Abstract
pRB activates transcription by a poorly understood mechanism that involves relieving negative regulation of the promoter specificity factor Sp1. We show here that MDM2 inhibits Sp1-mediated transcription, that MDM2 binds directly to Sp1 in vitro as well as in vivo, and that MDM2 inhibits the DNA-binding activity of Sp1. Forced expression of pRB relieves MDM2-mediated repression, and interaction of pRB with the MDM2-Sp1 complex releases Sp1 and restores DNA binding. These results suggest a model in which the opposing activities of MDM2 and pRB regulate Sp1 DNA-binding and transcriptional activity.
The oncogenic properties of MDM2 have been postulated to result from direct interaction with several cell cycle regulatory proteins. MDM2 interacts directly with the tumor-suppressor protein p53 (1) and blocks p53-mediated transactivation (2–7). In addition, MDM2 has been shown to target p53 for rapid degradation (8, 9). These observations have suggested a model in which MDM2 plays a critical role in controlling the extent and duration of the p53 response (10, 11). MDM2 also interacts with a second tumor suppressor protein, the retinoblastoma-associated protein pRB. This MDM2–pRB interaction results in inhibition of pRB growth-regulatory function (12, 13). Furthermore, MDM2 interacts with the activation domains of the S-phase-promoting transcription factors E2F1 and DP1, resulting in stimulation of E2F1/DP1 transcriptional activity (14). Taken together, these observations suggest that MDM2 not only relieves the proliferative block mediated by either p53 or pRB, but also promotes proliferation by stimulating the S phase inducing transcriptional activity of E2F1/DP1.
pRB can modulate transcriptional activation as well as transcriptional repression. One example of transcriptional activation mediated by pRB occurs through a poorly understood mechanism that involves promoter elements called retinoblastoma control elements. Retinoblastoma control elements are bound by Sp1 in vitro and are stimulated by Sp1 in vivo (15–17). Furthermore, coexpression of Sp1 and pRB results in “superactivation” of Sp1-mediated transcription (17). The mechanism by which pRB increases Sp1 activity is not yet clear. However, pRB has been shown to stimulate Sp1 activity by liberating Sp1 from an uncloned negative regulator called Sp1-I (18). In this report, we show that MDM2 inhibits transcriptional activation of Sp1 by binding to its C-terminal domain. Furthermore, pRB can counteract this inhibition by displacing Sp1 from MDM2, resulting in free Sp1, thus restoring Sp1 transcriptional activity.
Materials and Methods
Transfections and Chloramphenicol Acetyltransferase (CAT) Assay.
NIH 3T3 and the microcell hybrid 3T3(R1811)-7 (MDM2+) were transfected with Lipofectamine (GIBCO/BRL) according to the manufacture's protocol. CAT activity was measured by using a phase extraction procedure, as described (19). Values represent the average of duplicate dishes, and duplicate dishes showed <20% difference.
Gel Mobility-Shift Assay.
A probe containing three binding sites for Sp1 was labeled with 32P. Sp1 antibody (PEP2) was obtained from Santa Cruz Biotechnology as was the mutant oligonucleotide (AAT CGA TCG GTT CGC GGC GAG). Binding assays were performed with 10,000-cpm-labeled DNA fragment, 1× Shift buffer (20 mM Hepes, pH 7.6/50 mM KCl/1 mM EDTA/3 mM MgCl/1 mM DTT/10% glycerol), 0.1 μg of poly[d(IC)], 0.5% Nonidet P-40, plus designated amounts of in vitro-translated proteins. Components were allowed to bind for 20 min at room temperature before loading onto a 4% nondenaturing [1× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3)] acrylamide gel. Five microliters of in vitro-translated Sp1 was used in all lanes. Increasing amounts (3, 6, and 18 μl) of in vitro-translated MDM2 were added to the binding assay.
Pulldown Assays.
