Abstract
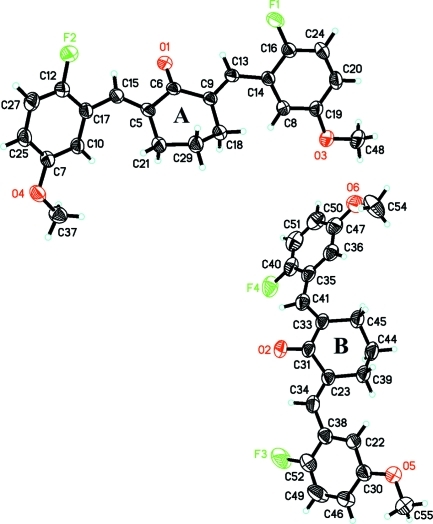
The title compound, C22H20F2O3, a derivative of curcumin, crystallized with two independent molecules in the asymmetric unit. The mean planes of the two 2-fluoro-5-methoxyphenyl groups are aligned at 24.88 (11)° in one molecule and 24.19 (15)° in the other. The dihedral angles between the mean plane of the penta-1,4-dien-3-one group and those of the two 2-fluoro-5-methoxyphenyl rings are 51.16 (11) and 49.16 (10)° in the first molecule, and 45.69 (15) and 54.00 (14)° in the second. The molecules adopt E configurations about the central olefinic bonds.
Related literature
For related structures, see: Liang et al. (2007 ▶); Zhao et al. (2009 ▶); Zhao, Yang, Liang et al. (2010 ▶). For background to and applications of related compounds, see: Aggarwal et al. (2003 ▶); Began et al. (1999 ▶); Ganesh & Aggarwal (2007) ▶; Liang et al.(2009 ▶); Zhao, Yang, Wang et al. (2010 ▶).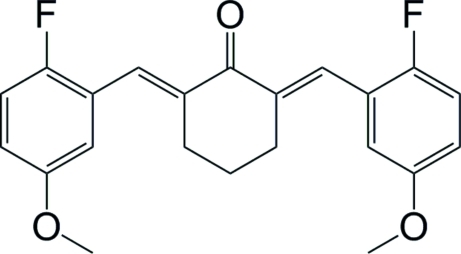
Experimental
Crystal data
C22H20F2O3
M r = 370.38
Triclinic,
a = 9.2334 (10) Å
b = 9.7601 (11) Å
c = 21.433 (2) Å
α = 90.195 (2)°
β = 100.568 (2)°
γ = 92.934 (2)°
V = 1896.1 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 273 K
0.10 × 0.10 × 0.10 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2004 ▶) T min = 0.990, T max = 0.990
10069 measured reflections
6634 independent reflections
3949 reflections with I > 2σ(I)
R int = 0.101
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.147
S = 1.00
6634 reflections
492 parameters
H-atom parameters not refined
Δρmax = 0.26 e Å−3
Δρmin = −0.27 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810048610/ng5065sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810048610/ng5065Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the Xinmiao Talent Project of Zhejiang Province (CLF). The use of the X-ray crystallographic service at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, and the valuable assistance of the staff is gratefully acknowledged.
supplementary crystallographic information
Comment
The title compound, (2E,6E)-2,6-bis(2-fluoro-5-hydroxybenzylidene)cyclohexanone (I), is one of mono-carbonyl analogues of curcumin designed and synthesized by our group.Curcumin reportedly possesses several pharmacological properties including anti-inflammatory, antimicrobial,antiviral, antifungal, antioxidant, chemosensitizing, radiosensitizing,and wound healing activities.Curcumin can suppress tumor initiation, promotion, and metastasis in experimental models. (Began, et al.1999;Ganesh et al.2007).Unlike most chemotherapeutic agents, curcumin has been reported to show almost nontoxicity. These compound have attracted more and more attention. (Aggarwal et al.2003). The need for curcumin-like compounds with improved bioavailability characteristics has led to the chemical synthesis of a series of analogues, using curcumin as the primary structure. In our previous study, a series of fluorine-containing, mono-carbonyl analogues of curcumin were designed and synthesized by the deletion of β-diketone moiety, and their bioactivities were evaluated (Liang et al., 2009; Zhao et al., 2010). Among those compounds, the cyclohexanone-containing analogues exhibited better anti-tumor properties and a wider anti-tumor spectrum than acetone- and cyclopentanone-containing analogues. As a continuation of our broad program of work on the synthesis and structural study of chalcones, the title chalcone derivative has been obtained and an X-ray diffraction study was carried out. Therefore, the structure of one of cyclohexanone-containing compounds (I), was further determined and analyzed using single-crystal X-ray diffraction. Accumulation of detailed structural and pharmacological data facilitated the explanation of the observed structure–activity relationships and modeling of new compounds with potential biological activity.
