Abstract
The absolute configuration of the title compound, C10H17NO3·H2O, was assigned from the synthesis. In the molecular structure, the central six-membered ring of the indolizine moiety adopts a chair conformation, with two atoms displaced by −0.578 (2) and 0.651 (1) Å from the plane of the other four atoms [maximum deviation 0.019 (2) Å] The conformation of the fused oxopyrrolidine ring is close to that of a flat envelope, with the flap atom displaced by 0.294 (1) Å from the plane through the remaining four atoms. In the crystal, one of the hydroxy groups is hydrogen-bonded to two water molecules, while the other hydroxy group exhibits an intermolecular hydrogen bond to the carbonyl O atom, resulting in a chain parallel to the b axis.
Related literature
For the uses of indolizine-based molecules, see: Weidner et al. (1989 ▶); Jaung & Jung (2003 ▶); Rotaru et al. (2005 ▶); Saeva & Luss (1988 ▶); Kelin et al. (2001 ▶). For biological activities of indolizines, see: Oslund et al. (2008 ▶); Asano et al. (2000 ▶); Tielmann & Hoenke (2006 ▶). For synthesis, see: Šafař et al. (2010 ▶). For ring-puckering and conformational analysis, see: Cremer & Pople (1975 ▶); Nardelli (1983 ▶). 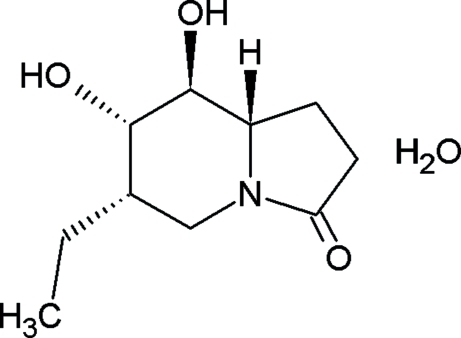
Experimental
Crystal data
C10H17NO3·H2O
M r = 217.26
Orthorhombic,

a = 7.1398 (3) Å
b = 7.3169 (2) Å
c = 20.8466 (9) Å
V = 1089.05 (7) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 298 K
0.55 × 0.25 × 0.09 mm
Data collection
Oxford Gemini R CCD diffractometer
Absorption correction: analytical (Clark & Reid, 1995 ▶) T min = 0.945, T max = 0.991
17428 measured reflections
1308 independent reflections
1161 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.029
wR(F 2) = 0.081
S = 1.06
1308 reflections
148 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.16 e Å−3
Δρmin = −0.14 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2006 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2001 ▶); software used to prepare material for publication: enCIFer (Allen et al., 2004 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810044855/fj2360sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810044855/fj2360Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O22—H22A⋯O21i | 0.82 (3) | 1.94 (3) | 2.7505 (19) | 173 (3) |
| O24—H24A⋯O22ii | 0.91 (3) | 2.07 (3) | 2.919 (2) | 155 (2) |
| O24—H24B⋯O23iii | 0.78 (3) | 2.14 (3) | 2.907 (2) | 167 (3) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
The authors thank the Grant Agency of the Slovak Republic (grant No. 1/0161/08) and the Structural Funds, Interreg IIIA, for financial support in purchasing the diffractometer. We also thank the Development Agency for support under contract No. APVV-0210-07.
supplementary crystallographic information
Comment
Indolizines based molecules are known for their use as synthetic dyes (Weidner et al., 1989; Jaung & Jung, 2003), fluorescent materials (Rotaru et al., 2005; Saeva & Luss, 1988) and also as key intermediates for the synthesis of indolizine based molecules (Kelin et al., 2001). Indolizines both synthetic and natural have also been ascribed with a number of useful biological activities (Oslund et al., 2008; Asano et al., 2000; Tielmann & Hoenke, 2006) such as antibacterial, antiviral, CNS depressants, anti-HIV, anti-cancer and have been used for treating cardiovascular ailments.