Glutathione S-transferase (GST) fusion proteins were prepared from bacteria by sonication in PBS containing 0.1% Triton X-100, 1 mM EDTA, 14 mM 2-mercaptoethanol, and protease inhibitors. Extracts were affinity purified by using glutathione-agarose beads (Sigma) and incubated with in vitro-translated and radiolabeled proteins in NETT buffer (20 mM Tris, pH 8.0/100 mM NaCl/1 mM EDTA/0.2% Triton-X-100). MDM2-maltose-binding protein (MBP) fusion proteins were prepared from bacteria by sonication in PBS containing 0.1% Triton X-100, 1 mM EDTA, 1 mM PMSF, 14 mM 2-mercaptoethanol, and protease inhibitors. Extracts were affinity purified by using amylose resin (New England Biolabs) according to the manufacturer's directions. 35S-labeled in vitro-translated proteins (TNT, Promega) were incubated with the fusion proteins in NETT at 4°C for 2 h with constant agitation. After extensive washes with NETT buffer, bound proteins were eluted by boiling and resolved by SDS/PAGE.
In vitro-translated MDM2 and in vitro-translated and 35S-labeled Sp1 were mixed and preincubated with an antibody against the N-terminal of MDM2 (Santa Cruz Biotechnology, N-20), and complexes were isolated by using protein A/G Sepharose (Santa Cruz Biotechnology). Binding and washing were performed using NETT. Purified complexes were incubated with GST coupled to pRB (GST-RB) or GST at 37°C for 20 min. Proteins bound to the beads were removed by centrifugation, and free proteins released into the supernatant were analyzed by PAGE. GST-RB, GST-RBΔC, and GST were generated by elution of glutathione beads with 5 mM reduced glutathione in 50 mM Tris, pH 8.0. Gels were enhanced by using NEN Entensify solutions and exposed to Kodak XAR-5 film or quantitated by using a PhosphorImager (Molecular Dynamics).
Immunoprecipitation and Western Analysis.
Mouse embryo fibroblasts were prepared from MDM2−/−; p53−/− (MDM2−/−) or MDM2+/+; p53−/− (MDM2+/+) embryos, and whole cell lysates were prepared by lysis in nuclear extraction buffer (50 mM Tris, pH 8.0/150 mM NaCl/1.0% Nonidet P-40 plus protease inhibitors). Cell extracts were incubated with the indicated MDM2 mAbs for 4 h at 4°C. Immune complexes were isolated with protein A/G agarose (Santa Cruz Biotechnology) and washed four times with lysis buffer. Bound proteins were analyzed by PAGE followed by Western blotting. For Western blot analysis, total cell lysate was prepared by detergent lysis of cells in TENN buffer (50 mM Tris, pH 8.0/5 mM EDTA/150 mM NaCl/0.5% Nonidet P-40) with protease inhibitors. Proteins were quantitated with the protein assay kit (Bio-Rad). Thirty micrograms of protein was resolved on an SDS/8.5% polyacrylamide gel, transferred to Immobilon membrane (Millipore), and detected with a polyclonal Sp1 antibody (Santa Cruz Biotechnology). The primary antibody was detected by using a horseradish peroxidase-conjugated secondary antibody and an enhanced chemiluminescent system (ECL-Renaissance, NEN).
Results
Overexpression of MDM2 Interferes with Sp1-Mediated Transactivation.