In this paper, we report the molecular and crystal structures of fluorine-containing, mono-carbonyl analogues of curcumin, (I). The molecule (I), consists of three ring systems, i.e., one cyclohexanone ring and two aryl rings. The central cyclohexanone ring has a distorted chair conformation, and molecular structures have an E-configuration towards the central olefinic bonds, exhibiting a butterfly-shaped geometry. The dihedral angle between the two terminal phenyl rings is 27.19 (13)°, and the two phenyl rings are twisted out of the plane of the central cyclohexanone on the two sides, respectively. Among these derivatives, some of them were reported of their crystal structures ( Liang et al., 2007; Zhao et al., 2009; Zhao et al., 2010).
Experimental
Cyclohexanone (7.5 mmol) was dissolved in ethanol (5 ml) and crushed KOH (15 mmol) was added. The flask was immersed in a bath of crushed ice and a solution of 2-fluoro-5-hydroxybenzaldehyde (15 mmol) in ethanol (5 mmol) was added. The reaction mixture was stirred at 300 K and completion of the reaction was monitored by thin-layer chromatography. Ice-cold water was added to the reaction mixture after 48 h and the yellow solid that separated was filtered off, washed with water and cold ethanol, dried and purified by column chromatography on silica gel (yield: 58.3%). Single crystals of the title compound were grown in a CH2Cl2/CH3OH mixture (5:2 v/v) by slow evaporation (mp 91.3-93.4 °C).
Yellow powder, 58.3% yield, mp 91.3-93.4°C. 1H-NMR (CDCl3) δ: 7.77 (2H, s, Ar-CH=C×2), 7.03 (2H, t, J=9.0Hz, Ar-H3×2), 6.83-6.87 (4H, m, Ar-H4,6×2), 3.80 (6H, s, Ar-OCH3×2), 2.81 (4H, t, J=5.4Hz, CH2-C-CH2), 1.78 (2H, m, >CH2). ESI-MS m/z: 371.0 (M+H)+, calcd for C22H20F2O3: 370.39.
Refinement
The H atoms were positioned geometrically (C—H = 0.93 and 0.96 Å) and refined as riding with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C).
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% displacement ellipsoids for the non-hydrogen atoms. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C22H20F2O3 | Z = 4 |
| Mr = 370.38 | F(000) = 776 |
| Triclinic, P1 | Dx = 1.297 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.2334 (10) Å | Cell parameters from 3025 reflections |
| b = 9.7601 (11) Å | θ = 2.3–23.1° |
| c = 21.433 (2) Å | µ = 0.10 mm−1 |
| α = 90.195 (2)° | T = 273 K |
| β = 100.568 (2)° | Block, colorless |
| γ = 92.934 (2)° | 0.10 × 0.10 × 0.10 mm |
| V = 1896.1 (4) Å3 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 6634 independent reflections |
| Radiation source: fine-focus sealed tube | 3949 reflections with I > 2σ(I) |
| graphite | Rint = 0.101 |
| φ and ω scans | θmax = 25.0°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2004) | h = −10→10 |
| Tmin = 0.990, Tmax = 0.990 | k = −11→11 |
| 10069 measured reflections | l = −18→25 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.057 | H-atom parameters not refined |
| wR(F2) = 0.147 | w = 1/[σ2(Fo2) + (0.055P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max = 0.015 |
