Abstract
In the title compound, [Al(C29H41N2)(C2H2N2P)2], the AlIII atom is coordinated by four N atoms from β-diketiminate and 1,2,4-diazaphospholide ligands in a slightly distorted tetrahedral fashion.
Related literature
For similar related 1,2,4-diazaphospholide complexes, see: Schmidpeter & Willhalm (1984 ▶); Cui et al. (2000 ▶); Ding et al. (2001 ▶); Kumar et al. (2004 ▶, 2005 ▶); Zheng et al. (2006 ▶); Wan et al. (2008 ▶); Pi et al. (2008 ▶, 2009 ▶).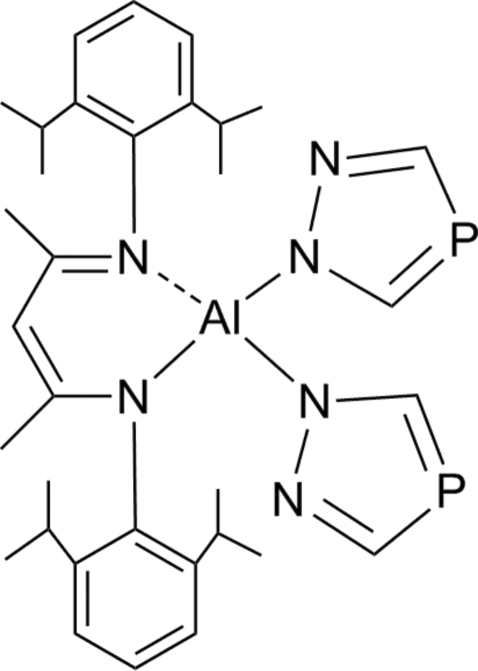
Experimental
Crystal data
[Al(C29H41N2)(C2H2N2P)2]
M r = 614.67
Triclinic,

a = 10.578 (4) Å
b = 12.578 (5) Å
c = 13.498 (5) Å
α = 92.059 (5)°
β = 98.766 (5)°
γ = 96.516 (5)°
V = 1760.8 (11) Å3
Z = 2
Mo Kα radiation
μ = 0.18 mm−1
T = 293 K
0.35 × 0.20 × 0.20 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.940, T max = 0.965
7337 measured reflections
6082 independent reflections
4238 reflections with I > 2σ(I)
R int = 0.037
Refinement
R[F 2 > 2σ(F 2)] = 0.075
wR(F 2) = 0.224
S = 1.02
6082 reflections
389 parameters
H-atom parameters constrained
Δρmax = 0.47 e Å−3
Δρmin = −0.56 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810049007/bq2246sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810049007/bq2246Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 20971058 and 20977042).
supplementary crystallographic information
Comment
Recently, the investigation of 1,2,4-diazaphospholide complexes has attracted considerable interest (Zheng et al., 2006-2009). On the other hand, aluminum hydride complexes with bulky beta-diketiminato ligand [HC(CMeNAr)2] AlH2 have been evidenced to be a reactive species (Roesky et al., 2000-2005). Herein, we report a centrosymmetric complex which was synthesized by the reaction of [HC(CMeNAr)2] AlH2 with 1H-1,2,4-diazaphosphole in hexane at room temperature. As illustrated in Fig. 1, the AlIII ion was coordinated by four nitrogen atoms of 2,6-iPr2C6H3NC(Me)C(H)C(Me)N and 1,2,4-diazaphospholide ligands. The two nitrogen atoms from the 2,6-iPr2C6H3NC(Me)C(H)C(Me)NH ligand form a six-member ring with the aluminum center, and the other two nitrogen atoms from the 1,2,4-diazaphospholide ligands coordinate to aluminum atom in a eta(1) mode. The four nitrogen atoms are arranged in a slightly distorted tetrahedral fashion. The plane of the six-membered ring C3—N2—Al is nearly perpendicular to the 1,2,4-diazaphospholide heterocycle rings.
Experimental
All manipulations were carried out under an argon atmosphere using standard Schlenk techniques. Hexane was dried over sodium and freshly distilled prior to use. 0.481 g [2,6-iPr2C6H3NC(Me)C(H)C(Me)N]AlH2 (1 eq.) and 0.172 g (2 eq.) 1,2,4-Dia-zaphosphole were dissolved in 20 ml toulent. The mixture was stirred for 24 h at room temperature and the solvent was then removed and dried in vacuo. The residua was extracted with 15 ml hexane and the solution was concentrated to about 5 ml to afford colorless crystals at -30°C for several days (yield: 0.32 g. 50%).