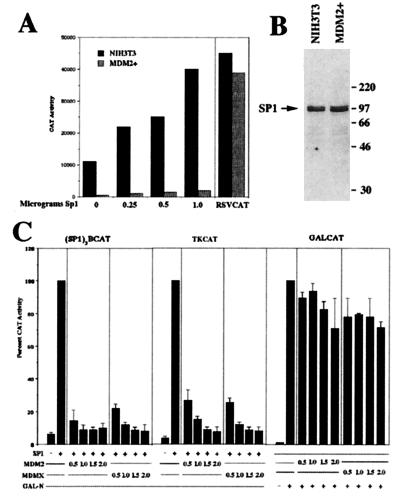
We previously identified a derivative rhabdomyosarcoma chromosome capable of inhibiting overt muscle differentiation when introduced, via microcell-mediated chromosome transfer, into the mouse myoblast cell line C2C12 (19). This derivative chromosome contains a region of amplified DNA originating from chromosome 12q13–14. Testing the amplified genes for the ability to inhibit muscle-specific gene expression indicated that over-expression of MDM2 interferes with MyoD function and consequently inhibits muscle differentiation. During characterization of microcell hybrids containing this derivative rhabdomyosarcoma chromosome (19), we observed a significantly lower level of transcriptional activity from Sp1-dependent promoter constructs when compared to parental cells. Fig. 1A shows a representative experiment with control NIH 3T3 cells and a hybrid with amplified MDM2 (MDM2+) transfected with the synthetic Sp1-dependent reporter construct (Sp1)3BCAT. The (Sp1)3BCAT reporter construct contains three copies of an Sp1 consensus site (GC box) and a TATA element (20). This lower level of basal activity from this reporter construct was not due to transfection differences between the cells because transfection of RSVCAT resulted in similar levels of CAT activity in both cell lines. Furthermore, forced expression of Sp1 resulted in a dose-dependent increase in activity from the Sp1-dependent promoter in control NIH 3T3 cells but only minimal induction in cells with amplified MDM2. However, at higher levels of transfected Sp1 we do observe a significant increase in (Sp1)3BCAT activity in cells with amplified MDM2, suggesting that the inhibitory activity in these cells can be overcome with higher levels of Sp1 (unpublished observations). Western blot analysis indicates that NIH 3T3 cells and the hybrid with amplified MDM2 express similar levels of Sp1 protein (Fig. 1B), indicating that amplification of MDM2 does not alter the expression of Sp1. These results suggested that MDM2 may be negatively regulating Sp1 activity. However, because most cells contain many different GC box-binding transcription factors (21) and the derivative chromosome contains several amplified genes in addition to MDM2 (19) that could potentially interfere with the activity of this promoter, we next tested directly whether MDM2 could interfere with transcriptional activation mediated by Sp1. Cotransfection of NIH 3T3 cells with an Sp1 expression vector, the synthetic Sp1-dependent reporter, and increasing amounts of an MDM2 expression vector indicates that forced expression of MDM2 interferes with Sp1-activated transcription (Fig. 1C). Furthermore, forced expression of MDM2 also interferes with Sp1-mediated activation of the minimal HSVTK promoter, indicating that MDM2 inhibits Sp1-mediated transactivation of naturally occurring Sp1-activated promoters (22). In addition, forced expression of the MDM2 related protein MDMX also inhibits Sp1-activated transcription on both the synthetic Sp1 reporter and on the HSVTK reporter, indicating that this activity is conserved between MDM2 and MDMX. To control for nonspecific inhibition, we assayed a fusion protein (GAL-N), generated by fusing the activation domain of MyoD to the DNA-binding domain of GAL4, for activity on the GAL4-dependent reporter GALCAT. We chose the activation domain of MyoD because it does not interact with MDM2 (T.J.P. and M.J.T., unpublished observations), pRB (23), nor Sp1 (24). Forced expression of either MDM2 or MDMX does not result in a significant reduction in CAT activity from the GALCAT reporter activated by GAL-N (Fig. 1C), indicating that the inhibition of Sp1 activity by either MDM2 or MDMX is specific to promoters activated by Sp1.
Figure 1.
Inhibition of Sp1 activity by MDM2. (A) Transcriptional activity of endogenous Sp1 in cells with amplified MDM2. NIH 3T3 cells and a hybrid with amplified MDM2 (MDM2+) were transfected with the synthetic Sp1-dependent promoter construct (Sp1)3BCAT (20) and increasing amounts of an Sp1-dependent expression vector cytomegalovirus (CMV)-Sp1. RSVCAT was used to control for transfection efficiency between cell lines. CAT activity is expressed as cpm above background. Values represent the average of duplicate dishes, with <20% difference between duplicate dishes. (B) Expression of endogenous Sp1 is not affected by MDM2 amplification. Western blot analysis, using an antibody against Sp1, on cell extracts from NIH 3T3 cells and a hybrid with amplified MDM2 (MDM2+). (C) Inhibition of Sp1 activity by MDM2 and MDMX. NIH 3T3 cells were transfected with the Sp1-dependent reporter (Sp1)3BCAT, HSVTKCAT (TKCAT), or GALCAT. Cells were cotransfected with either an Sp1 expression vector or an GAL-N expression vector in the presence of increasing amounts of an MDM2 or MDMX expression vector. CAT activity was measured as in A above and is expressed as percentage of activated levels.
MDM2 Binds to the Zinc Plus D Domain of Sp1.
