Abstract
The structure of the title compound, C16H19N3O4S, shows the planes described by the thiophene and the pyrroles are twisted by 17.06 (4)°. Additionally, the structure shows the azomethine bond adopts the E configuration, while the pyrrole is disordered as a heterocycle flip [occupancy ratio 0.729 (5):0.271 (5)]. The three-dimensional network is well packed and involves N–H⋯O hydrogen bonding and π–π stacking [centroid–centroid distance = 4.294 (8) Å].
Related literature
For our on-going research on conjugated azomethines, see: Dufresne & Skene (2008 ▶). For bond lengths in comparable azomethines, see: Skene et al. (2006 ▶); Dufresne & Skene (2010 ▶).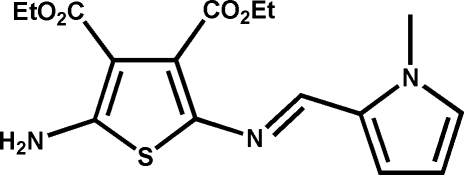
Experimental
Crystal data
C16H19N3O4S
M r = 349.40
Monoclinic,

a = 8.8212 (18) Å
b = 9.0799 (18) Å
c = 21.793 (4) Å
β = 97.50 (3)°
V = 1730.6 (6) Å3
Z = 4
Cu Kα radiation
μ = 1.89 mm−1
T = 123 K
0.17 × 0.16 × 0.15 mm
Data collection
Bruker SMART 6000 diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick,1996 ▶) T min = 0.710, T max = 0.762
20876 measured reflections
3367 independent reflections
3046 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.116
S = 1.07
3367 reflections
267 parameters
32 restraints
H-atom parameters constrained
Δρmax = 0.32 e Å−3
Δρmin = −0.54 e Å−3
Data collection: SMART (Bruker, 2003 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶) and ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: UdMX (Marris, 2004 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810046775/bh2321sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810046775/bh2321Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1B⋯O3i | 0.88 | 2.09 | 2.925 (3) | 157 |
Symmetry code: (i)  .
.
Acknowledgments
NSERC Canada is thanked for DG and RTI grants allowing this work to be performed in addition to CFI for additional equipment funding. SD also thanks NSERC for a graduate scholarship. WGS acknowledges both the Alexander von Humboldt Foundation and the RSC for a JWT Jones Travelling fellowships, allowing the completion of this manuscript.
supplementary crystallographic information
Comment
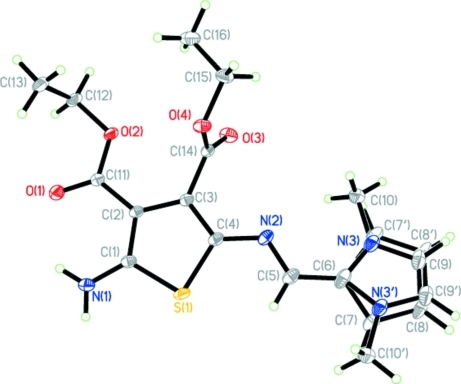
During our on-going research relating to conjugated azomethines (Dufresne & Skene, 2008), we prepared the title compound. The structure is given in figure 1. The pyrrole is disordered. The occupation factor was found to be 73% for the antiperiplanar heterocycle. The salient feature of the resolved structure is assigning the absolute isomer of the azomethine, which is not readily possible by other means. The E isomer was found and the crystal symmetry was P21/c. Neither solvent nor counter-ions were found in the structure.
A major point of interest is the azomethine bond. The bond lengths for N2—C4, N2—C5 and C5—C6 are 1.372 (2), 1.292 (2) and 1.424 (2) Å, respectively. These are similar to comparable azomethines (Skene et al., 2006 and Dufresne & Skene, 2010) whose homologue lengths are 1.381 (3), 1.283 (3) and 1.426 (3) Å.
We found that the heterocycles of the title compound are not coplanar, according to angle between the mean planes described by them. The angle between these planes was found to be 17.06 (4)°. This is in contrast to an analogous thiophene-azomethine compound (Skene et al., 2006) whose mean plane angle is 7.25 (11)°.