Refinement
All H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms with C—H distances of 0.93–0.96 Å, and Uiso(H) = 1.2–1.5 times of those of their parent atoms.
Figures
Fig. 1.
The structure of the title complex with the atom numbering scheme. The thermal displacements are drawn at the 30% probability level. Hydrogen atoms are omitted for clarity.
Crystal data
| [Al(C29H41N2)(C2H2N2P)2] | Z = 2 |
| Mr = 614.67 | F(000) = 656 |
| Triclinic, P1 | Dx = 1.159 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.578 (4) Å | Cell parameters from 872 reflections |
| b = 12.578 (5) Å | θ = 3.4–25.6° |
| c = 13.498 (5) Å | µ = 0.18 mm−1 |
| α = 92.059 (5)° | T = 293 K |
| β = 98.766 (5)° | Sheet, yellow |
| γ = 96.516 (5)° | 0.35 × 0.20 × 0.20 mm |
| V = 1760.8 (11) Å3 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 6082 independent reflections |
| Radiation source: fine-focus sealed tube | 4238 reflections with I > 2σ(I) |
| graphite | Rint = 0.037 |
| phi and ω scans | θmax = 25.0°, θmin = 1.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −12→10 |
| Tmin = 0.940, Tmax = 0.965 | k = −11→14 |
| 7337 measured reflections | l = −16→12 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.075 | w = 1/[σ2(Fo2) + (0.1531P)2 + ] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.224 | (Δ/σ)max < 0.001 |
| S = 1.01 | Δρmax = 0.47 e Å−3 |
| 6082 reflections | Δρmin = −0.56 e Å−3 |
| 389 parameters |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Al1 | 0.22665 (7) | 0.79946 (6) | 0.78731 (6) | 0.0419 (3) | |
| P1 | −0.16571 (10) | 0.71060 (10) | 0.62146 (10) | 0.0880 (4) | |
| P2 | 0.18913 (13) | 1.08771 (11) | 0.97295 (11) | 0.1047 (5) | |
| N1 | 0.0608 (2) | 0.7524 (2) | 0.7248 (2) | 0.0574 (6) | |
| N2 | −0.0133 (3) | 0.7202 (4) | 0.7910 (3) | 0.1033 (13) | |
| N3 | 0.2190 (2) | 0.9204 (2) | 0.86843 (18) | 0.0537 (6) | |
| N4 | 0.3412 (2) | 0.9627 (2) | 0.9076 (2) | 0.0652 (7) | |
| N5 | 0.3149 (2) | 0.70050 (18) | 0.86114 (16) | 0.0479 (6) | |
| N6 | 0.3367 (2) | 0.83042 (17) | 0.69477 (16) | 0.0435 (5) | |
| C1 | −0.1375 (5) | 0.6917 (5) | 0.7425 (4) | 0.125 (2) | |
| H1 | −0.2025 | 0.6632 | 0.7766 | 0.149* | |
| C2 | −0.0057 (4) | 0.7533 (3) | 0.6309 (3) | 0.0762 (10) | |
| H2 | 0.0327 | 0.7753 | 0.5763 | 0.091* | |
| C3 | 0.1292 (3) | 0.9771 (3) | 0.8954 (3) | 0.0717 (9) | |
| H3 | 0.0415 | 0.9581 | 0.8738 | 0.086* | |
| C4 | 0.3387 (4) | 1.0485 (3) | 0.9637 (3) | 0.0806 (11) | |
| H4 | 0.4141 | 1.0871 | 0.9975 | 0.097* | |