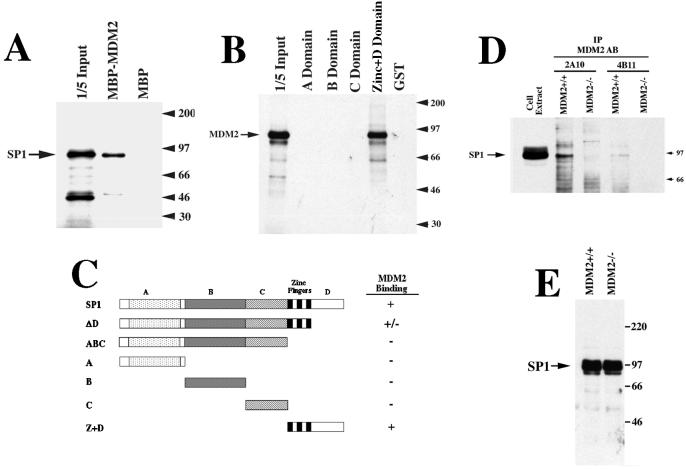
MDM2 has been shown to interfere with transcriptional activation through a direct interaction with the p53 activation domain (1–7). Therefore, we determined whether MDM2 could interact directly with Sp1. Fig. 2A shows that in vitro synthesized Sp1 binds to a MBP-MDM2 fusion protein but not to MBP alone. In addition, using glutathione-S-transferase-Sp1 (GST-Sp1) fusion proteins, we show that in vitro synthesized MDM2 binds to the zinc finger plus D domain of Sp1 (Fig. 2B). This is a specific interaction because MDM2 does not bind to the A, B, or C domains of Sp1 or to GST alone. A schematic representation of these and additional Sp1 deletion mutants is shown in Fig. 2C. To determine whether MDM2-Sp1 complexes could be detected in vivo, we used coimmunoprecipitation from MDM2+/+ or MDM2−/− mouse embryo fibroblasts. Fig. 2D shows that Sp1 protein can be immunoprecipitated with two different MDM2 antibodies from the MDM2+/+ cells and not from the MDM2−/− cells, indicating that the presence of wild-type MDM2 is necessary for coimmunoprecipitation of Sp1 with the MDM2 antibodies. Furthermore, the MDM2+/+ and MDM2−/− cells express similar levels of Sp1 protein (Fig. 2E), indicating that mutation of MDM2 does not alter expression of Sp1. We conclude that MDM2-Sp1 complexes can be detected in vivo.
Figure 2.
Physical association between MDM2 and Sp1 in vitro and in vivo. (A) In vitro-translated Sp1 binds to MBP-MDM2. Radiolabeled Sp1 was incubated with MBP-MDM2 alone, and visualized after PAGE. (B) In vitro-translated MDM2 binds to GST-Sp1. Radiolabedled MDM2 was incubated with different GST-Sp1 constructs and subjected to a GST-puldown assay. GST-A, -B, and -C domain fusion proteins, as well as the GST-zinc + D domain were described previously (34). (C) Schematic representation of the Sp1 deletion constructs used in the MBP or GST pulldown assays. (D) Coimmunoprecipitation of Sp1 with two different MDM2 antibodies. Cell extracts from MDM2+/+ or MDM−/− cells were subjected to immunoprecipitation with the MDM2 antibodies 2A10 and 4B11 (5); bound proteins were subjected to immunoblot analysis with an antibody against Sp1. Sp1 immunoreactivity is detected only in cells that contain an intact MDM2 gene. (E) Expression of endogenous Sp1 is similar in MDM2+/+ and MDM2−/− cells. Western blot hybridization, using an antibody against Sp1, on the cell extracts used in D above.
pRB Restores Sp1 Activity in Cells with Amplified MDM2.