| 6634 reflections | Δρmax = 0.26 e Å−3 |
| 492 parameters | Δρmin = −0.27 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0102 (12) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.64586 (15) | −0.51316 (13) | 0.03944 (7) | 0.0840 (4) | |
| O1 | 0.59989 (17) | −0.11281 (16) | −0.08404 (7) | 0.0687 (4) | |
| O4 | 0.94423 (17) | 0.60048 (16) | −0.09902 (8) | 0.0769 (5) | |
| F2 | 0.94845 (17) | 0.08313 (16) | −0.19582 (7) | 0.0938 (5) | |
| C5 | 0.7533 (2) | 0.0813 (2) | −0.04416 (10) | 0.0525 (5) | |
| C6 | 0.6674 (2) | −0.0492 (2) | −0.03706 (10) | 0.0521 (5) | |
| C7 | 0.9400 (2) | 0.4676 (2) | −0.12065 (11) | 0.0602 (6) | |
| C8 | 0.5428 (2) | −0.2508 (2) | 0.13952 (10) | 0.0577 (6) | |
| H8 | 0.5357 | −0.1562 | 0.1412 | 0.069* | |
| C9 | 0.6657 (2) | −0.1030 (2) | 0.02818 (10) | 0.0513 (5) | |
| C10 | 0.8601 (2) | 0.3605 (2) | −0.09952 (10) | 0.0585 (6) | |
| H10 | 0.8033 | 0.3770 | −0.0689 | 0.070* | |
| O3 | 0.4590 (2) | −0.25806 (18) | 0.23530 (8) | 0.0859 (5) | |
| C12 | 0.9460 (3) | 0.2116 (2) | −0.17015 (11) | 0.0666 (6) | |
| C13 | 0.6103 (2) | −0.2322 (2) | 0.03145 (10) | 0.0561 (5) | |
| H13 | 0.5820 | −0.2785 | −0.0072 | 0.067* | |
| C14 | 0.5881 (2) | −0.3100 (2) | 0.08760 (10) | 0.0541 (5) | |
| C15 | 0.7815 (2) | 0.1087 (2) | −0.10238 (11) | 0.0593 (6) | |
| H15 | 0.7441 | 0.0433 | −0.1336 | 0.071* | |
| C16 | 0.6001 (2) | −0.4513 (2) | 0.08892 (11) | 0.0631 (6) | |
| C17 | 0.8626 (2) | 0.2266 (2) | −0.12325 (10) | 0.0576 (6) | |
| C18 | 0.7318 (2) | −0.0142 (2) | 0.08455 (10) | 0.0603 (6) | |
| H18A | 0.7671 | −0.0722 | 0.1202 | 0.072* | |
| H18B | 0.6561 | 0.0409 | 0.0961 | 0.072* | |
| C19 | 0.5085 (2) | −0.3297 (2) | 0.18834 (11) | 0.0633 (6) | |
| C20 | 0.5209 (3) | −0.4707 (3) | 0.18760 (12) | 0.0742 (7) | |
| H20 | 0.4980 | −0.5238 | 0.2208 | 0.089* | |
| C21 | 0.8076 (3) | 0.1678 (2) | 0.01397 (10) | 0.0664 (6) | |
| H21A | 0.7292 | 0.2237 | 0.0222 | 0.080* | |
| H21B | 0.8891 | 0.2288 | 0.0068 | 0.080* | |
| C22 | 0.5255 (3) | 0.7126 (3) | 0.62792 (12) | 0.0742 (7) | |
| H22 | 0.5377 | 0.6186 | 0.6268 | 0.089* | |
| C23 | 0.3185 (3) | 0.5673 (3) | 0.51074 (11) | 0.0715 (7) | |
| C24 | 0.5675 (3) | −0.5308 (3) | 0.13702 (13) | 0.0727 (7) | |
| H24 | 0.5766 | −0.6252 | 0.1358 | 0.087* | |
| C25 | 1.0243 (3) | 0.4454 (3) | −0.16668 (12) | 0.0723 (7) | |
| H25 | 1.0792 | 0.5179 | −0.1805 | 0.087* | |
| O5 | 0.6789 (2) | 0.7139 (2) | 0.72684 (9) | 0.1000 (6) | |
| C27 | 1.0266 (3) | 0.3162 (3) | −0.19182 (12) | 0.0780 (7) | |
| H27 | 1.0820 | 0.3002 | −0.2230 | 0.094* | |
| O2 | 0.2913 (2) | 0.5903 (2) | 0.39992 (8) | 0.1026 (6) | |
| C29 | 0.8584 (2) | 0.0796 (2) | 0.07149 (11) | 0.0682 (6) | |
| H29A | 0.9384 | 0.0251 | 0.0638 | 0.082* | |
| H29B | 0.8948 | 0.1382 | 0.1083 | 0.082* | |
| C30 | 0.5948 (3) | 0.7889 (3) | 0.68033 (13) | 0.0799 (7) | |
| C31 | 0.2607 (3) | 0.5211 (3) | 0.44426 (12) | 0.0769 (7) | |
| F4 | −0.1739 (2) | 0.4179 (2) | 0.28990 (8) | 0.1210 (6) | |
| C33 | 0.1666 (3) | 0.3919 (3) | 0.43095 (12) | 0.0733 (7) | |