| C5 | 0.5225 (3) | 0.6473 (3) | 0.9408 (3) | 0.0738 (10) | |
| H5A | 0.5084 | 0.6655 | 1.0076 | 0.111* | |
| H5B | 0.6123 | 0.6638 | 0.9364 | 0.111* | |
| H5C | 0.4967 | 0.5722 | 0.9253 | 0.111* | |
| C6 | 0.4445 (3) | 0.7107 (2) | 0.8674 (2) | 0.0504 (7) | |
| C7 | 0.5112 (3) | 0.7783 (2) | 0.8092 (2) | 0.0525 (7) | |
| H7 | 0.6000 | 0.7900 | 0.8291 | 0.063* | |
| C8 | 0.4642 (3) | 0.8307 (2) | 0.7260 (2) | 0.0493 (7) | |
| C9 | 0.5580 (3) | 0.8893 (3) | 0.6683 (3) | 0.0737 (10) | |
| H9A | 0.5349 | 0.8682 | 0.5983 | 0.111* | |
| H9B | 0.6432 | 0.8721 | 0.6920 | 0.111* | |
| H9C | 0.5562 | 0.9651 | 0.6778 | 0.111* | |
| C10 | 0.2540 (3) | 0.6173 (3) | 0.9171 (2) | 0.0582 (8) | |
| C11 | 0.2390 (3) | 0.6407 (3) | 1.0157 (3) | 0.0725 (10) | |
| C12 | 0.1862 (4) | 0.5554 (5) | 1.0671 (4) | 0.0997 (16) | |
| H12 | 0.1770 | 0.5676 | 1.1339 | 0.120* | |
| C13 | 0.1485 (5) | 0.4568 (5) | 1.0230 (5) | 0.1080 (17) | |
| H13 | 0.1132 | 0.4027 | 1.0593 | 0.130* | |
| C14 | 0.1618 (4) | 0.4355 (4) | 0.9258 (4) | 0.0977 (14) | |
| H14 | 0.1370 | 0.3667 | 0.8966 | 0.117* | |
| C15 | 0.2122 (3) | 0.5161 (3) | 0.8695 (3) | 0.0715 (10) | |
| C16 | 0.2732 (4) | 0.7513 (4) | 1.0672 (3) | 0.0849 (12) | |
| H16 | 0.3084 | 0.7988 | 1.0194 | 0.102* | |
| C17 | 0.3773 (6) | 0.7541 (6) | 1.1615 (3) | 0.138 (2) | |
| H17A | 0.3416 | 0.7163 | 1.2133 | 0.207* | |
| H17B | 0.4057 | 0.8272 | 1.1849 | 0.207* | |
| H17C | 0.4491 | 0.7207 | 1.1450 | 0.207* | |
| C18 | 0.1544 (5) | 0.7959 (5) | 1.0947 (4) | 0.1133 (16) | |
| H18A | 0.0886 | 0.7905 | 1.0366 | 0.170* | |
| H18B | 0.1766 | 0.8697 | 1.1178 | 0.170* | |
| H18C | 0.1231 | 0.7555 | 1.1468 | 0.170* | |
| C19 | 0.2203 (4) | 0.4905 (3) | 0.7606 (3) | 0.0836 (11) | |
| H19 | 0.2347 | 0.5587 | 0.7289 | 0.100* | |
| C20 | 0.0958 (6) | 0.4301 (5) | 0.7050 (5) | 0.142 (2) | |
| H20A | 0.0760 | 0.3648 | 0.7371 | 0.213* | |
| H20B | 0.1056 | 0.4137 | 0.6368 | 0.213* | |
| H20C | 0.0271 | 0.4737 | 0.7059 | 0.213* | |
| C21 | 0.3319 (6) | 0.4291 (5) | 0.7488 (5) | 0.149 (2) | |
| H21A | 0.4112 | 0.4722 | 0.7761 | 0.224* | |
| H21B | 0.3313 | 0.4118 | 0.6789 | 0.224* | |
| H21C | 0.3242 | 0.3642 | 0.7839 | 0.224* | |
| C22 | 0.2938 (3) | 0.8573 (2) | 0.5916 (2) | 0.0476 (6) | |
| C23 | 0.2616 (3) | 0.9594 (2) | 0.5707 (2) | 0.0548 (7) | |
| C24 | 0.2151 (4) | 0.9782 (3) | 0.4717 (3) | 0.0702 (9) | |
| H24 | 0.1928 | 1.0458 | 0.4561 | 0.084* | |
| C25 | 0.2012 (4) | 0.8998 (4) | 0.3964 (3) | 0.0782 (10) | |