Figure 2 shows the H-bonding occurring within the lattice. Only one H-bonding was found between N1—H1B···O3ii with an angle of 157.1° and a distance of 2.925 (3) Å between the nitrogen and the oxygen. Hydrogen bonding and π-stacking are the driving forces for the overall assembly. π-stacking was found to take place between the pyrroles as seen in Figure 3.
Experimental
1-Methyl-2-pyrrole-carboxaldehyde and 2,5-diamino-thiophene-3,4-dicarboxylic acid diethyl ester were mixed in anhydrous 2-propanol with a catalytic amount of TFA and refluxed for 12 h. The reaction was then purified by flash chromatography to afford the title compound as a yellow solid. Single crystals were obtained by slow evaporation of an acetone solution.
Refinement
C-bonded H atoms were placed in calculated positions (C—H = 0.93–0.98 Å) and included in the refinement in the riding-model approximation, with Uiso(H) = 1.2-1.5 Ueq(C). The protons on the amino group were placed in calculated positions (N—H = 0.88 Å) and included in the refinement in the riding-model approximation, with Uiso(H) = 1.2 Ueq(N). During the refinement, evidence came that the structure was disordered as an inversion of terminal heterocycles. We first tried to fix each part to half of the weight and then let it vary to the optimized proportion of 73:27. We were forced to add constraints to the minor counterpart so it looks like the major one. We used fixed similar temperature factors, as well as distances and angles restraints with every disordered atom.
Figures
Fig. 1.
ORTEP representation of the title molecule with the numbering scheme adopted (Farrugia, 1997). The disorder on the pyrrole unit is represented by prime symbols. Ellipsoids drawn at 30% probability level.
Fig. 2.
Supramolecular structure showing the intermolecular H-bonding giving the structural arrangement. Disorder has been omitted for clarity. Dashed lines indicate hydrogen bonds. [Symmetry codes: (i) 1 - x, -1/2 + y, 1/2 - z; (ii) 1 - x, 1/2 + y, 1/2 - z; (iii) x, 1 + y, z.]
Fig. 3.
The three-dimensional network demonstrating the π-stacking in the lattice. Disorder has been omitted for clarity.
Crystal data
| C16H19N3O4S | F(000) = 736 |
| Mr = 349.40 | Dx = 1.341 Mg m−3 |
| Monoclinic, P21/c | Melting point: 404(2) K |
| Hall symbol: -P 2ybc | Cu Kα radiation, λ = 1.54178 Å |
| a = 8.8212 (18) Å | Cell parameters from 10603 reflections |
| b = 9.0799 (18) Å | θ = 4.1–71.3° |
| c = 21.793 (4) Å | µ = 1.89 mm−1 |
| β = 97.50 (3)° | T = 123 K |
| V = 1730.6 (6) Å3 | Block, yellow |
| Z = 4 | 0.17 × 0.16 × 0.15 mm |