| C34 | 0.3691 (3) | 0.6978 (3) | 0.51894 (12) | 0.0803 (7) | |
| H34 | 0.3588 | 0.7492 | 0.4821 | 0.096* | |
| C35 | −0.0142 (3) | 0.2610 (3) | 0.34497 (12) | 0.0787 (8) | |
| C36 | 0.0179 (3) | 0.1235 (3) | 0.35708 (12) | 0.0826 (8) | |
| H36 | 0.1025 | 0.1030 | 0.3856 | 0.099* | |
| C37 | 0.8455 (3) | 0.6307 (3) | −0.05740 (13) | 0.0797 (7) | |
| H37A | 0.7462 | 0.6047 | −0.0775 | 0.120* | |
| H37B | 0.8534 | 0.7272 | −0.0478 | 0.120* | |
| H37C | 0.8706 | 0.5804 | −0.0188 | 0.120* | |
| C38 | 0.4382 (3) | 0.7727 (3) | 0.57692 (13) | 0.0776 (7) | |
| C39 | 0.3083 (3) | 0.4666 (3) | 0.56284 (12) | 0.0808 (7) | |
| H39A | 0.3055 | 0.5166 | 0.6018 | 0.097* | |
| H39B | 0.3959 | 0.4139 | 0.5699 | 0.097* | |
| C40 | −0.1396 (4) | 0.2852 (4) | 0.30224 (14) | 0.0935 (9) | |
| C41 | 0.0852 (3) | 0.3759 (3) | 0.37287 (13) | 0.0840 (8) | |
| H41 | 0.0928 | 0.4493 | 0.3459 | 0.101* | |
| O6 | −0.0522 (3) | −0.1187 (2) | 0.33634 (11) | 0.1141 (7) | |
| F3 | 0.3415 (3) | 0.97700 (19) | 0.53269 (11) | 0.1524 (9) | |
| C44 | 0.1740 (3) | 0.3700 (3) | 0.54768 (12) | 0.0921 (9) | |
| H44A | 0.0863 | 0.4217 | 0.5457 | 0.111* | |
| H44B | 0.1757 | 0.3042 | 0.5816 | 0.111* | |
| C45 | 0.1653 (3) | 0.2928 (3) | 0.48495 (12) | 0.0846 (8) | |
| H45A | 0.2485 | 0.2349 | 0.4879 | 0.102* | |
| H45B | 0.0756 | 0.2343 | 0.4766 | 0.102* | |
| C46 | 0.5792 (4) | 0.9275 (3) | 0.68372 (16) | 0.0975 (9) | |
| H46 | 0.6264 | 0.9786 | 0.7190 | 0.117* | |
| C47 | −0.0755 (3) | 0.0179 (4) | 0.32694 (15) | 0.0913 (9) | |
| C48 | 0.3954 (4) | −0.3343 (3) | 0.28049 (13) | 0.1087 (10) | |
| H48A | 0.3209 | −0.3989 | 0.2589 | 0.163* | |
| H48B | 0.3517 | −0.2730 | 0.3060 | 0.163* | |
| H48C | 0.4704 | −0.3826 | 0.3072 | 0.163* | |
| C49 | 0.4926 (4) | 0.9889 (3) | 0.6342 (2) | 0.1229 (12) | |
| H49 | 0.4792 | 1.0825 | 0.6358 | 0.147* | |
| C50 | −0.2011 (4) | 0.0478 (4) | 0.28457 (16) | 0.1095 (11) | |
| H50 | −0.2637 | −0.0229 | 0.2643 | 0.131* | |
| C51 | −0.2339 (4) | 0.1829 (5) | 0.27229 (16) | 0.1145 (11) | |
| H51 | −0.3189 | 0.2037 | 0.2441 | 0.137* | |
| C52 | 0.4259 (4) | 0.9131 (3) | 0.58215 (17) | 0.1020 (9) | |
| C54 | 0.0799 (4) | −0.1549 (4) | 0.3739 (2) | 0.1360 (14) | |
| H54A | 0.1614 | −0.1118 | 0.3581 | 0.204* | |
| H54B | 0.0861 | −0.2527 | 0.3726 | 0.204* | |
| H54C | 0.0830 | −0.1252 | 0.4169 | 0.204* | |
| C55 | 0.7647 (4) | 0.7839 (3) | 0.77809 (15) | 0.1175 (11) | |
| H55A | 0.8265 | 0.8535 | 0.7630 | 0.176* | |
| H55B | 0.8252 | 0.7208 | 0.8039 | 0.176* | |
| H55C | 0.7019 | 0.8258 | 0.8029 | 0.176* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.1075 (10) | 0.0526 (8) | 0.1018 (11) | 0.0057 (7) | 0.0447 (9) | −0.0015 (7) |
| O1 | 0.0905 (11) | 0.0540 (10) | 0.0632 (10) | −0.0054 (8) | 0.0210 (9) | −0.0035 (8) |
| O4 | 0.0902 (11) | 0.0533 (10) | 0.0936 (12) | −0.0070 (8) | 0.0364 (10) | 0.0036 (8) |
| F2 | 0.1283 (12) | 0.0732 (10) | 0.0938 (11) | 0.0039 (9) | 0.0572 (9) | −0.0062 (8) |
| C5 | 0.0541 (12) | 0.0410 (12) | 0.0645 (14) | 0.0075 (9) | 0.0151 (10) | 0.0029 (10) |