| H25 | 0.1699 | 0.9140 | 0.3306 | 0.094* | |
| C26 | 0.2336 (4) | 0.8011 (3) | 0.4187 (3) | 0.0776 (10) | |
| H26 | 0.2243 | 0.7484 | 0.3672 | 0.093* | |
| C27 | 0.2802 (3) | 0.7761 (3) | 0.5160 (2) | 0.0613 (8) | |
| C28 | 0.2762 (4) | 1.0491 (3) | 0.6497 (3) | 0.0691 (9) | |
| H28 | 0.3124 | 1.0225 | 0.7140 | 0.083* | |
| C29 | 0.3710 (4) | 1.1436 (3) | 0.6248 (3) | 0.0898 (12) | |
| H29A | 0.4510 | 1.1183 | 0.6161 | 0.135* | |
| H29B | 0.3860 | 1.1975 | 0.6789 | 0.135* | |
| H29C | 0.3348 | 1.1737 | 0.5641 | 0.135* | |
| C30 | 0.1480 (5) | 1.0861 (4) | 0.6605 (3) | 0.0971 (14) | |
| H30A | 0.1175 | 1.1232 | 0.6023 | 0.146* | |
| H30B | 0.1579 | 1.1336 | 0.7193 | 0.146* | |
| H30C | 0.0869 | 1.0252 | 0.6669 | 0.146* | |
| C31 | 0.3132 (4) | 0.6642 (3) | 0.5355 (3) | 0.0788 (10) | |
| H31 | 0.3299 | 0.6582 | 0.6084 | 0.095* | |
| C32 | 0.2037 (7) | 0.5800 (4) | 0.4934 (5) | 0.143 (2) | |
| H32A | 0.1274 | 0.5943 | 0.5193 | 0.214* | |
| H32B | 0.2253 | 0.5107 | 0.5125 | 0.214* | |
| H32C | 0.1885 | 0.5813 | 0.4215 | 0.214* | |
| C33 | 0.4358 (7) | 0.6444 (5) | 0.4945 (6) | 0.163 (3) | |
| H33A | 0.4168 | 0.6331 | 0.4228 | 0.244* | |
| H33B | 0.4681 | 0.5822 | 0.5232 | 0.244* | |
| H33C | 0.4996 | 0.7056 | 0.5117 | 0.244* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Al1 | 0.0409 (4) | 0.0430 (5) | 0.0436 (4) | 0.0117 (3) | 0.0059 (3) | 0.0077 (3) |
| P1 | 0.0608 (6) | 0.0820 (7) | 0.1125 (9) | 0.0055 (5) | −0.0134 (5) | 0.0134 (6) |
| P2 | 0.0950 (8) | 0.0948 (9) | 0.1276 (10) | 0.0281 (7) | 0.0282 (7) | −0.0450 (8) |
| N1 | 0.0461 (13) | 0.0545 (15) | 0.0702 (16) | 0.0116 (11) | 0.0003 (11) | 0.0041 (12) |
| N2 | 0.069 (2) | 0.160 (4) | 0.084 (2) | 0.013 (2) | 0.0102 (17) | 0.052 (2) |
| N3 | 0.0525 (13) | 0.0565 (15) | 0.0545 (13) | 0.0158 (12) | 0.0100 (11) | −0.0011 (11) |
| N4 | 0.0556 (15) | 0.0683 (18) | 0.0710 (17) | 0.0125 (13) | 0.0084 (12) | −0.0148 (14) |
| N5 | 0.0490 (13) | 0.0486 (13) | 0.0482 (12) | 0.0112 (11) | 0.0077 (10) | 0.0143 (10) |
| N6 | 0.0479 (12) | 0.0416 (12) | 0.0439 (12) | 0.0139 (10) | 0.0088 (9) | 0.0069 (9) |
| C1 | 0.087 (3) | 0.152 (5) | 0.157 (5) | 0.031 (3) | 0.061 (3) | 0.084 (4) |
| C2 | 0.071 (2) | 0.086 (3) | 0.069 (2) | 0.005 (2) | 0.0050 (17) | 0.0152 (19) |
| C3 | 0.0625 (19) | 0.077 (2) | 0.082 (2) | 0.0228 (18) | 0.0210 (16) | −0.0035 (18) |
| C4 | 0.073 (2) | 0.079 (3) | 0.085 (2) | 0.0090 (19) | 0.0056 (18) | −0.029 (2) |
| C5 | 0.062 (2) | 0.081 (2) | 0.083 (2) | 0.0280 (18) | 0.0020 (17) | 0.0361 (19) |