Due to the diverse properties of indolizine derivatives, the structure of the title compound, (I), has been determined as part of our study of the conformational changes caused by different substituents at various positions on the indolizine ring system. The absolute configuration was established by synthesis and is depicted in the scheme and figure. The expected stereochemistry of atoms C5, C6, C7 and C8 was confirmed as S, S, S and S, respectively (Fig. 1). The central six-membered ring is not planar and adopts a chair conformation (Cremer & Pople, 1975). A calculation of least-squares planes shows that this ring is puckered in such a manner that the four atoms C5, C6, C8 and C9 are coplanar to within 0.019 (2) Å, while atoms N1 and C7 are displaced from this plane on opposite sides, with out-of-plane displacements of -0.578 (2) and 0.651 (1) Å, respectively. The oxopyrrolidine ring attached to the indolizine ring system has flat-envelope conformation with atom C4 on the flap (Nardelli, 1983). The deviation of atom C4 from the mean plane of the remaining four atoms N1/C2/C3/C5 is 0.294 (1) Å. The N1—C5 and N1—C9 bonds are approximately equivalent and both are much longer than the N1—C2 bond. Atom N1 is sp2-hybridized, as evidenced by the sum of the valence angles around it (358.4 (2)°). These data are consistent with conjugation of the lone-pair electrons on N1 with the adjacent carbonyl C2═O21. The H atoms (H24A and H24B) of the water molecule form O—H···O intermolecular hydrogen bonds with the O atoms (O22 and O23) of both present hydroxy groups, which may, in part, influence the molecular configuration. There have been observed also another intermolecular O—H···O hydrogen bonds, in which carbonyl oxygen O21 participates as acceptor and atom O22 as donator (Table 1). All the interactions demonstrated were found by PLATON (Spek, 2009).
Experimental
The title compound 6S,7S,8S,8aS)-6-ethyl-7,8-dihydroxyhexahydroindolizin-3(2H)-one monohydrate was prepared according literature procedures of Šafař et al. (2010).
Refinement
All H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H distances in the range 0.93–0.98 Å and O—H distance 0.85 Å and Uiso set at 1.2Ueq of the parent atom. The absolute configuration could not be reliably determined for this compound using Mo radiation, and has been assigned according to the synthesis; 907 total Friedel pairs have been merged. Due to the absence of anomalous dispersion the Flack parameter was not refined
Figures
Fig. 1.
Molecular structure of (I) with the atomic numbering scheme. Displacement ellipsoids are drawn at the 50% probability level (Brandenburg, 2001).
Fig. 2.
A packing of the molecule of (I), viewed along the b axis.
Crystal data
| C10H17NO3·H2O | F(000) = 472 |
| Mr = 217.26 | Dx = 1.325 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 10421 reflections |
| a = 7.1398 (3) Å | θ = 3.4–29.5° |
| b = 7.3169 (2) Å | µ = 0.10 mm−1 |
| c = 20.8466 (9) Å | T = 298 K |
| V = 1089.05 (7) Å3 | Prism, colourless |
| Z = 4 | 0.55 × 0.25 × 0.09 mm |