Data collection
| Bruker SMART 6000 diffractometer | 3367 independent reflections |
| Radiation source: Rotating Anode | 3046 reflections with I > 2σ(I) |
| Montel 200 optics | Rint = 0.034 |
| Detector resolution: 5.5 pixels mm-1 | θmax = 72.0°, θmin = 4.1° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (SADABS; Sheldrick,1996) | k = −11→11 |
| Tmin = 0.710, Tmax = 0.762 | l = −26→25 |
| 20876 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.116 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0845P)2 + 0.153P] where P = (Fo2 + 2Fc2)/3 |
| 3367 reflections | (Δ/σ)max < 0.001 |
| 267 parameters | Δρmax = 0.32 e Å−3 |
| 32 restraints | Δρmin = −0.54 e Å−3 |
| 0 constraints |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | 0.45470 (4) | 0.43388 (4) | 0.275627 (17) | 0.02979 (14) | |
| O1 | 0.80431 (13) | 0.22781 (12) | 0.16548 (5) | 0.0338 (3) | |
| O2 | 0.89746 (11) | 0.11974 (11) | 0.25566 (5) | 0.0280 (2) | |
| O3 | 0.73230 (12) | 0.04262 (11) | 0.38224 (5) | 0.0347 (3) | |
| O4 | 0.89935 (11) | 0.22984 (11) | 0.38638 (5) | 0.0288 (2) | |
| N1 | 0.56568 (15) | 0.42094 (15) | 0.16734 (6) | 0.0352 (3) | |
| H1A | 0.6308 | 0.3913 | 0.1426 | 0.042* | |
| H1B | 0.4906 | 0.4809 | 0.1534 | 0.042* | |
| N2 | 0.50239 (13) | 0.31869 (14) | 0.39473 (6) | 0.0304 (3) | |
| C1 | 0.58092 (15) | 0.37474 (15) | 0.22635 (6) | 0.0255 (3) | |
| C2 | 0.69216 (14) | 0.28057 (14) | 0.25579 (6) | 0.0217 (3) | |
| C3 | 0.67137 (15) | 0.25520 (14) | 0.31923 (6) | 0.0227 (3) | |
| C4 | 0.54893 (15) | 0.32748 (16) | 0.33720 (7) | 0.0268 (3) | |
| C5 | 0.39981 (16) | 0.40771 (17) | 0.41036 (8) | 0.0328 (3) | |
| H5 | 0.3630 | 0.4830 | 0.3820 | 0.039* | |
| C6 | 0.33869 (17) | 0.39978 (19) | 0.46758 (8) | 0.0377 (4) | |
| C11 | 0.80092 (15) | 0.20966 (14) | 0.22090 (6) | 0.0231 (3) | |
| C12 | 1.01267 (19) | 0.04672 (18) | 0.22452 (8) | 0.0370 (4) | |
| H12A | 0.9660 | 0.0140 | 0.1830 | 0.044* | |
| H12B | 1.0502 | −0.0417 | 0.2484 | 0.044* | |
| C13 | 1.14477 (19) | 0.1469 (2) | 0.21800 (9) | 0.0468 (5) | |
| H13A | 1.1091 | 0.2309 | 0.1918 | 0.070* | |
| H13B | 1.2228 | 0.0928 | 0.1990 | 0.070* | |
| H13C | 1.1887 | 0.1824 | 0.2589 | 0.070* | |
| C14 | 0.76941 (15) | 0.16206 (14) | 0.36484 (6) | 0.0229 (3) | |
| C15 | 1.00158 (18) | 0.15225 (19) | 0.43389 (7) | 0.0357 (4) | |
| H15A | 0.9401 | 0.0962 | 0.4608 | 0.043* | |
| H15B | 1.0642 | 0.2248 | 0.4600 | 0.043* | |
| C16 | 1.1047 (2) | 0.0488 (2) | 0.40530 (9) | 0.0498 (5) | |
| H16A | 1.0429 | −0.0259 | 0.3811 | 0.075* | |
| H16B | 1.1741 | 0.0005 | 0.4380 | 0.075* | |