| C6 | 0.0485 (15) | 0.0505 (16) | 0.0531 (15) | 0.0147 (13) | 0.0034 (12) | 0.0080 (13) |
| C7 | 0.0419 (14) | 0.0551 (17) | 0.0626 (17) | 0.0149 (13) | 0.0074 (12) | 0.0071 (14) |
| C8 | 0.0491 (15) | 0.0446 (15) | 0.0570 (16) | 0.0123 (13) | 0.0119 (12) | 0.0037 (12) |
| C9 | 0.0579 (19) | 0.086 (3) | 0.084 (2) | 0.0121 (18) | 0.0261 (17) | 0.026 (2) |
| C10 | 0.0487 (16) | 0.066 (2) | 0.0618 (18) | 0.0156 (15) | 0.0032 (13) | 0.0298 (15) |
| C11 | 0.0610 (19) | 0.100 (3) | 0.0609 (19) | 0.0191 (19) | 0.0094 (15) | 0.0404 (19) |
| C12 | 0.080 (3) | 0.142 (5) | 0.087 (3) | 0.028 (3) | 0.023 (2) | 0.069 (3) |
| C13 | 0.086 (3) | 0.109 (4) | 0.132 (4) | 0.008 (3) | 0.016 (3) | 0.073 (4) |
| C14 | 0.081 (3) | 0.078 (3) | 0.136 (4) | 0.012 (2) | 0.009 (3) | 0.057 (3) |
| C15 | 0.0608 (19) | 0.060 (2) | 0.094 (3) | 0.0116 (17) | 0.0055 (18) | 0.0335 (19) |
| C16 | 0.083 (2) | 0.124 (4) | 0.0481 (18) | 0.015 (2) | 0.0081 (17) | 0.019 (2) |
| C17 | 0.114 (4) | 0.229 (7) | 0.066 (3) | 0.032 (4) | −0.004 (3) | 0.000 (4) |
| C18 | 0.112 (4) | 0.148 (5) | 0.089 (3) | 0.034 (3) | 0.029 (3) | 0.012 (3) |
| C19 | 0.098 (3) | 0.0468 (19) | 0.105 (3) | 0.0131 (19) | 0.010 (2) | 0.0077 (19) |
| C20 | 0.137 (5) | 0.130 (5) | 0.139 (5) | −0.030 (4) | −0.008 (4) | 0.003 (4) |
| C21 | 0.143 (5) | 0.138 (5) | 0.175 (6) | 0.058 (4) | 0.028 (4) | −0.031 (4) |
| C22 | 0.0482 (14) | 0.0521 (16) | 0.0449 (14) | 0.0108 (13) | 0.0095 (11) | 0.0087 (12) |
| C23 | 0.0603 (17) | 0.0542 (17) | 0.0519 (16) | 0.0130 (14) | 0.0088 (13) | 0.0122 (13) |
| C24 | 0.078 (2) | 0.072 (2) | 0.064 (2) | 0.0208 (19) | 0.0079 (16) | 0.0246 (17) |
| C25 | 0.086 (2) | 0.099 (3) | 0.0493 (18) | 0.018 (2) | 0.0036 (16) | 0.0194 (19) |
| C26 | 0.092 (3) | 0.091 (3) | 0.0501 (18) | 0.016 (2) | 0.0098 (17) | −0.0016 (18) |
| C27 | 0.072 (2) | 0.0618 (19) | 0.0533 (17) | 0.0107 (16) | 0.0168 (14) | 0.0032 (14) |
| C28 | 0.094 (2) | 0.0515 (18) | 0.0631 (19) | 0.0244 (18) | 0.0038 (17) | 0.0110 (15) |
| C29 | 0.108 (3) | 0.060 (2) | 0.095 (3) | 0.006 (2) | −0.002 (2) | 0.013 (2) |
| C30 | 0.122 (4) | 0.087 (3) | 0.097 (3) | 0.052 (3) | 0.033 (3) | 0.012 (2) |
| C31 | 0.111 (3) | 0.059 (2) | 0.069 (2) | 0.017 (2) | 0.017 (2) | −0.0058 (16) |
| C32 | 0.202 (6) | 0.065 (3) | 0.139 (5) | −0.008 (4) | −0.023 (4) | −0.014 (3) |
| C33 | 0.169 (6) | 0.097 (4) | 0.255 (8) | 0.072 (4) | 0.089 (6) | 0.026 (5) |
Geometric parameters (Å, °)
| Al1—N1 | 1.848 (3) | C16—H16 | 0.9800 |