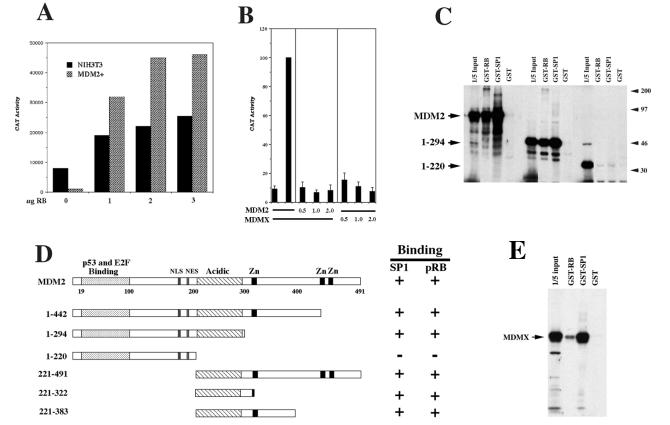
Because pRB has been shown to stimulate Sp1 transcriptional activity in transfected cells and because it interacts with MDM2 in vivo (12, 13), we next tested whether forced expression of pRB could restore Sp1 activity in cells overexpressing MDM2. NIH 3T3 cells and a hybrid with amplified MDM2 were transfected with the Sp1-dependent reporter construct and increasing amounts of an pRB expression vector. Fig. 3A shows that forced expression of pRB induces expression of the Sp1-dependent reporter in the cells with amplified MDM2 to levels comparable to NIH 3T3 cells. This result suggests that pRB can counteract the negative regulation of Sp1 activity mediated by MDM2 resulting in superactivation of the Sp1-dependent promoter. However, because MDM2 has been shown to interact directly with pRB, and pRB has been shown to positively regulate Sp1 activity, we determined whether MDM2 could interfere with Sp1 activity in the absence of pRB. For this analysis we transfected SAOS2 human osteosarcoma (pRB−/−) cells with the Sp1-dependent promoter and increasing amounts of either MDM2 or MDMX expression vectors. Fig. 3B indicates that both MDM2 and MDMX inhibit Sp1-mediated transcription of the Sp1-dependent promoter, indicating that MDM2 and MDMX can inhibit Sp1 activity in the absence of pRB.
Figure 3.
Functional interaction between MDM2 and pRB. (A) pRB restores Sp1 activity in cells with amplified MDM2. NIH 3T3 cells and a hybrid with amplified MDM2 (MDM2+) were transfected with the synthetic Sp1-dependent promoter construct (Sp1)3BCAT and increasing amounts of CMV-RB. CAT activity is expressed as cpm above background. Values represent the average of duplicate dishes, with <20% difference between duplicates. (B) MDM2 inhibits Sp1 activity in the absence of pRB. SAOS2 cells were transfected with the Sp1-dependent promoter construct (Sp1)3BCAT, the Sp1 expression vector CMVSp1, and increasing amounts of either CMV-MDM2 or CMV-MDMX. CAT activity was measured as in Fig. 1 and is expressed as percentage of activated levels. (C) In vitro-translated MDM2 binds to GST-RB and GST-Sp1. Radiolabeled, full-length MDM2 and the two MDM2 deletion mutants 1–294 and 1–220 were incubated with GST-Sp1 (zinc + D domain) or GST-RB (pRB amino acids 379–928) and subjected to a GST-pulldown assay. (D) Schematic representation of additional MDM2 deletion mutants were analyzed using the GST pulldown assay. (E) In vitro-translated MDMX binds to GST-RB and to GST-Sp1. Radiolabeled MDMX was incubated with GST-RB and GST-Sp1 and subjected to a GST-pulldown assay.
Previous studies have demonstrated that MDM2 binds to pRB in vitro as well as in vivo (12, 13). Therefore, to identify the region of MDM2 responsible for interaction with Sp1 and pRB, we generated a series of MDM2 deletion mutants and tested them for binding to GST-Sp1 and GST-RB. Fig. 3C shows that full length MDM2, and the deletion mutant 1–294, binds to both GST-Sp1 and to GST-RB, while the mutant 1–220 does not bind to either. Analysis of additional deletion mutants indicates that the central region of MDM2, containing amino acids 221 to 294, is both necessary and sufficient for binding to both Sp1 and pRB (Fig. 3D). This observation is consistent with a previous report that showed that pRB binds to MDM2 in a region between amino acids 273 and 321 (13). Furthermore, MDMX binds to both Sp1 and pRB by using the GST pulldown assay (Fig. 3E), indicating that this activity is conserved between MDM2 and MDMX.
MDM2 and pRB Regulate the Sequence-Specific DNA-Binding Activity of Sp1.