| C6 | 0.0572 (12) | 0.0434 (12) | 0.0591 (14) | 0.0080 (10) | 0.0183 (11) | −0.0030 (10) |
| C7 | 0.0629 (13) | 0.0544 (15) | 0.0648 (14) | 0.0012 (11) | 0.0163 (11) | 0.0092 (11) |
| C8 | 0.0638 (13) | 0.0511 (13) | 0.0594 (14) | −0.0038 (10) | 0.0160 (11) | 0.0065 (11) |
| C9 | 0.0541 (11) | 0.0437 (12) | 0.0594 (13) | 0.0057 (9) | 0.0180 (10) | 0.0009 (10) |
| C10 | 0.0642 (13) | 0.0559 (14) | 0.0602 (14) | 0.0032 (11) | 0.0237 (11) | 0.0092 (11) |
| O3 | 0.1237 (14) | 0.0769 (12) | 0.0651 (11) | −0.0099 (10) | 0.0422 (10) | 0.0060 (9) |
| C12 | 0.0820 (15) | 0.0563 (15) | 0.0671 (15) | 0.0027 (12) | 0.0283 (13) | 0.0022 (12) |
| C13 | 0.0669 (13) | 0.0447 (13) | 0.0606 (13) | 0.0033 (10) | 0.0222 (11) | −0.0028 (10) |
| C14 | 0.0573 (12) | 0.0472 (13) | 0.0603 (14) | 0.0004 (10) | 0.0178 (10) | 0.0058 (10) |
| C15 | 0.0664 (13) | 0.0504 (13) | 0.0651 (15) | 0.0049 (10) | 0.0222 (11) | 0.0045 (11) |
| C16 | 0.0705 (14) | 0.0533 (14) | 0.0684 (15) | 0.0007 (11) | 0.0213 (12) | 0.0024 (12) |
| C17 | 0.0617 (13) | 0.0541 (14) | 0.0598 (14) | 0.0024 (10) | 0.0189 (11) | 0.0093 (11) |
| C18 | 0.0670 (13) | 0.0535 (13) | 0.0616 (14) | 0.0004 (11) | 0.0158 (11) | 0.0040 (11) |
| C19 | 0.0717 (14) | 0.0625 (16) | 0.0565 (14) | −0.0056 (12) | 0.0162 (11) | 0.0068 (12) |
| C20 | 0.0850 (16) | 0.0688 (18) | 0.0674 (16) | −0.0083 (13) | 0.0134 (13) | 0.0227 (13) |
| C21 | 0.0795 (15) | 0.0544 (14) | 0.0632 (15) | −0.0096 (11) | 0.0108 (12) | 0.0057 (11) |
| C22 | 0.0987 (18) | 0.0635 (16) | 0.0681 (17) | 0.0113 (14) | 0.0334 (14) | 0.0018 (13) |
| C23 | 0.0922 (17) | 0.0703 (18) | 0.0614 (16) | 0.0245 (14) | 0.0337 (13) | 0.0114 (12) |
| C24 | 0.0839 (16) | 0.0494 (14) | 0.0856 (18) | 0.0006 (12) | 0.0185 (14) | 0.0153 (13) |
| C25 | 0.0761 (15) | 0.0691 (17) | 0.0772 (17) | −0.0067 (13) | 0.0311 (13) | 0.0148 (13) |
| O5 | 0.1470 (17) | 0.0775 (13) | 0.0736 (13) | −0.0049 (12) | 0.0182 (12) | 0.0008 (11) |
| C27 | 0.0853 (17) | 0.084 (2) | 0.0748 (17) | 0.0039 (14) | 0.0420 (14) | 0.0101 (14) |
| O2 | 0.1667 (18) | 0.0881 (14) | 0.0632 (11) | 0.0080 (13) | 0.0474 (12) | 0.0147 (10) |
| C29 | 0.0729 (14) | 0.0639 (15) | 0.0650 (15) | −0.0100 (12) | 0.0087 (12) | 0.0079 (12) |
| C30 | 0.0995 (19) | 0.0662 (18) | 0.0807 (19) | 0.0013 (15) | 0.0351 (16) | 0.0068 (15) |
| C31 | 0.1073 (19) | 0.0776 (18) | 0.0566 (15) | 0.0314 (15) | 0.0355 (14) | 0.0123 (13) |
| F4 | 0.1282 (13) | 0.1394 (17) | 0.1007 (12) | 0.0539 (12) | 0.0215 (10) | 0.0341 (11) |
| C33 | 0.0932 (17) | 0.0785 (19) | 0.0570 (16) | 0.0277 (15) | 0.0305 (14) | 0.0111 (13) |
| C34 | 0.1038 (19) | 0.080 (2) | 0.0666 (17) | 0.0287 (16) | 0.0325 (14) | 0.0184 (14) |
| C35 | 0.0879 (18) | 0.098 (2) | 0.0588 (15) | 0.0259 (17) | 0.0311 (14) | 0.0082 (15) |
| C36 | 0.0878 (18) | 0.099 (2) | 0.0680 (17) | 0.0251 (17) | 0.0271 (14) | 0.0042 (16) |
| C37 | 0.0767 (15) | 0.0667 (17) | 0.101 (2) | 0.0059 (13) | 0.0291 (15) | −0.0043 (14) |
