Abstract
Tissue-engineered in vitro models have the potential to be used for investigating inflammation in the complex microenvironment found in vivo. We have developed an in vitro model of hepatic tissue that facilitates real-time monitoring of endothelium activation in liver tissue. This was achieved by creating a layered coculture model in which hepatocytes were embedded in collagen gel and a reporter clone of endothelial cells, which synthesizes green fluorescent protein in response to nuclear factor-kappa B (NF-κB) activation, was overlaid on top of the gel. The efficacy of our approach was established by monitoring in real time the dynamics of NF-κB-regulated fluorescence in response to tumor necrosis factor α. Our studies revealed that endothelial cells in coculture with hepatocytes exhibited a similar NF-κB-mediated fluorescence to both pulse and step stimulation of lipopolysaccharide. By contrast, endothelial cells in monoculture displayed enhanced NF-κB-regulated fluorescence to step in comparison to pulse lipopolysaccharide stimulation. The NF-κB-mediated fluorescence correlated with endothelial cell expression of NF-κB-regulated genes such as intercellular adhesion molecule 1, vascular cell adhesion molecule 1, and E-Selectin, as well as with leukocyte adhesion. These findings suggest that our model provides a powerful platform for investigating hepatic endothelium activation in real time.
Introduction
Inflammation is the basic response of a tissue for repairing damage caused by injury or infection. However, if inflammation is not resolved it can cause further damage.1 In response to injury, different cell types in a tissue exhibit dynamic transcriptional patterns that ultimately regulate levels of growth factors, cytokines, and chemokines in a spatiotemporal fashion for the activation of an inflammatory response. Identifying mechanisms at cellular and tissue level that regulate inflammation will facilitate development of superior treatment strategies. In the liver, there are a number of diseased states in which inflammation is implicated, including sepsis,2 ischemia/reperfusion injury,3 steatohepatitis,4 fibrosis,5 and certain drug-induced injury.6 Tissue-engineered models of liver that capture the complex signaling microenvironment found in vivo can provide an important platform for investigating hepatic inflammation. These models have the potential of bridging the gap between cell culture and animal experiments, and also provide surrogate models for human studies.
Nuclear factor-kappa B (NF-κB) is the central transcription factor implicated in regulation of several inflammation-associated genes.7 Previous studies with animals have established elevated levels of NF-κB activation in liver in response to various insults, including endotoxin8 and viral infection.9 However, localizing the NF-κB activation patterns in individual cell types was complicated by other confounding factors such as stress associated with isolating cells from the liver, which resulted in an artifactual elevation of NF-κB activation.8 Since NF-κB is involved in expression of several cytokines, chemokines, and adhesion molecules,10 which ultimately regulate the dynamic microenvironment of liver tissue, development of in vitro models that facilitate identifying activation patterns of NF-κB in individual cell types under complex multicellular environments is highly desirable.
Traditional methods for gene expression analysis include reverse transcription polymerase chain reaction (RT-PCR), DNA microarray, and northern blot. These methods are essentially destructive and not well suited for measuring dynamics of gene expression patterns. Further, in tissue models that employ multiple cell types there is an additional complexity of separating individual cell types to localize the cellular source of gene expression. Fluorescent reporters provide a nondestructive method for measuring gene expression in live cells.11–14 These clones are prepared by transfecting cells with plasmid DNA that encodes for easily detectable proteins such as green fluorescent protein (GFP) under the regulation of a specific transcription factor. When the desired transcription factor gets activated, it leads to expression of GFP and the fluorescence level increases, thereby enabling real-time measurement of transcriptional activity in live cells.
In this work, we have combined tissue engineering with a fluorescent reporter clone for creating a dynamic system that enables real-time investigation of endothelium activation in a hepatic tissue model. We have developed and integrated a NF-κB reporter clone of primary rat endothelial cells in our previously described organotypical model of liver tissue.15 The model consists of hepatocytes embedded in collagen gel and a NF-κB reporter clone of endothelial cells overlaid on top of the gel. The model was exposed to tumor necrosis factor α (TNFα) and fluorescence was measured temporally in endothelial cells for evaluating dynamics of NF-κB-regulated gene expression in endothelium of hepatic tissue under inflammatory conditions. Our studies with this model revealed a differential NF-κB response of endothelial cells to pulse and step of lipopolysaccharide (LPS), depending on whether they were in coculture with hepatocytes or in monoculture. Finally, we were able to correlate NF-κB-mediated fluorescence to expression of adhesion molecules on endothelial cells and their adhesion to leukocytes.
Materials and Methods
Materials
Williams E basal medium, collagenase, and LPS were purchased from Sigma-Aldrich (St. Louis, MO). Epidermal growth factor (EGF), penicillin-streptomycin, geneticin, and CM-DiI were obtained from Invitrogen Life Technologies (Carlsbad, CA). Glucagon was acquired from Lilly (Indianapolis, IN), insulin was purchased from Squibb (Princeton, NJ), and hydrocortisone from Upjohn (Kalamazoo, MI). MCDB-131-complete medium was obtained from VEC Technologies (Rensselaer, NY). TNFα was purchased from R&D Systems (Minneapolis, MN).