Data collection
| Oxford Gemini R CCD diffractometer | 1308 independent reflections |
| Radiation source: fine-focus sealed tube | 1161 reflections with I > 2σ(I) |
| graphite | Rint = 0.028 |
| Detector resolution: 10.4340 pixels mm-1 | θmax = 26.4°, θmin = 4.1° |
| Rotation method data acquisition using ω and φ scans | h = −8→8 |
| Absorption correction: analytical (Clark & Reid, 1995) | k = −9→9 |
| Tmin = 0.945, Tmax = 0.991 | l = −26→26 |
| 17428 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.081 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0511P)2 + 0.0923P] where P = (Fo2 + 2Fc2)/3 |
| 1308 reflections | (Δ/σ)max < 0.001 |
| 148 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.14 e Å−3 |
Special details
| Experimental. face-indexed (Oxford Diffraction, 2006) |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C2 | 0.7971 (2) | 0.5950 (2) | 0.21098 (8) | 0.0301 (4) | |
| C3 | 0.7338 (3) | 0.7182 (2) | 0.26470 (8) | 0.0347 (4) | |
| H3B | 0.8405 | 0.7649 | 0.2884 | 0.042* | |
| H3A | 0.6524 | 0.6529 | 0.2941 | 0.042* | |
| C4 | 0.6288 (3) | 0.8728 (3) | 0.23202 (9) | 0.0454 (5) | |
| H4B | 0.6617 | 0.9895 | 0.2510 | 0.054* | |
| H4A | 0.4946 | 0.8553 | 0.2358 | 0.054* | |
| C5 | 0.6903 (2) | 0.8652 (2) | 0.16126 (8) | 0.0314 (4) | |
| H5A | 0.5790 | 0.8736 | 0.1339 | 0.038* | |
| C6 | 0.8300 (3) | 1.0102 (2) | 0.14059 (8) | 0.0331 (4) | |
| H6A | 0.9284 | 1.0227 | 0.1731 | 0.040* | |
| C7 | 0.9168 (3) | 0.9620 (2) | 0.07610 (9) | 0.0359 (4) | |
| H7A | 0.8175 | 0.9664 | 0.0437 | 0.043* | |
| C8 | 1.0011 (2) | 0.7695 (3) | 0.07524 (8) | 0.0341 (4) | |
| H8A | 1.0387 | 0.7430 | 0.0310 | 0.041* | |
| C9 | 0.8502 (3) | 0.6316 (2) | 0.09368 (8) | 0.0339 (4) | |
| H9B | 0.7529 | 0.6298 | 0.0612 | 0.041* | |
| H9A | 0.9043 | 0.5102 | 0.0963 | 0.041* | |
| C10 | 1.1765 (3) | 0.7517 (3) | 0.11756 (10) | 0.0399 (4) | |
| H10B | 1.2577 | 0.8557 | 0.1098 | 0.048* | |
| H10A | 1.1390 | 0.7552 | 0.1623 | 0.048* | |
| C11 | 1.2854 (3) | 0.5778 (3) | 0.10545 (10) | 0.0514 (5) | |
| H11C | 1.3930 | 0.5743 | 0.1331 | 0.062* | |
| H11B | 1.3255 | 0.5745 | 0.0615 | 0.062* | |
| H11A | 1.2070 | 0.4741 | 0.1141 | 0.062* | |
| N1 | 0.7703 (2) | 0.68207 (18) | 0.15522 (7) | 0.0292 (3) | |
| O21 | 0.8643 (2) | 0.44131 (16) | 0.21682 (6) | 0.0420 (3) | |
| O22 | 0.7283 (2) | 1.17774 (17) | 0.13508 (8) | 0.0473 (4) | |
| H22A | 0.776 (4) | 1.257 (4) | 0.1571 (12) | 0.057* | |
| O23 | 1.0557 (2) | 1.0928 (2) | 0.05882 (8) | 0.0565 (4) | |
| H23A | 1.015 (4) | 1.177 (4) | 0.0304 (13) | 0.068* | |
| O24 | 0.9507 (2) | 1.3468 (3) | −0.03262 (8) | 0.0559 (4) | |
| H24A | 1.013 (4) | 1.312 (4) | −0.0684 (13) | 0.067* | |
| H24B | 0.851 (4) | 1.369 (4) | −0.0451 (13) | 0.067* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C2 | 0.0274 (8) | 0.0256 (8) | 0.0373 (9) | −0.0042 (7) | −0.0022 (7) | −0.0007 (7) |
| C3 | 0.0365 (9) | 0.0329 (8) | 0.0346 (9) | −0.0024 (8) | 0.0014 (7) | −0.0006 (7) |
| C4 | 0.0523 (11) | 0.0395 (10) | 0.0444 (11) | 0.0134 (10) | 0.0139 (9) | 0.0010 (9) |