| C38 | 0.0963 (18) | 0.0647 (17) | 0.0798 (19) | 0.0160 (14) | 0.0339 (15) | 0.0086 (14) |
| C39 | 0.0998 (18) | 0.089 (2) | 0.0597 (15) | 0.0095 (16) | 0.0302 (14) | 0.0125 (13) |
| C40 | 0.097 (2) | 0.122 (3) | 0.0701 (19) | 0.041 (2) | 0.0291 (17) | 0.0216 (19) |
| C41 | 0.107 (2) | 0.088 (2) | 0.0671 (18) | 0.0394 (17) | 0.0333 (16) | 0.0119 (15) |
| O6 | 0.1315 (18) | 0.0947 (18) | 0.1233 (18) | 0.0054 (14) | 0.0425 (15) | −0.0041 (13) |
| F3 | 0.196 (2) | 0.0803 (13) | 0.1656 (19) | 0.0409 (13) | −0.0178 (16) | 0.0237 (12) |
| C44 | 0.113 (2) | 0.104 (2) | 0.0666 (17) | 0.0101 (18) | 0.0360 (15) | 0.0221 (15) |
| C45 | 0.1054 (19) | 0.088 (2) | 0.0659 (17) | 0.0096 (15) | 0.0288 (14) | 0.0173 (14) |
| C46 | 0.123 (2) | 0.071 (2) | 0.099 (2) | −0.0025 (18) | 0.0245 (19) | −0.0082 (17) |
| C47 | 0.091 (2) | 0.110 (3) | 0.081 (2) | 0.011 (2) | 0.0364 (17) | −0.0005 (19) |
| C48 | 0.162 (3) | 0.104 (2) | 0.0699 (18) | −0.014 (2) | 0.0522 (19) | 0.0138 (16) |
| C49 | 0.146 (3) | 0.058 (2) | 0.162 (4) | 0.013 (2) | 0.020 (3) | −0.005 (2) |
| C50 | 0.101 (2) | 0.137 (3) | 0.093 (2) | 0.001 (2) | 0.026 (2) | −0.006 (2) |
| C51 | 0.098 (2) | 0.158 (4) | 0.088 (2) | 0.019 (3) | 0.0149 (19) | 0.012 (2) |
| C52 | 0.125 (2) | 0.070 (2) | 0.111 (2) | 0.0194 (19) | 0.016 (2) | 0.0157 (19) |
| C54 | 0.109 (3) | 0.091 (3) | 0.218 (4) | 0.025 (2) | 0.049 (3) | 0.038 (3) |
| C55 | 0.166 (3) | 0.104 (3) | 0.076 (2) | −0.010 (2) | 0.009 (2) | −0.0054 (18) |
Geometric parameters (Å, °)
| F1—C16 | 1.362 (3) | O2—C31 | 1.234 (3) |
| O1—C6 | 1.231 (2) | C29—H29A | 0.9700 |
| O4—C7 | 1.372 (3) | C29—H29B | 0.9700 |
| O4—C37 | 1.427 (3) | C30—C46 | 1.371 (4) |
| F2—C12 | 1.371 (3) | C31—C33 | 1.490 (4) |
| C5—C15 | 1.346 (3) | F4—C40 | 1.364 (4) |
| C5—C6 | 1.489 (3) | C33—C41 | 1.335 (3) |
| C5—C21 | 1.495 (3) | C33—C45 | 1.512 (3) |
| C6—C9 | 1.498 (3) | C34—C38 | 1.464 (4) |
| C7—C10 | 1.375 (3) | C34—H34 | 0.9300 |
| C7—C25 | 1.387 (3) | C35—C40 | 1.370 (4) |
| C8—C19 | 1.375 (3) | C35—C36 | 1.404 (4) |
| C8—C14 | 1.392 (3) | C35—C41 | 1.465 (4) |
| C8—H8 | 0.9300 | C36—C47 | 1.387 (4) |
| C9—C13 | 1.343 (3) | C36—H36 | 0.9300 |
| C9—C18 | 1.501 (3) | C37—H37A | 0.9600 |
| C10—C17 | 1.404 (3) | C37—H37B | 0.9600 |
| C10—H10 | 0.9300 | C37—H37C | 0.9600 |
| O3—C19 | 1.381 (3) | C38—C52 | 1.386 (4) |
| O3—C48 | 1.416 (3) | C39—C44 | 1.504 (4) |
| C12—C27 | 1.366 (3) | C39—H39A | 0.9700 |
| C12—C17 | 1.385 (3) | C39—H39B | 0.9700 |
| C13—C14 | 1.465 (3) | C40—C51 | 1.368 (5) |
| C13—H13 | 0.9300 | C41—H41 | 0.9300 |
| C14—C16 | 1.389 (3) | O6—C47 | 1.370 (4) |
| C15—C17 | 1.457 (3) | O6—C54 | 1.395 (4) |
| C15—H15 | 0.9300 | F3—C52 | 1.368 (3) |
| C16—C24 | 1.361 (3) | C44—C45 | 1.526 (4) |
| C18—C29 | 1.515 (3) | C44—H44A | 0.9700 |
| C18—H18A | 0.9700 | C44—H44B | 0.9700 |
| C18—H18B | 0.9700 | C45—H45A | 0.9700 |
| C19—C20 | 1.387 (3) | C45—H45B | 0.9700 |
| C20—C24 | 1.378 (3) | C46—C49 | 1.367 (4) |
| C20—H20 | 0.9300 | C46—H46 | 0.9300 |
| C21—C29 | 1.522 (3) | C47—C50 | 1.381 (4) |
| C21—H21A | 0.9700 | C48—H48A | 0.9600 |