| Al1—N6 | 1.855 (2) | C17—H17A | 0.9600 |
| Al1—N3 | 1.858 (3) | C17—H17B | 0.9600 |
| Al1—N5 | 1.867 (2) | C17—H17C | 0.9600 |
| P1—C1 | 1.646 (6) | C18—H18A | 0.9600 |
| P1—C2 | 1.700 (4) | C18—H18B | 0.9600 |
| P2—C3 | 1.711 (4) | C18—H18C | 0.9600 |
| P2—C4 | 1.731 (4) | C19—C21 | 1.506 (7) |
| N1—N2 | 1.320 (4) | C19—C20 | 1.518 (7) |
| N1—C2 | 1.354 (4) | C19—H19 | 0.9800 |
| N2—C1 | 1.378 (6) | C20—H20A | 0.9600 |
| N3—C3 | 1.336 (4) | C20—H20B | 0.9600 |
| N3—N4 | 1.360 (4) | C20—H20C | 0.9600 |
| N4—C4 | 1.301 (4) | C21—H21A | 0.9600 |
| N5—C6 | 1.352 (4) | C21—H21B | 0.9600 |
| N5—C10 | 1.460 (4) | C21—H21C | 0.9600 |
| N6—C8 | 1.350 (4) | C22—C23 | 1.393 (4) |
| N6—C22 | 1.461 (3) | C22—C27 | 1.397 (4) |
| C1—H1 | 0.9300 | C23—C24 | 1.390 (4) |
| C2—H2 | 0.9300 | C23—C28 | 1.503 (4) |
| C3—H3 | 0.9300 | C24—C25 | 1.370 (5) |
| C4—H4 | 0.9300 | C24—H24 | 0.9300 |
| C5—C6 | 1.502 (4) | C25—C26 | 1.356 (5) |
| C5—H5A | 0.9600 | C25—H25 | 0.9300 |
| C5—H5B | 0.9600 | C26—C27 | 1.391 (5) |
| C5—H5C | 0.9600 | C26—H26 | 0.9300 |
| C6—C7 | 1.384 (4) | C27—C31 | 1.511 (5) |
| C7—C8 | 1.379 (4) | C28—C30 | 1.509 (6) |
| C7—H7 | 0.9300 | C28—C29 | 1.546 (6) |
| C8—C9 | 1.498 (4) | C28—H28 | 0.9800 |
| C9—H9A | 0.9600 | C29—H29A | 0.9600 |
| C9—H9B | 0.9600 | C29—H29B | 0.9600 |
| C9—H9C | 0.9600 | C29—H29C | 0.9600 |
| C10—C11 | 1.389 (5) | C30—H30A | 0.9600 |
| C10—C15 | 1.398 (5) | C30—H30B | 0.9600 |
| C11—C12 | 1.407 (6) | C30—H30C | 0.9600 |
| C11—C16 | 1.513 (6) | C31—C32 | 1.504 (7) |
| C12—C13 | 1.347 (7) | C31—C33 | 1.527 (7) |
| C12—H12 | 0.9300 | C31—H31 | 0.9800 |
| C13—C14 | 1.361 (7) | C32—H32A | 0.9600 |
| C13—H13 | 0.9300 | C32—H32B | 0.9600 |
| C14—C15 | 1.395 (5) | C32—H32C | 0.9600 |
| C14—H14 | 0.9300 | C33—H33A | 0.9600 |
| C15—C19 | 1.511 (6) | C33—H33B | 0.9600 |
| C16—C18 | 1.523 (6) | C33—H33C | 0.9600 |
| C16—C17 | 1.548 (6) | ||
| N1—Al1—N6 | 111.60 (12) | C16—C17—H17C | 109.5 |
| N1—Al1—N3 | 107.50 (11) | H17A—C17—H17C | 109.5 |
| N6—Al1—N3 | 110.79 (11) | H17B—C17—H17C | 109.5 |
| N1—Al1—N5 | 116.84 (12) | C16—C18—H18A | 109.5 |
| N6—Al1—N5 | 99.77 (10) | C16—C18—H18B | 109.5 |
| N3—Al1—N5 | 110.22 (11) | H18A—C18—H18B | 109.5 |
| C1—P1—C2 | 86.8 (2) | C16—C18—H18C | 109.5 |
| C3—P2—C4 | 85.27 (17) | H18A—C18—H18C | 109.5 |
| N2—N1—C2 | 112.7 (3) | H18B—C18—H18C | 109.5 |
| N2—N1—Al1 | 110.9 (2) | C21—C19—C15 | 112.2 (4) |
| C2—N1—Al1 | 135.9 (3) | C21—C19—C20 | 110.1 (4) |
| N1—N2—C1 | 109.3 (3) | C15—C19—C20 | 112.1 (4) |