| C5 | 0.0312 (8) | 0.0270 (8) | 0.0359 (9) | 0.0044 (7) | −0.0026 (7) | −0.0004 (7) |
| C6 | 0.0355 (9) | 0.0251 (8) | 0.0387 (9) | −0.0008 (8) | −0.0106 (7) | 0.0016 (7) |
| C7 | 0.0325 (9) | 0.0376 (9) | 0.0375 (9) | −0.0057 (8) | −0.0061 (8) | 0.0108 (8) |
| C8 | 0.0354 (9) | 0.0404 (9) | 0.0265 (8) | −0.0020 (8) | 0.0011 (7) | 0.0010 (7) |
| C9 | 0.0378 (9) | 0.0327 (9) | 0.0312 (9) | −0.0021 (8) | 0.0004 (7) | −0.0064 (7) |
| C10 | 0.0323 (9) | 0.0396 (10) | 0.0478 (11) | −0.0019 (9) | −0.0005 (8) | 0.0035 (9) |
| C11 | 0.0504 (12) | 0.0576 (13) | 0.0462 (12) | 0.0150 (11) | −0.0002 (10) | 0.0036 (10) |
| N1 | 0.0316 (7) | 0.0230 (6) | 0.0330 (7) | 0.0003 (6) | 0.0019 (6) | −0.0007 (6) |
| O21 | 0.0514 (8) | 0.0282 (6) | 0.0465 (7) | 0.0091 (6) | −0.0047 (7) | 0.0021 (6) |
| O22 | 0.0560 (9) | 0.0234 (6) | 0.0624 (9) | 0.0034 (7) | −0.0181 (7) | −0.0010 (6) |
| O23 | 0.0418 (8) | 0.0553 (9) | 0.0726 (10) | −0.0098 (8) | −0.0016 (7) | 0.0328 (8) |
| O24 | 0.0482 (9) | 0.0650 (10) | 0.0546 (9) | 0.0009 (9) | 0.0021 (7) | 0.0157 (8) |
Geometric parameters (Å, °)
| C2—O21 | 1.228 (2) | C7—H7A | 0.9800 |
| C2—N1 | 1.339 (2) | C8—C9 | 1.525 (2) |
| C2—C3 | 1.507 (2) | C8—C10 | 1.538 (3) |
| C3—C4 | 1.518 (3) | C8—H8A | 0.9800 |
| C3—H3B | 0.9700 | C9—N1 | 1.452 (2) |
| C3—H3A | 0.9700 | C9—H9B | 0.9700 |
| C4—C5 | 1.540 (2) | C9—H9A | 0.9700 |
| C4—H4B | 0.9700 | C10—C11 | 1.513 (3) |
| C4—H4A | 0.9700 | C10—H10B | 0.9700 |
| C5—N1 | 1.462 (2) | C10—H10A | 0.9700 |
| C5—C6 | 1.519 (2) | C11—H11C | 0.9600 |
| C5—H5A | 0.9800 | C11—H11B | 0.9600 |
| C6—O22 | 1.429 (2) | C11—H11A | 0.9600 |
| C6—C7 | 1.522 (3) | O22—H22A | 0.82 (3) |
| C6—H6A | 0.9800 | O23—H23A | 0.90 (3) |
| C7—O23 | 1.425 (2) | O24—H24A | 0.91 (3) |
| C7—C8 | 1.532 (3) | O24—H24B | 0.78 (3) |
| O21—C2—N1 | 125.27 (16) | C8—C7—H7A | 107.9 |
| O21—C2—C3 | 126.23 (16) | C9—C8—C7 | 109.13 (14) |
| N1—C2—C3 | 108.50 (14) | C9—C8—C10 | 112.01 (14) |
| C2—C3—C4 | 105.09 (15) | C7—C8—C10 | 113.01 (15) |
| C2—C3—H3B | 110.7 | C9—C8—H8A | 107.5 |
| C4—C3—H3B | 110.7 | C7—C8—H8A | 107.5 |
| C2—C3—H3A | 110.7 | C10—C8—H8A | 107.5 |
| C4—C3—H3A | 110.7 | N1—C9—C8 | 109.40 (14) |
| H3B—C3—H3A | 108.8 | N1—C9—H9B | 109.8 |
| C3—C4—C5 | 105.20 (14) | C8—C9—H9B | 109.8 |
| C3—C4—H4B | 110.7 | N1—C9—H9A | 109.8 |
| C5—C4—H4B | 110.7 | C8—C9—H9A | 109.8 |
| C3—C4—H4A | 110.7 | H9B—C9—H9A | 108.2 |
| C5—C4—H4A | 110.7 | C11—C10—C8 | 113.21 (17) |
| H4B—C4—H4A | 108.8 | C11—C10—H10B | 108.9 |
| N1—C5—C6 | 111.06 (13) | C8—C10—H10B | 108.9 |
| N1—C5—C4 | 103.10 (13) | C11—C10—H10A | 108.9 |
| C6—C5—C4 | 115.70 (15) | C8—C10—H10A | 108.9 |
| N1—C5—H5A | 108.9 | H10B—C10—H10A | 107.7 |
| C6—C5—H5A | 108.9 | C10—C11—H11C | 109.5 |
| C4—C5—H5A | 108.9 | C10—C11—H11B | 109.5 |
| O22—C6—C5 | 106.74 (14) | H11C—C11—H11B | 109.5 |
| O22—C6—C7 | 109.57 (15) | C10—C11—H11A | 109.5 |
| C5—C6—C7 | 110.85 (14) | H11C—C11—H11A | 109.5 |
| O22—C6—H6A | 109.9 | H11B—C11—H11A | 109.5 |
| C5—C6—H6A | 109.9 | C2—N1—C9 | 126.13 (14) |
| C7—C6—H6A | 109.9 | C2—N1—C5 | 114.67 (13) |