| C21—H21B | 0.9700 | C48—H48B | 0.9600 |
| C22—C30 | 1.381 (3) | C48—H48C | 0.9600 |
| C22—C38 | 1.385 (3) | C49—C52 | 1.366 (5) |
| C22—H22 | 0.9300 | C49—H49 | 0.9300 |
| C23—C34 | 1.334 (4) | C50—C51 | 1.383 (5) |
| C23—C31 | 1.486 (4) | C50—H50 | 0.9300 |
| C23—C39 | 1.502 (3) | C51—H51 | 0.9300 |
| C24—H24 | 0.9300 | C54—H54A | 0.9600 |
| C25—C27 | 1.372 (3) | C54—H54B | 0.9600 |
| C25—H25 | 0.9300 | C54—H54C | 0.9600 |
| O5—C30 | 1.383 (3) | C55—H55A | 0.9600 |
| O5—C55 | 1.386 (3) | C55—H55B | 0.9600 |
| C27—H27 | 0.9300 | C55—H55C | 0.9600 |
| C7—O4—C37 | 116.94 (17) | C23—C31—C33 | 120.4 (2) |
| C15—C5—C6 | 116.80 (19) | C41—C33—C31 | 117.1 (2) |
| C15—C5—C21 | 125.17 (19) | C41—C33—C45 | 125.2 (3) |
| C6—C5—C21 | 117.96 (19) | C31—C33—C45 | 117.6 (2) |
| O1—C6—C5 | 120.62 (19) | C23—C34—C38 | 130.1 (2) |
| O1—C6—C9 | 120.33 (18) | C23—C34—H34 | 115.0 |
| C5—C6—C9 | 119.04 (19) | C38—C34—H34 | 115.0 |
| O4—C7—C10 | 124.4 (2) | C40—C35—C36 | 117.3 (3) |
| O4—C7—C25 | 115.3 (2) | C40—C35—C41 | 120.1 (3) |
| C10—C7—C25 | 120.3 (2) | C36—C35—C41 | 122.6 (3) |
| C19—C8—C14 | 121.1 (2) | C47—C36—C35 | 120.6 (3) |
| C19—C8—H8 | 119.4 | C47—C36—H36 | 119.7 |
| C14—C8—H8 | 119.4 | C35—C36—H36 | 119.7 |
| C13—C9—C6 | 116.31 (19) | O4—C37—H37A | 109.5 |
| C13—C9—C18 | 124.71 (19) | O4—C37—H37B | 109.5 |
| C6—C9—C18 | 118.90 (18) | H37A—C37—H37B | 109.5 |
| C7—C10—C17 | 121.2 (2) | O4—C37—H37C | 109.5 |
| C7—C10—H10 | 119.4 | H37A—C37—H37C | 109.5 |
| C17—C10—H10 | 119.4 | H37B—C37—H37C | 109.5 |
| C19—O3—C48 | 118.0 (2) | C52—C38—C22 | 115.7 (3) |
| C27—C12—F2 | 118.2 (2) | C52—C38—C34 | 120.7 (3) |
| C27—C12—C17 | 124.2 (2) | C22—C38—C34 | 123.5 (2) |
| F2—C12—C17 | 117.6 (2) | C23—C39—C44 | 112.5 (2) |
| C9—C13—C14 | 128.9 (2) | C23—C39—H39A | 109.1 |
| C9—C13—H13 | 115.6 | C44—C39—H39A | 109.1 |
| C14—C13—H13 | 115.6 | C23—C39—H39B | 109.1 |
| C16—C14—C8 | 116.4 (2) | C44—C39—H39B | 109.1 |
| C16—C14—C13 | 120.4 (2) | H39A—C39—H39B | 107.8 |
| C8—C14—C13 | 122.94 (19) | F4—C40—C51 | 118.3 (3) |
| C5—C15—C17 | 129.0 (2) | F4—C40—C35 | 118.5 (3) |
| C5—C15—H15 | 115.5 | C51—C40—C35 | 123.2 (3) |
| C17—C15—H15 | 115.5 | C33—C41—C35 | 130.2 (3) |
| C24—C16—F1 | 118.5 (2) | C33—C41—H41 | 114.9 |
| C24—C16—C14 | 123.2 (2) | C35—C41—H41 | 114.9 |
| F1—C16—C14 | 118.3 (2) | C47—O6—C54 | 118.4 (3) |
| C12—C17—C10 | 115.72 (19) | C39—C44—C45 | 112.5 (2) |
| C12—C17—C15 | 120.5 (2) | C39—C44—H44A | 109.1 |
| C10—C17—C15 | 123.8 (2) | C45—C44—H44A | 109.1 |
| C9—C18—C29 | 112.16 (18) | C39—C44—H44B | 109.1 |
| C9—C18—H18A | 109.2 | C45—C44—H44B | 109.1 |
| C29—C18—H18A | 109.2 | H44A—C44—H44B | 107.8 |
| C9—C18—H18B | 109.2 | C33—C45—C44 | 110.8 (2) |
| C29—C18—H18B | 109.2 | C33—C45—H45A | 109.5 |
| H18A—C18—H18B | 107.9 | C44—C45—H45A | 109.5 |
| C8—C19—O3 | 114.9 (2) | C33—C45—H45B | 109.5 |
| C8—C19—C20 | 120.6 (2) | C44—C45—H45B | 109.5 |
| O3—C19—C20 | 124.4 (2) | H45A—C45—H45B | 108.1 |
| C24—C20—C19 | 119.0 (2) | C49—C46—C30 | 118.6 (3) |
| C24—C20—H20 | 120.5 | C49—C46—H46 | 120.7 |