| H16C | 1.1644 | 0.1040 | 0.3782 | 0.075* | |
| N3 | 0.3667 (7) | 0.3033 (4) | 0.5110 (3) | 0.0300 (10) | 0.729 (5) |
| C7 | 0.2273 (5) | 0.5042 (6) | 0.4841 (2) | 0.0307 (9) | 0.729 (5) |
| H7 | 0.1852 | 0.5864 | 0.4609 | 0.037* | 0.729 (5) |
| C8 | 0.1955 (9) | 0.4567 (9) | 0.5423 (3) | 0.0351 (13) | 0.729 (5) |
| H8 | 0.1274 | 0.5019 | 0.5670 | 0.042* | 0.729 (5) |
| C9 | 0.2812 (8) | 0.3328 (7) | 0.5569 (3) | 0.0339 (12) | 0.729 (5) |
| H9 | 0.2807 | 0.2761 | 0.5935 | 0.041* | 0.729 (5) |
| C10 | 0.4701 (3) | 0.1768 (3) | 0.51050 (11) | 0.0425 (7) | 0.729 (5) |
| H10A | 0.4547 | 0.1301 | 0.4696 | 0.064* | 0.729 (5) |
| H10B | 0.4484 | 0.1055 | 0.5420 | 0.064* | 0.729 (5) |
| H10C | 0.5763 | 0.2104 | 0.5196 | 0.064* | 0.729 (5) |
| N83 | 0.2589 (11) | 0.4768 (12) | 0.5004 (4) | 0.0224 (17) | 0.271 (5) |
| C87 | 0.369 (2) | 0.2593 (12) | 0.5158 (9) | 0.027 (2) | 0.271 (5) |
| H87 | 0.4264 | 0.1731 | 0.5101 | 0.033* | 0.271 (5) |
| C88 | 0.290 (2) | 0.2937 (17) | 0.5684 (7) | 0.026 (2) | 0.271 (5) |
| H88 | 0.2875 | 0.2365 | 0.6048 | 0.031* | 0.271 (5) |
| C89 | 0.221 (2) | 0.426 (2) | 0.5544 (8) | 0.028 (3) | 0.271 (5) |
| H89 | 0.1559 | 0.4746 | 0.5792 | 0.033* | 0.271 (5) |
| C90 | 0.2156 (6) | 0.6219 (7) | 0.4747 (2) | 0.0302 (15) | 0.271 (5) |
| H90A | 0.3063 | 0.6850 | 0.4773 | 0.045* | 0.271 (5) |
| H90B | 0.1400 | 0.6666 | 0.4982 | 0.045* | 0.271 (5) |
| H90C | 0.1713 | 0.6113 | 0.4312 | 0.045* | 0.271 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0183 (2) | 0.0303 (2) | 0.0411 (2) | 0.00671 (12) | 0.00480 (14) | 0.00178 (13) |
| O1 | 0.0328 (6) | 0.0391 (6) | 0.0299 (5) | 0.0073 (4) | 0.0059 (4) | −0.0022 (4) |
| O2 | 0.0241 (5) | 0.0257 (5) | 0.0357 (5) | 0.0080 (4) | 0.0101 (4) | 0.0037 (4) |
| O3 | 0.0277 (5) | 0.0292 (5) | 0.0458 (6) | −0.0060 (4) | −0.0010 (5) | 0.0100 (5) |
| O4 | 0.0195 (5) | 0.0273 (5) | 0.0379 (6) | −0.0020 (4) | −0.0026 (4) | −0.0003 (4) |
| N1 | 0.0293 (7) | 0.0422 (8) | 0.0333 (7) | 0.0123 (5) | 0.0008 (5) | 0.0051 (5) |
| N2 | 0.0206 (6) | 0.0346 (7) | 0.0376 (7) | −0.0001 (5) | 0.0096 (5) | −0.0028 (5) |
| C1 | 0.0188 (6) | 0.0236 (7) | 0.0334 (7) | −0.0004 (5) | 0.0009 (5) | −0.0015 (5) |
| C2 | 0.0170 (6) | 0.0183 (6) | 0.0296 (7) | −0.0003 (5) | 0.0028 (5) | −0.0015 (5) |
| C3 | 0.0172 (6) | 0.0200 (6) | 0.0310 (7) | −0.0017 (5) | 0.0037 (5) | −0.0005 (5) |
| C4 | 0.0181 (6) | 0.0264 (7) | 0.0363 (7) | 0.0000 (5) | 0.0053 (5) | −0.0001 (5) |
| C5 | 0.0225 (7) | 0.0314 (7) | 0.0461 (9) | −0.0030 (5) | 0.0107 (6) | −0.0042 (6) |