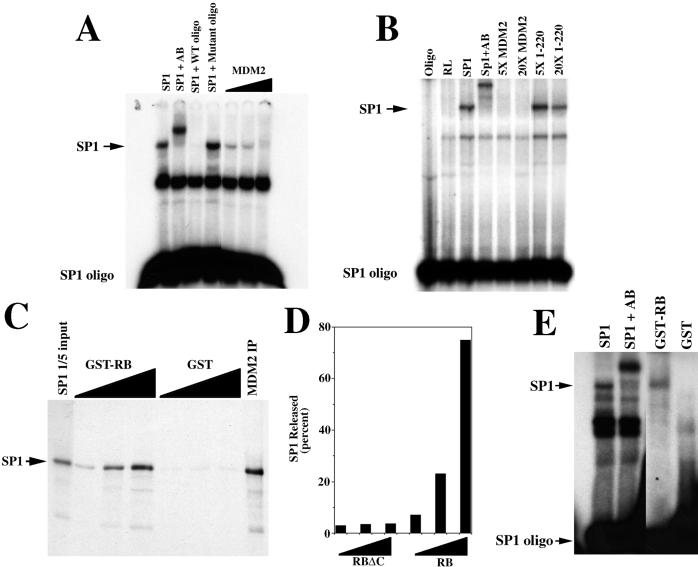
Because MDM2 binds to Sp1 in the region that contains the zinc finger domain, one potential mechanism by which MDM2 could inhibit Sp1 activity is through inhibition of DNA binding. To test this possibility, we measured Sp1 DNA-binding activity in the presence of MDM2 using a gel mobility shift assay. Fig. 4A shows that adding increasing amounts of in vitro-translated MDM2 inhibits DNA binding of in vitro-translated Sp1. In control lanes, we show that the Sp1-dependent band can be supershifted with an Sp1 antibody, and that the Sp1-dependent band can be competed away by excess wild-type but not mutant oligonucleotide. In addition, an MDM2 deletion mutant that does not interact with Sp1 does not interfere with the Sp1-DNA complex (Fig. 4B). Furthermore, this is a specific inhibition of Sp1 DNA binding, because in vitro-translated MDM2 does not inhibit the DNA-binding activity of either MyoD or CREB (unpublished observations). These results suggest that MDM2 interferes with Sp1-activated transcription by inhibiting Sp1 DNA binding.
Figure 4.
MDM2 inhibits the DNA-binding activity of Sp1, and pRB releases Sp1 from MDM2. (A) Gel mobility-shift assay showing that in vitro-translated MDM2 inhibits the DNA-binding activity of in vitro-translated Sp1. Control lanes show that adding an Sp1-specific antibody (AB) to the reaction results in a supershift of the Sp1-dependent band, and that wild-type (WT) but not mutant oligonucleotide (oligo) interferes with the same Sp1-dependent band. (B) Gel mobility shift assay showing that 5- (5×) and 20- (20×) fold excess of the N-terminal mutant (1–220) of MDM2 does not interfere with the DNA-binding activity of in vitro-translated Sp1. Control lanes show that adding an Sp1-specific antibody (AB) to the reaction results in a supershift of the Sp1-dependent band. The amount of Sp1, full-length MDM2, and the N-terminal mutant of MDM2 (1–220) was determined by 35S incorporation. (C) pRB releases Sp1 from MDM2. In vitro-translated and radiolabeled Sp1 was immunoprecipitated with in vitro-translated MDM2 (MDM2 IP). The MDM2-Sp1 immunoprecipitate was incubated with increasing amounts of either GST-RB or GST alone and re-immunoprecipitated, and proteins in the supernatant were separated by gel electrophoresis. (D) The C terminus of pRB is required for Sp1 release. In vitro-translated and radiolabeled Sp1 was immunoprecipitated with in vitro-translated MDM2, as in C above. The MDM2-Sp1 immunoprecipitate was incubated with increasing amounts (25, 125, and 500 ng) of either GST-RB (379–928) or GSTRBΔC (379–824) and re-immunoprecipitated. Released Sp1 was quantitated by using a PhosphorImager after SDS/PAGE. Values represent the amount of Sp1 released, expressed as the percentage of Sp1 immunoprecipitated by MDM2. (E) Released Sp1 can bind DNA. In vitro-translated Sp1 was incubated with in vitro-translated MDM2 and immunoprecipitated and released as in C above, except that the released Sp1 was subjected to gel mobility shift analysis. A control lane shows a supershift of the Sp1 band with the Sp1-specific antibody.
