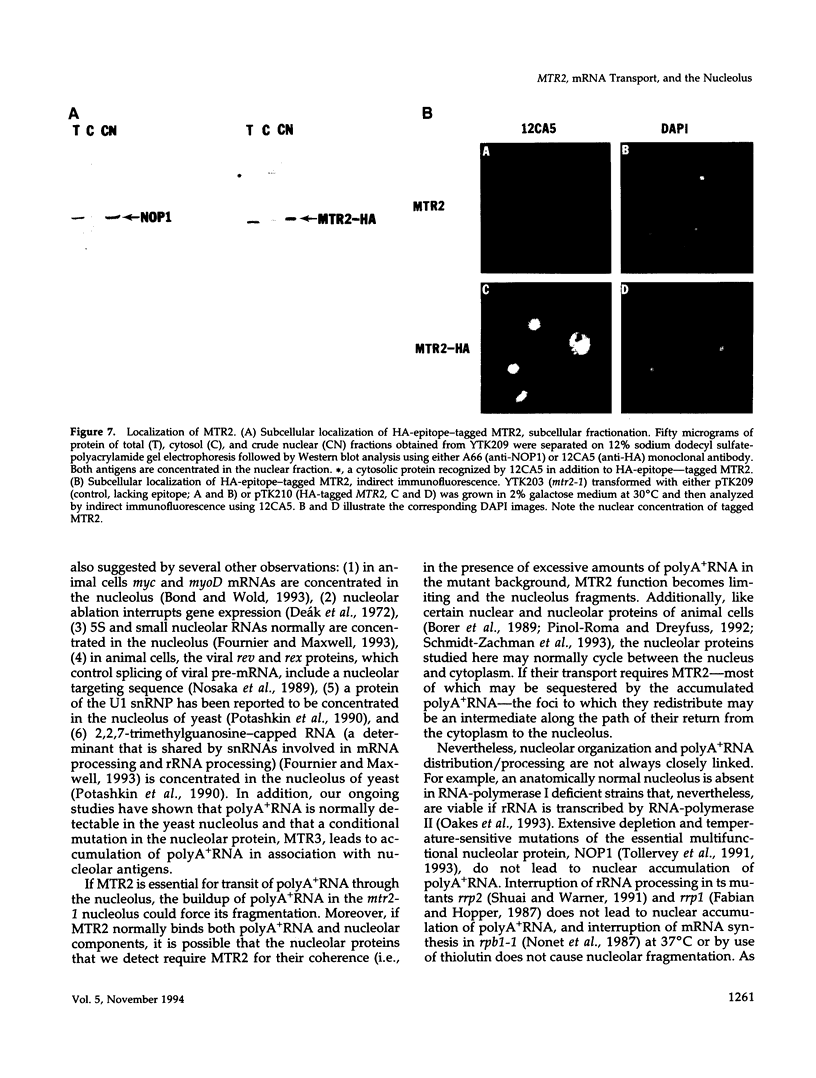
We have identified a set of genes that affect mRNA transport (mtr) from the nucleus to the cytoplasm of Saccharomyces cerevisiae. One of these genes, MTR2, has been cloned and shown to encode a novel 21-kDa nuclear protein that is essential for vegetative growth. MTR2 shows limited homology to a protein implicated in plasmid DNA transfer in Escherichia coli. PolyA+RNA accumulates within the nucleus of mtr2-1 in two to three foci at 37 degrees C. mRNA, tRNA, and rRNA synthesis continue as do pre-mRNA splicing, tRNA processing, and rRNA export at 37 degrees C. Under these conditions the polyA tail length increases, and protein synthesis is progressively inhibited. Nucleolar antigens also redistribute to two to three nuclear foci at 37 degrees C, and this redistribution depends on ongoing transcription by RNA polymerase II. Surprisingly, these foci coincide with the sites of polyA+RNA accumulation. Comparable colocalization and dependance on RNA polymerase II transcription is seen for the mtr1-1 mutant. The disorganization of the nucleolus thus depends on mRNA accumulation in these mutants. We discuss the possible functions of MTR2 and the yeast nucleolus in mRNA export.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Clark M. W., Vijayraghavan U., Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol Gen Genet. 1990 Oct;224(1):72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988 Jul;107(1):17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P., Dabauvalle M. C., Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J Cell Biol. 1992 Sep;118(6):1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond V. C., Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol. 1993 Jun;13(6):3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R. A., Lehner C. F., Eppenberger H. M., Nigg E. A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989 Feb 10;56(3):379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Boyd A. C., Archer J. A., Sherratt D. J. Characterization of the ColE1 mobilization region and its protein products. Mol Gen Genet. 1989 Jun;217(2-3):488–498. doi: 10.1007/BF02464922. [DOI] [PubMed] [Google Scholar]
- Deák I., Sidebottom E., Harris H. Further experiments on the role of the nucleolus in the expression of structural genes. J Cell Sci. 1972 Sep;11(2):379–391. doi: 10.1242/jcs.11.2.379. [DOI] [PubMed] [Google Scholar]
- Elliott D. J., Bowman D. S., Abovich N., Fay F. S., Rosbash M. A yeast splicing factor is localized in discrete subnuclear domains. EMBO J. 1992 Oct;11(10):3731–3736. doi: 10.1002/j.1460-2075.1992.tb05458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian G. R., Hopper A. K. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J Bacteriol. 1987 Apr;169(4):1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Traglia H. M., Dunst R. W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990 Aug;111(2):309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Chen S., Hitomi M., Jacobs E., Kumagai C., Liang S., Schneiter R., Singleton D., Wisniewska J., Tartakoff A. M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994 Aug;126(3):649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Goldfarb D., Spitz L. M., Tartakoff A. M., Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993 Jul;12(7):2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Zhao Y., Tartakoff A. M. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M., Stevens A., McCammon M., Douglas M. G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993 Jan;13(1):341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989 May 19;57(4):573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991 Jul 26;66(2):347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor B., Bonneaud N., Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol. 1991 Jun;11(6):3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan C., Copeland C. S., Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992 Mar;116(6):1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka T., Siomi H., Adachi Y., Ishibashi M., Kubota S., Maki M., Hatanaka M. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9798–9802. doi: 10.1073/pnas.86.24.9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M. W., Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol Cell Biol. 1993 Apr;13(4):2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992 Feb 20;355(6362):730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Potashkin J. A., Derby R. J., Spector D. L. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol Cell Biol. 1990 Jul;10(7):3524–3534. doi: 10.1128/mcb.10.7.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Chang T. H., Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993 Oct;7(10):1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M. S., Dargemont C., Kühn L. C., Nigg E. A. Nuclear export of proteins: the role of nuclear retention. Cell. 1993 Aug 13;74(3):493–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Warner J. R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991 Sep 25;19(18):5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Chiang A., Sadler I. Mutations that alter both localization and production of a yeast nuclear protein. Genes Dev. 1988 Jun;2(6):707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- Smitt W. W., Vlak J. M., Molenaar I., Rozijn T. H. Nucleolar function of the dense crescent in the yeast nucleus. A biochemical and ultrastructural study. Exp Cell Res. 1973 Aug;80(2):313–321. doi: 10.1016/0014-4827(73)90302-9. [DOI] [PubMed] [Google Scholar]
- Tipper D. J. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol. 1973 Oct;116(1):245–256. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993 Feb 12;72(3):443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973 Feb 25;248(4):1412–1416. [PubMed] [Google Scholar]
- Warner J. R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989 Jun;53(2):256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]