| C6 | 0.0253 (8) | 0.0436 (9) | 0.0470 (10) | −0.0086 (7) | 0.0152 (7) | −0.0144 (8) |
| C11 | 0.0198 (6) | 0.0196 (6) | 0.0298 (7) | −0.0015 (5) | 0.0033 (5) | −0.0019 (5) |
| C12 | 0.0344 (8) | 0.0303 (8) | 0.0499 (9) | 0.0153 (6) | 0.0185 (7) | 0.0049 (6) |
| C13 | 0.0295 (8) | 0.0536 (11) | 0.0611 (11) | 0.0137 (7) | 0.0200 (8) | 0.0188 (9) |
| C14 | 0.0176 (6) | 0.0231 (6) | 0.0283 (6) | −0.0012 (5) | 0.0045 (5) | −0.0016 (5) |
| C15 | 0.0284 (7) | 0.0422 (8) | 0.0336 (8) | 0.0025 (6) | −0.0072 (6) | 0.0008 (6) |
| C16 | 0.0381 (10) | 0.0591 (11) | 0.0499 (10) | 0.0204 (8) | −0.0031 (8) | 0.0037 (8) |
| N3 | 0.0230 (11) | 0.032 (2) | 0.0357 (16) | 0.001 (2) | 0.0083 (9) | −0.005 (2) |
| C7 | 0.0216 (19) | 0.030 (3) | 0.041 (3) | 0.0043 (13) | 0.0066 (15) | −0.0019 (16) |
| C8 | 0.028 (2) | 0.040 (3) | 0.041 (3) | −0.0014 (19) | 0.015 (2) | −0.012 (2) |
| C9 | 0.0328 (18) | 0.044 (4) | 0.026 (2) | 0.000 (3) | 0.0054 (17) | 0.003 (2) |
| C10 | 0.0417 (14) | 0.0451 (14) | 0.0425 (13) | 0.0156 (11) | 0.0120 (10) | 0.0147 (10) |
| N83 | 0.018 (4) | 0.019 (4) | 0.029 (4) | 0.007 (3) | −0.003 (3) | 0.007 (3) |
| C87 | 0.035 (4) | 0.012 (5) | 0.036 (4) | −0.003 (4) | 0.007 (3) | 0.000 (4) |
| C88 | 0.032 (4) | 0.026 (6) | 0.021 (5) | 0.008 (4) | 0.010 (4) | 0.010 (3) |
| C89 | 0.031 (7) | 0.031 (8) | 0.023 (4) | 0.000 (5) | 0.009 (4) | 0.008 (4) |
| C90 | 0.030 (3) | 0.033 (4) | 0.029 (3) | 0.008 (2) | 0.006 (2) | 0.008 (2) |
Geometric parameters (Å, °)
| S1—C1 | 1.7301 (15) | C13—H13b | 0.98 |
| S1—C4 | 1.7703 (15) | C13—H13c | 0.98 |
| O1—C11 | 1.2232 (17) | C15—C16 | 1.499 (2) |
| O2—C11 | 1.3407 (16) | C15—H15a | 0.99 |
| O2—C12 | 1.4537 (17) | C15—H15b | 0.99 |
| O3—C14 | 1.2080 (17) | C16—H16a | 0.98 |
| O4—C14 | 1.3313 (16) | C16—H16b | 0.98 |
| O4—C15 | 1.4622 (17) | C16—H16c | 0.98 |
| N1—C1 | 1.3428 (19) | N3—C9 | 1.354 (7) |
| N1—H1a | 0.88 | N3—C10 | 1.468 (4) |
| N1—H1b | 0.88 | C7—C8 | 1.402 (7) |
| N2—C5 | 1.2918 (19) | C7—H7 | 0.95 |
| N2—C4 | 1.3715 (19) | C8—C9 | 1.369 (5) |
| C1—C2 | 1.3933 (18) | C8—H8 | 0.95 |
| C2—C3 | 1.4368 (18) | C9—H9 | 0.95 |
| C2—C11 | 1.4503 (18) | C10—H10a | 0.98 |
| C3—C4 | 1.3643 (19) | C10—H10b | 0.98 |
| C3—C14 | 1.4923 (18) | C10—H10c | 0.98 |
| C5—C6 | 1.424 (2) | N83—C89 | 1.346 (15) |
| C5—H5 | 0.95 | N83—C90 | 1.463 (8) |
| C6—N83 | 1.276 (12) | C87—C88 | 1.450 (18) |
| C6—N3 | 1.290 (5) | C87—H87 | 0.95 |
| C6—C7 | 1.445 (5) | C88—C89 | 1.361 (11) |
| C6—C87 | 1.652 (14) | C88—H88 | 0.95 |
| C12—C13 | 1.499 (2) | C89—H89 | 0.95 |
| C12—H12a | 0.99 | C90—H90a | 0.98 |