Because pRB and Sp1 bind to MDM2 in the same central region (Fig 3D), it is possible that pRB reverses MDM2 inhibition by displacing Sp1 from MDM2. To test this possibility, we coimmunoprecipitated in vitro-translated Sp1 with in vitro-translated MDM2 and tested whether pRB could release Sp1 from the complex. Fig. 4C shows that incubation of the MDM2-Sp1 coimmunoprecipitate with increasing amounts of GST-RB (amino acids 379–928) releases Sp1 into the supernatant. This is a specific effect of the C terminus of pRB, because incubation of the MDM2-Sp1 complex with a C-terminal truncation of pRB that does not interact with MDM2, GSTRBΔC (amino acids 379–864), did not release Sp1 (Fig. 4D). Furthermore, the Sp1 that is released from the MDM2 coimmunoprecipitate is now able to bind DNA (Fig. 4E). These results suggest that Sp1 and pRB compete for binding to MDM2, and that the DNA-binding activity of Sp1 is regulated by the competing activities of MDM2 and pRB.
Discussion
Sp1 was one of the first mammalian transcription factors to be characterized (25). Sp1 is a member of a family of proteins with highly related zinc-finger domains that bind to GC or GT boxes in the regulatory regions of many housekeeping as well as tissue-specific genes (21, 26). Gene knockout of the mouse Sp1 locus indicates that Sp1-null embryos are severely retarded in growth, die early in embryogenesis, and show a broad range of phenotypic abnormalities (27). Surprisingly, the expression of many putative target genes, including cell cycle-regulated genes, is not affected in the Sp1−/− embryos. Because Sp1 is a member of a large family of zinc finger transcription factors that bind to similar DNA sequences (21), the continued expression of these putative Sp1 targets may be due to functional redundancy within the Sp1 family. It will be of interest to determine if other Sp1 family members are regulated by MDM2 and pRB.
It is well established that transcriptional regulation can result from physical interaction between diverse transcription factors. Previous reports have indicated that pRB positively regulates Sp1 activity (15–17). However, the mechanism by which pRB regulates Sp1 activity is poorly understood and may involve an uncharacterized pRB-binding protein called Sp1-I (18). In addition, a cathepsin-like protease has been shown to be involved in the rapid degradation of both Sp1 and pRB (28, 29). Whether MDM2 is involved in regulating this activity is currently not known. However, we have not observed a significant difference in the levels of Sp1 protein either in cells with amplified MDM2 or in cells with deleted MDM2, suggesting that MDM2 does not regulate the levels of Sp1 protein. In addition, pRB has been shown to regulate Sp1 activity indirectly by interacting with TAFII250, which interacts with TAFII110, which in turn interacts with Sp1 to stimulate transcription (30). The interaction between TAFII110 and Sp1 occurs in one of the glutamine rich transactivation domains (domain B) located in the N-terminal half of Sp1. Because MDM2 interacts with the zinc plus D domain of Sp1, these observations suggest that pRB regulates Sp1 activity by at least two distinct mechanisms. The first involves facilitation of the TBP–Sp1 interaction via a glutamine rich activation domain of Sp1 via TAFII250 and TAFII110. The second involves negative regulation of Sp1 transcriptional activity by MDM2 and reversal of this inhibition by pRB. Our results suggest a model in which Sp1 transcriptional activity is regulated by physical interaction with MDM2, and that this interaction inhibits DNA binding (Fig. 5). Furthermore, pRB can reverse the inhibitory effects of MDM2 by physically interacting with the MDM2-Sp1 complex and releasing free Sp1, thus restoring DNA binding and transcriptional activation. Amplification and overexpression of MDM2 occurs in approximately 30% of human sarcomas (1). Our model predicts that in cells with amplified MDM2, pRB becomes limiting and as a consequence, transcription from genes dependent on Sp1 activity is reduced. Because Sp1 has been implicated in the expression of genes involved in both terminal differentiation, e.g., human cardiac actin (31), as well as S-phase, e.g., dihydrofolate reductase (32) and thymidine kinase (33), it will be interesting to determine whether the opposing activities of pRB and MDM2 mediate the switch from active proliferation to cell cycle arrest during terminal differentiation by modulating Sp1 activity.