| C3—N3—N4 | 113.3 (3) | C21—C19—H19 | 107.4 |
| C3—N3—Al1 | 138.0 (2) | C15—C19—H19 | 107.4 |
| N4—N3—Al1 | 108.65 (18) | C20—C19—H19 | 107.4 |
| C4—N4—N3 | 109.9 (3) | C19—C20—H20A | 109.5 |
| C6—N5—C10 | 118.2 (2) | C19—C20—H20B | 109.5 |
| C6—N5—Al1 | 117.43 (19) | H20A—C20—H20B | 109.5 |
| C10—N5—Al1 | 124.27 (18) | C19—C20—H20C | 109.5 |
| C8—N6—C22 | 118.4 (2) | H20A—C20—H20C | 109.5 |
| C8—N6—Al1 | 117.63 (18) | H20B—C20—H20C | 109.5 |
| C22—N6—Al1 | 123.90 (17) | C19—C21—H21A | 109.5 |
| N2—C1—P1 | 116.9 (3) | C19—C21—H21B | 109.5 |
| N2—C1—H1 | 121.6 | H21A—C21—H21B | 109.5 |
| P1—C1—H1 | 121.6 | C19—C21—H21C | 109.5 |
| N1—C2—P1 | 114.1 (3) | H21A—C21—H21C | 109.5 |
| N1—C2—H2 | 122.9 | H21B—C21—H21C | 109.5 |
| P1—C2—H2 | 122.9 | C23—C22—C27 | 121.4 (3) |
| N3—C3—P2 | 114.3 (3) | C23—C22—N6 | 120.7 (2) |
| N3—C3—H3 | 122.8 | C27—C22—N6 | 117.8 (3) |
| P2—C3—H3 | 122.8 | C24—C23—C22 | 117.8 (3) |
| N4—C4—P2 | 117.2 (3) | C24—C23—C28 | 119.2 (3) |
| N4—C4—H4 | 121.4 | C22—C23—C28 | 123.0 (3) |
| P2—C4—H4 | 121.4 | C25—C24—C23 | 121.7 (3) |
| C6—C5—H5A | 109.5 | C25—C24—H24 | 119.2 |
| C6—C5—H5B | 109.5 | C23—C24—H24 | 119.2 |
| H5A—C5—H5B | 109.5 | C26—C25—C24 | 119.4 (3) |
| C6—C5—H5C | 109.5 | C26—C25—H25 | 120.3 |
| H5A—C5—H5C | 109.5 | C24—C25—H25 | 120.3 |
| H5B—C5—H5C | 109.5 | C25—C26—C27 | 122.3 (3) |
| N5—C6—C7 | 123.2 (3) | C25—C26—H26 | 118.9 |
| N5—C6—C5 | 119.6 (3) | C27—C26—H26 | 118.9 |
| C7—C6—C5 | 117.2 (3) | C26—C27—C22 | 117.5 (3) |
| C8—C7—C6 | 129.0 (3) | C26—C27—C31 | 119.4 (3) |
| C8—C7—H7 | 115.5 | C22—C27—C31 | 123.2 (3) |
| C6—C7—H7 | 115.5 | C23—C28—C30 | 111.5 (3) |
| N6—C8—C7 | 122.1 (3) | C23—C28—C29 | 110.2 (3) |
| N6—C8—C9 | 119.2 (3) | C30—C28—C29 | 110.5 (3) |
| C7—C8—C9 | 118.8 (3) | C23—C28—H28 | 108.2 |
| C8—C9—H9A | 109.5 | C30—C28—H28 | 108.2 |
| C8—C9—H9B | 109.5 | C29—C28—H28 | 108.2 |
| H9A—C9—H9B | 109.5 | C28—C29—H29A | 109.5 |
| C8—C9—H9C | 109.5 | C28—C29—H29B | 109.5 |
| H9A—C9—H9C | 109.5 | H29A—C29—H29B | 109.5 |
| H9B—C9—H9C | 109.5 | C28—C29—H29C | 109.5 |
| C11—C10—C15 | 121.9 (3) | H29A—C29—H29C | 109.5 |
| C11—C10—N5 | 119.3 (3) | H29B—C29—H29C | 109.5 |
| C15—C10—N5 | 118.8 (3) | C28—C30—H30A | 109.5 |
| C10—C11—C12 | 116.5 (4) | C28—C30—H30B | 109.5 |
| C10—C11—C16 | 123.5 (3) | H30A—C30—H30B | 109.5 |
| C12—C11—C16 | 120.1 (4) | C28—C30—H30C | 109.5 |
| C13—C12—C11 | 122.2 (5) | H30A—C30—H30C | 109.5 |
| C13—C12—H12 | 118.9 | H30B—C30—H30C | 109.5 |