| C12—H12b | 0.99 | C90—H90b | 0.98 |
| C13—H13a | 0.98 | C90—H90c | 0.98 |
| C1—S1—C4 | 91.41 (7) | O3—C14—O4 | 124.09 (13) |
| C11—O2—C12 | 116.43 (11) | O3—C14—C3 | 124.09 (12) |
| C14—O4—C15 | 116.74 (11) | O4—C14—C3 | 111.73 (11) |
| C1—N1—H1A | 120 | O4—C15—C16 | 111.07 (13) |
| C1—N1—H1B | 120 | O4—C15—H15A | 109.4 |
| H1A—N1—H1B | 120 | C16—C15—H15A | 109.4 |
| C5—N2—C4 | 120.54 (14) | O4—C15—H15B | 109.4 |
| N1—C1—C2 | 127.53 (13) | C16—C15—H15B | 109.4 |
| N1—C1—S1 | 120.36 (11) | H15A—C15—H15B | 108 |
| C2—C1—S1 | 112.11 (11) | C15—C16—H16A | 109.5 |
| C1—C2—C3 | 111.76 (12) | C15—C16—H16B | 109.5 |
| C1—C2—C11 | 120.36 (12) | H16A—C16—H16B | 109.5 |
| C3—C2—C11 | 127.55 (12) | C15—C16—H16C | 109.5 |
| C4—C3—C2 | 113.85 (12) | H16A—C16—H16C | 109.5 |
| C4—C3—C14 | 119.55 (13) | H16B—C16—H16C | 109.5 |
| C2—C3—C14 | 126.60 (12) | C6—N3—C9 | 109.6 (4) |
| C3—C4—N2 | 125.24 (13) | C6—N3—C10 | 125.8 (4) |
| C3—C4—S1 | 110.85 (11) | C9—N3—C10 | 124.5 (4) |
| N2—C4—S1 | 123.90 (11) | C8—C7—C6 | 104.3 (4) |
| N2—C5—C6 | 123.90 (16) | C8—C7—H7 | 127.9 |
| N2—C5—H5 | 118.1 | C6—C7—H7 | 127.9 |
| C6—C5—H5 | 118.1 | C9—C8—C7 | 107.0 (5) |
| N83—C6—N3 | 91.6 (4) | C9—C8—H8 | 126.5 |
| N83—C6—C5 | 139.8 (4) | C7—C8—H8 | 126.5 |
| N3—C6—C5 | 128.3 (2) | N3—C9—C8 | 109.6 (5) |
| N3—C6—C7 | 109.5 (3) | N3—C9—H9 | 125.2 |
| C5—C6—C7 | 122.2 (2) | C8—C9—H9 | 125.2 |
| N83—C6—C87 | 97.0 (7) | C6—N83—C89 | 121.2 (1) |
| C5—C6—C87 | 123.2 (6) | C6—N83—C90 | 114.4 (7) |
| C7—C6—C87 | 114.0 (6) | C89—N83—C90 | 124.4 (1) |
| O1—C11—O2 | 122.99 (12) | C88—C87—C6 | 106.4 (9) |
| O1—C11—C2 | 124.12 (13) | C88—C87—H87 | 126.8 |
| O2—C11—C2 | 112.89 (11) | C6—C87—H87 | 126.8 |
| O2—C12—C13 | 111.53 (14) | C89—C88—C87 | 104.90 (11) |
| O2—C12—H12A | 109.3 | C89—C88—H88 | 127.5 |
| C13—C12—H12A | 109.3 | C87—C88—H88 | 127.5 |
| O2—C12—H12B | 109.3 | N83—C89—C88 | 110.30 (13) |
| C13—C12—H12B | 109.3 | N83—C89—H89 | 124.9 |
| H12A—C12—H12B | 108 | C88—C89—H89 | 124.9 |
| C12—C13—H13A | 109.5 | N83—C90—H90A | 109.5 |
| C12—C13—H13B | 109.5 | N83—C90—H90B | 109.5 |
| H13A—C13—H13B | 109.5 | H90A—C90—H90B | 109.5 |
| C12—C13—H13C | 109.5 | N83—C90—H90C | 109.5 |
| H13A—C13—H13C | 109.5 | H90A—C90—H90C | 109.5 |
| H13B—C13—H13C | 109.5 | H90B—C90—H90C | 109.5 |
| C4—S1—C1—N1 | 179.29 (13) | C2—C3—C14—O4 | 77.00 (16) |
| C4—S1—C1—C2 | −1.27 (11) | C14—O4—C15—C16 | 86.08 (17) |
| N1—C1—C2—C3 | −179.63 (13) | N83—C6—N3—C9 | 8.3 (7) |
| S1—C1—C2—C3 | 0.98 (14) | C5—C6—N3—C9 | −178.0 (4) |
| N1—C1—C2—C11 | −5.7 (2) | C7—C6—N3—C9 | 0.4 (6) |