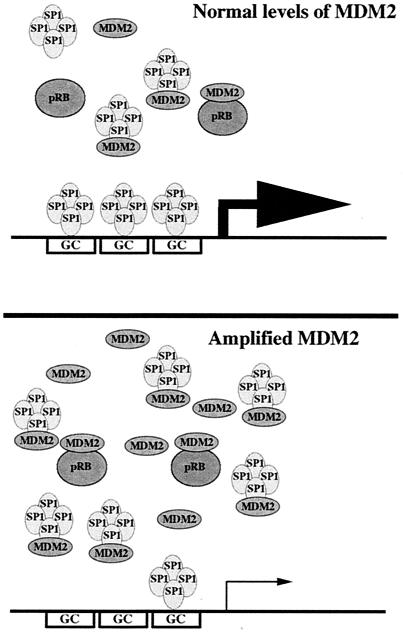
Figure 5.
Model for regulation of Sp1-mediated transcription by the competing activities of MDM2 and pRB. During normal growth of wild-type cells, the level of transcriptionally active Sp1 is established by the levels of MDM2 and pRB. However, in cells where MDM2 has been amplified, pRB becomes limiting and as a consequence more Sp1 is bound to MDM2 and thus lowering gene expression from Sp1-dependent promoters.
Acknowledgments
We thank S. Jones for providing the MDM2−/− mouse embryo fibroblast cultures, B. Vogelstein for the human MDM2 expression vector, R. Tjian for the mouse Sp1 expression vector and the Sp1 reporter construct, J. Horowitz for the GST-Sp1 plasmids, and B. Kaelin for the GST-RB plasmids. This work was supported by a grant from the National Institutes of Health, AR44553.
Abbreviations
- MBP
maltose-binding protein
- GST
glutathione S-transferase
- CMV
cytomegalovirus
- CAT
chloramphenicol acetyltransferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 2.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 3.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 4.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haines D S, Landers J E, Engle L J, George D L. Mol Cell Biol. 1994;14:1171–1178. doi: 10.1128/mcb.14.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 10.Lane D P, Hall P A. Trends Biochem Sci. 1997;22:372–374. doi: 10.1016/s0968-0004(97)01119-5. [DOI] [PubMed] [Google Scholar]
- 11.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Nature (London) 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 14.Martin K, Trouche D, Hagemeier C, Sorensen T S, La-Thangue N B, Kouzarides T. Nature (London) 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim S J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udvadia A J, Rogers K T, Higgins P D, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udvadia A J, Templeton D J, Horowitz J M. Proc Natl Acad Sci USA. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y H, Grunwald S, Chiu R. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiddler T A, Smith L, Tapscott S J, Thayer M J. Mol Cell Biol. 1996;16:5048–5057. doi: 10.1128/mcb.16.9.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pascal E, Tjian R. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 21.Philipsen S, Suske G. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones K A, Yamamoto K R, Tjian R. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 24.Biesiada E, Hamamori Y, Kedes L, Sartorelli V. Mol Cell Biol. 1999;19:2577–2584. doi: 10.1128/mcb.19.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dynan W S, Tjian R. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 26.Turner J, Crossley M. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 27.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 28.Nishinaka T, Fu Y H, Chen L I, Yokoyama K, Chiu R. Biochim Biophys Acta. 1997;1351:274–286. doi: 10.1016/s0167-4781(96)00210-2. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y H, Nishinaka T, Yokoyama K, Chiu R. FEBS Lett. 1998;421:89–93. doi: 10.1016/s0014-5793(97)01541-x. [DOI] [PubMed] [Google Scholar]
- 30.Shao Z, Ruppert S, Robbins P D. Proc Natl Acad Sci USA. 1995;92:3115–3119. doi: 10.1073/pnas.92.8.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartorelli V, Webster K A, Kedes L. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- 32.Lin S Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlseder J, Rotheneder H, Wintersberger E. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata Y, Kim H G, Rogers K T, Udvadia A J, Horowitz J M. J Biol Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]