| C11—C12—H12 | 118.9 | C32—C31—C27 | 112.0 (4) |
| C12—C13—C14 | 120.7 (4) | C32—C31—C33 | 110.7 (5) |
| C12—C13—H13 | 119.7 | C27—C31—C33 | 111.0 (4) |
| C14—C13—H13 | 119.7 | C32—C31—H31 | 107.6 |
| C13—C14—C15 | 120.5 (5) | C27—C31—H31 | 107.6 |
| C13—C14—H14 | 119.8 | C33—C31—H31 | 107.6 |
| C15—C14—H14 | 119.8 | C31—C32—H32A | 109.5 |
| C14—C15—C10 | 118.2 (4) | C31—C32—H32B | 109.5 |
| C14—C15—C19 | 118.9 (4) | H32A—C32—H32B | 109.5 |
| C10—C15—C19 | 122.9 (3) | C31—C32—H32C | 109.5 |
| C11—C16—C18 | 111.3 (4) | H32A—C32—H32C | 109.5 |
| C11—C16—C17 | 112.8 (4) | H32B—C32—H32C | 109.5 |
| C18—C16—C17 | 109.8 (4) | C31—C33—H33A | 109.5 |
| C11—C16—H16 | 107.6 | C31—C33—H33B | 109.5 |
| C18—C16—H16 | 107.6 | H33A—C33—H33B | 109.5 |
| C17—C16—H16 | 107.6 | C31—C33—H33C | 109.5 |
| C16—C17—H17A | 109.5 | H33A—C33—H33C | 109.5 |
| C16—C17—H17B | 109.5 | H33B—C33—H33C | 109.5 |
| H17A—C17—H17B | 109.5 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BQ2246).
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst.32, 115–119.
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). APEX2 and SAINT Bruker AXS inc., Madison, Wisconsin, USA.
- Cui, C. M., Roesky, H. W., Hao, H. J., Schmidt, H. G. & Noletmeyer, M. (2000). Angew. Chem. Int. Ed.39, 1815–1817. [DOI] [PubMed]
- Ding, Y. Q., Roesky, H. W., Noletmeyer, M. & Schmidt, H. G. (2001). Organometallics, 20, 1190–1194.
- Kumar, S. S., Singh, S., Hongjun, F., Roesky, H. W. & Magull, J. (2005). Inorg. Chem.44, 1199–1201. [DOI] [PubMed]
- Kumar, S. S., Singh, S., Hongjun, F., Roesky, H. W., Vidovic, D. & Magull, J. (2004). Organometallics, 23, 6327–6329.
- Pi, C. F., Wan, L., Gu, Y. Y., Zheng, W. J., Wu, H. Y., Weng, L. H., Chen, Z. X. & Wu, L. M. (2008). Inorg. Chem.47, 9739–9741. [DOI] [PubMed]
- Pi, C. F., Wan, L., Liu, W. P., Pan, Z. F., Wu, H. Y., Wang, Y. H., Zheng, W. J., Weng, L. H., Chen, Z. X. & Wu, L. M. (2009). Inorg. Chem 48, 2967–2975. [DOI] [PubMed]
- Schmidpeter, A. & Willhalm, A. (1984). Angew. Chem. Int. Ed.23, 903–904.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Wan, L., Pi, C. F., Zhang, L., Zheng, W. J., Weng, L. H., Chen, Z. X. & Zhang, Y. (2008). Chem. Commun. pp. 2266–2268. [DOI] [PubMed]
- Zheng, W. J., Zhang, G. Z. & Fan, K. N. (2006). Organometallics, 25, 1548–1550.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810049007/bq2246sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810049007/bq2246Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