| C19—C20—H20 | 120.5 | C30—C46—H46 | 120.7 |
| C5—C21—C29 | 111.31 (19) | O6—C47—C50 | 116.0 (3) |
| C5—C21—H21A | 109.4 | O6—C47—C36 | 124.1 (3) |
| C29—C21—H21A | 109.4 | C50—C47—C36 | 119.9 (3) |
| C5—C21—H21B | 109.4 | O3—C48—H48A | 109.5 |
| C29—C21—H21B | 109.4 | O3—C48—H48B | 109.5 |
| H21A—C21—H21B | 108.0 | H48A—C48—H48B | 109.5 |
| C30—C22—C38 | 121.5 (3) | O3—C48—H48C | 109.5 |
| C30—C22—H22 | 119.3 | H48A—C48—H48C | 109.5 |
| C38—C22—H22 | 119.3 | H48B—C48—H48C | 109.5 |
| C34—C23—C31 | 116.4 (2) | C46—C49—C52 | 120.1 (3) |
| C34—C23—C39 | 125.6 (2) | C46—C49—H49 | 119.9 |
| C31—C23—C39 | 117.9 (2) | C52—C49—H49 | 119.9 |
| C16—C24—C20 | 119.6 (2) | C47—C50—C51 | 120.1 (4) |
| C16—C24—H24 | 120.2 | C47—C50—H50 | 120.0 |
| C20—C24—H24 | 120.2 | C51—C50—H50 | 120.0 |
| C27—C25—C7 | 119.9 (2) | C40—C51—C50 | 119.0 (3) |
| C27—C25—H25 | 120.1 | C40—C51—H51 | 120.5 |
| C7—C25—H25 | 120.1 | C50—C51—H51 | 120.5 |
| C30—O5—C55 | 118.4 (2) | C49—C52—F3 | 119.2 (3) |
| C12—C27—C25 | 118.7 (2) | C49—C52—C38 | 123.1 (3) |
| C12—C27—H27 | 120.7 | F3—C52—C38 | 117.7 (3) |
| C25—C27—H27 | 120.7 | O6—C54—H54A | 109.5 |
| C18—C29—C21 | 110.31 (18) | O6—C54—H54B | 109.5 |
| C18—C29—H29A | 109.6 | H54A—C54—H54B | 109.5 |
| C21—C29—H29A | 109.6 | O6—C54—H54C | 109.5 |
| C18—C29—H29B | 109.6 | H54A—C54—H54C | 109.5 |
| C21—C29—H29B | 109.6 | H54B—C54—H54C | 109.5 |
| H29A—C29—H29B | 108.1 | O5—C55—H55A | 109.5 |
| C46—C30—O5 | 124.6 (3) | O5—C55—H55B | 109.5 |
| C46—C30—C22 | 121.0 (3) | H55A—C55—H55B | 109.5 |
| O5—C30—C22 | 114.4 (2) | O5—C55—H55C | 109.5 |
| O2—C31—C23 | 119.6 (3) | H55A—C55—H55C | 109.5 |
| O2—C31—C33 | 119.9 (2) | H55B—C55—H55C | 109.5 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG5065).
References
- Aggarwal, B. B., Kumar, A. & Bharti, A. C. (2003). Anticancer Res 23, 363–398. [PubMed]
- Began, G., Sudharshan, E., Sankar, K. U. A. & Appu, R. G. (1999). J. Agric. Food Chem.47, 4992–4997. [DOI] [PubMed]
- Bruker (2004). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Ganesh, J. C. & Aggarwal, B. B. (2007). Clin. Immunol.27, 19–35. [DOI] [PubMed]
- Liang, G., Shao, L. L., Wang, Y., Zhao, C. G., Chu, Y. H., Xiao, J., Zhao, Y., Li, X. K. & Yang, S. L. (2009). Bioorg. Med. Chem 17, 2623–2631. [DOI] [PubMed]
- Liang, G., Tian, J.-L., Zhao, C.-G. & Li, X.-K. (2007). Acta Cryst. E63, o3630.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhao, C. G., Yang, J., Huang, Y., Liang, G. & Li, X. K. (2009). Z. Kristallogr. New Cryst. Struct.224, 337–338.
- Zhao, C. G., Yang, J., Liang, D. L., Tang, Q. Q., Zhang, Y., Liang, G. & Li, X. K. (2010). Chin. J. Org. Chem.30, 289–294.
- Zhao, C. G., Yang, J., Wang, Y., Liang, D. L., Yang, X. Y., Wu, J. Z., Wu, X. P., Yang, S. L., Li, X. K. & Liang, G. (2010). Bioorg. Med. Chem 18, 2388–2393. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810048610/ng5065sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810048610/ng5065Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report