| S1—C1—C2—C11 | 174.91 (9) | C87—C6—N3—C9 | −126 (7) |
| C1—C2—C3—C4 | −0.01 (16) | N83—C6—N3—C10 | −174.1 (7) |
| C11—C2—C3—C4 | −173.40 (12) | C5—C6—N3—C10 | −0.4 (7) |
| C1—C2—C3—C14 | −179.28 (12) | C7—C6—N3—C10 | 178.1 (5) |
| C11—C2—C3—C14 | 7.3 (2) | C87—C6—N3—C10 | 52 (6) |
| C2—C3—C4—N2 | 178.30 (12) | N83—C6—C7—C8 | −23.70 (19) |
| C14—C3—C4—N2 | −2.4 (2) | N3—C6—C7—C8 | 0.3 (5) |
| C2—C3—C4—S1 | −0.93 (15) | C5—C6—C7—C8 | 178.9 (4) |
| C14—C3—C4—S1 | 178.40 (9) | C87—C6—C7—C8 | 7.2 (1) |
| C5—N2—C4—C3 | 169.49 (14) | C6—C7—C8—C9 | −1.0 (7) |
| C5—N2—C4—S1 | −11.4 (2) | C6—N3—C9—C8 | −1.1 (8) |
| C1—S1—C4—C3 | 1.25 (11) | C10—N3—C9—C8 | −178.7 (6) |
| C1—S1—C4—N2 | −177.99 (13) | C7—C8—C9—N3 | 1.3 (9) |
| C4—N2—C5—C6 | 175.94 (14) | N3—C6—N83—C89 | −9.00 (14) |
| N2—C5—C6—N83 | 166.7 (8) | C5—C6—N83—C89 | 178.70 (11) |
| N2—C5—C6—N3 | −3.6 (4) | C7—C6—N83—C89 | 148 (3) |
| N2—C5—C6—C7 | 178.1 (3) | C87—C6—N83—C89 | −3.40 (15) |
| N2—C5—C6—C87 | −10.9 (9) | N3—C6—N83—C90 | 170.4 (7) |
| C12—O2—C11—O1 | 2.11 (19) | C5—C6—N83—C90 | −2.00 (13) |
| C12—O2—C11—C2 | −178.63 (12) | C7—C6—N83—C90 | −32.20 (15) |
| C1—C2—C11—O1 | 0.9 (2) | C87—C6—N83—C90 | 176.0 (1) |
| C3—C2—C11—O1 | 173.81 (13) | N83—C6—C87—C88 | 0.70 (15) |
| C1—C2—C11—O2 | −178.33 (11) | N3—C6—C87—C88 | 47 (6) |
| C3—C2—C11—O2 | −5.44 (19) | C5—C6—C87—C88 | 179.20 (11) |
| C11—O2—C12—C13 | 80.18 (17) | C7—C6—C87—C88 | −9.20 (17) |
| C15—O4—C14—O3 | 0.2 (2) | C6—C87—C88—C89 | 2(2) |
| C15—O4—C14—C3 | 176.97 (11) | C6—N83—C89—C88 | 5(2) |
| C4—C3—C14—O3 | 74.56 (18) | C90—N83—C89—C88 | −174.20 (14) |
| C2—C3—C14—O3 | −106.21 (17) | C87—C88—C89—N83 | −4(2) |
| C4—C3—C14—O4 | −102.23 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.88 | 2.09 | 2.925 (3) | 157 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BH2321).
References
- Bruker (2003). SMART. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2004). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Dufresne, S. & Skene, W. G. (2008). J. Org. Chem.73, 3859–3866. [DOI] [PubMed]
- Dufresne, S. & Skene, W. G. (2010). Acta Cryst. E66, o3027. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Marris, T. (2004). UdMX Université de Montréal, Montréal, Québec, Canada.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Skene, W. G., Dufresne, S., Trefz, T. & Simard, M. (2006). Acta Cryst. E62, o2382–o2384.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810046775/bh2321sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810046775/bh2321Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report