| O23—C7—C6 | 110.54 (16) | C9—N1—C5 | 117.56 (13) |
| O23—C7—C8 | 109.97 (14) | C6—O22—H22A | 110.7 (18) |
| C6—C7—C8 | 112.57 (14) | C7—O23—H23A | 113.7 (16) |
| O23—C7—H7A | 107.9 | H24A—O24—H24B | 104 (3) |
| C6—C7—H7A | 107.9 | ||
| O21—C2—C3—C4 | −169.29 (17) | C6—C7—C8—C10 | −68.92 (19) |
| N1—C2—C3—C4 | 11.57 (19) | C7—C8—C9—N1 | −55.28 (18) |
| C2—C3—C4—C5 | −17.83 (19) | C10—C8—C9—N1 | 70.64 (19) |
| C3—C4—C5—N1 | 17.48 (19) | C9—C8—C10—C11 | 69.5 (2) |
| C3—C4—C5—C6 | −103.96 (16) | C7—C8—C10—C11 | −166.77 (16) |
| N1—C5—C6—O22 | 167.96 (14) | O21—C2—N1—C9 | −14.3 (3) |
| C4—C5—C6—O22 | −74.98 (18) | C3—C2—N1—C9 | 164.90 (15) |
| N1—C5—C6—C7 | 48.69 (18) | O21—C2—N1—C5 | −179.17 (16) |
| C4—C5—C6—C7 | 165.76 (15) | C3—C2—N1—C5 | −0.02 (19) |
| O22—C6—C7—O23 | 65.90 (18) | C8—C9—N1—C2 | −108.10 (19) |
| C5—C6—C7—O23 | −176.56 (14) | C8—C9—N1—C5 | 56.43 (19) |
| O22—C6—C7—C8 | −170.69 (13) | C6—C5—N1—C2 | 113.25 (16) |
| C5—C6—C7—C8 | −53.14 (19) | C4—C5—N1—C2 | −11.28 (19) |
| O23—C7—C8—C9 | −179.85 (15) | C6—C5—N1—C9 | −53.04 (19) |
| C6—C7—C8—C9 | 56.42 (18) | C4—C5—N1—C9 | −177.56 (15) |
| O23—C7—C8—C10 | 54.81 (19) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O22—H22A···O21i | 0.82 (3) | 1.94 (3) | 2.7505 (19) | 173 (3) |
| O24—H24A···O22ii | 0.91 (3) | 2.07 (3) | 2.919 (2) | 155 (2) |
| O24—H24B···O23iii | 0.78 (3) | 2.14 (3) | 2.907 (2) | 167 (3) |
Symmetry codes: (i) x, y+1, z; (ii) x+1/2, −y+5/2, −z; (iii) x−1/2, −y+5/2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2360).
References
- Allen, F. H., Johnson, O., Shields, G. P., Smith, B. R. & Towler, M. (2004). J. Appl. Cryst.37, 335–338.
- Asano, N., Nash, R. J., Molyneux, R. J. & Fleet, G. W. J. (2000). Tetrahedron Asymmetry, 11, 1645–1680.
- Brandenburg, K. (2001). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887–897.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1362.
- Jaung, J. Y. & Jung, Y. S. (2003). Bull. Korean Chem. Soc.24, 1565–1566.
- Kelin, A. V., Sromek, A. W. & Gevorgyan, V. (2001). J. Am. Chem. Soc.123, 2074–2075. [DOI] [PubMed]
- Nardelli, M. (1983). Acta Cryst. C39, 1141–1142.
- Oslund, R. C., Cermak, N. & Gelb, M. H. (2008). J. Med. Chem.51, 4708–4714. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2006). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Abingdon, Oxfordshire, England.
- Rotaru, A. V., Druta, I. D., Oeser, T. & Muller, T. J. J. (2005). Helv. Chim. Acta, 88, 1798–1812.
- Saeva, F. D. & Luss, H. R. (1988). J. Org. Chem.53, 1804–1806.
- Šafař, P., Žúžiová, J., Marchalín, Š., Prónayová, N., Švorc, Ľ., Vrábel, V., Comesse, S. & Daich, A. (2010). Tetrahedron Asymmetry, 21, 623–630.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tielmann, P. & Hoenke, C. (2006). Tetrahedron Lett.47, 261–265.
- Weidner, C. H., Wadsworth, D. H., Bender, S. L. & Beltman, D. J. (1989). J. Org. Chem.54, 3660–3664.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810044855/fj2360sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810044855/fj2360Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