Construction of NF-κB reporter clone of endothelial cells
NF-κB reporter plasmid consisted of multiple response elements upstream of destabilized GFP gene that encodes for destabilized enhanced green fluorescent protein (d2EGFP) reporter protein. The details of plasmid construction are described elsewhere.11–12 Primary rat heart microvessel endothelial cells (RHMEC) were purchased from VEC Technologies and maintained in MCDB-131 medium supplemented with 10% fetal bovine serum, 10 ng/mL EGF, 1 μg/mL hydrocortisone, 200 μg/mL EndoGro, 90 μg/mL heparin, and 1% antimycotic solution. RHMEC were cultured in a humidified incubator maintained at 37°C and 5% CO2. Early passage endothelial cells (2.5 million) were electroporated with NF-κB reporter plasmid (10 μg) using a BTX Electro Cell Manipulator 600 (Biotechnology and Experimental Research, San Diego, CA) at 280 V and 960 μF. Cells were selected for plasmid integration by addition of geneticin to a final concentration of 700 μg/mL. Clones that grew in the selection media were harvested using cloning rings. The clone that exhibited maximum shift in fluorescence (Supplementary Fig. S1, available online at www.liebertonline.com/ten) upon stimulation with TNFα was used in all the experiments.
Isolation and culture of hepatocytes
Hepatocytes were isolated from female Lewis rats (Charles River Laboratories, Wilmington, MA) weighing 180–200 g. Hepatocytes were isolated by a two-step collagenase perfusion technique originally described by Seglen16 and modified by Dunn et al.17 Isolation yields ranged from 200 to 300 million hepatocytes per rat with viabilities ranging from 85% to 95%, and purity was >95%. All animals were treated in accordance with National Research Council guidelines, and the studies were approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital.
The culture medium for hepatocytes consisted of Williams E supplemented with 20 ng/mL EGF, 14 ng/mL glucagon, 0.5 U/mL insulin, 7.5 μg/mL hydrocortisone, 100 U/mL penicillin, and 100 μg/mL streptomycin. Type I collagen was prepared by extracting acid-soluble collagen from rat tail tendons as previously described.18 Hepatocytes were suspended in the ice-cold culture medium at a concentration of 2 × 106 cells/mL. Collagen solution was prepared on ice by mixing nine parts of type I collagen (1.25 mg/mL) with one part of 10 × Dulbecco's modified Eagle's medium (phenol red free). For embedding hepatocytes in collagen gel, cell suspension was mixed with collagen solution in a 1:1 ratio by volume. Typically, 500 μL of this mixture was introduced in one well of a 12-well plate. The cell suspension was allowed to gel at 37°C for 90 min and then the hepatocyte culture medium was added on top of the gel.
Monoculture and coculture of endothelial cells with hepatocytes
For preparing layered tissue model, endothelial cells were trypsinized and suspended at a concentration of 2 × 106 cells/mL in a medium prepared by mixing hepatocyte and endothelial cell culture media at a ratio of 1:1 by volume. Typically, 500 μL of cell suspension was introduced on top of the gel, 1 day after embedding of hepatocytes in collagen gel. In another set of experiments, collagen gel was prepared by mixing collagen solution and the hepatocyte culture medium at 1:1 ratio by volume as described above (in previous section), except in this case no hepatocytes were present. Next, 500 μL of endothelial cell suspension was introduced on top of the gel. In all the experiments, both cocultures and monocultures of endothelial cells were maintained in the initial suspension medium for 24 h. Next, both cocultures and monocultures were washed with phosphate-buffered saline (PBS) once, and then maintained in the hepatocyte culture medium with medium changes every alternate day. All the experiments were conducted between days 4–6 of the start of culture.
Time lapse imaging using fluorescence microscope
The layered tissue model was exposed to TNFα (10 ng/mL) prepared in the hepatocyte culture medium. At the beginning of time lapse imaging, phase-contrast images of hepatocytes and endothelial cells were acquired by focusing the microscope to different planes in the layered tissue model. During time-lapse imaging, the microscope was focused on the endothelial cell plane. Images were acquired at 1 h interval for the duration of experiment using Zeiss 200 M microscope (Carl Zeiss, Thornwood, NY) fitted with incubation chamber and humidifier. The phase-contrast and fluorescence images were captured using a CCD camera (Carl Zeiss) and Zeiss imaging software (Axiovision LE). Unless otherwise noted, the same CCD camera and Zeiss imaging software were used throughout for capturing images. The autofocus feature was selected during image acquisition, which facilitated maintaining focus on the endothelium plane. Mean fluorescence intensity of the images was quantified using Image J software (National Institute of Health, Bethesda, MD). Mean fluorescence was normalized to values between 0 and 1 using the intensity levels recorded at 1 and 12 h, respectively.
Fluorescence response for pulse and step treatment of LPS
Cocultures and monocultures were exposed to LPS (100 ng/mL) prepared in the hepatocyte culture medium. Cells were exposed to two types of stimulation profiles—2-h pulse and step treatment. In the case of pulse stimulation, cells were treated with the LPS-containing medium for 2 h. At the end of 2 h, the LPS-containing medium was removed and cells were washed three times with PBS (1 × ). Next, the fresh hepatocyte culture medium was added and cells were maintained in that medium for rest of the duration of the experiment. For step treatment, the LPS-containing medium was added on top of the cells. In this case, cells were maintained throughout in the LPS-containing medium for the entire duration of the experiment. In the case of control, cells were exposed to the hepatocyte culture medium for the entire duration of the experiment. For measuring NF-κB-mediated fluorescence response, images were acquired at 9 h for all three conditions—control, pulse, and step. In monocultures and cocultures, fluorescence intensity (quantified using Image J software) for control and pulse treatment was expressed as percentage of step stimulation.
Real-time PCR
At 6 h after introduction of LPS, both monocultures and cocultures were treated with collagenase prepared at a concentration of 1 mg/mL in PBS. Cocultures and monocultures were incubated with collagenase at 37°C for 5 and 30 min, respectively. Shorter incubation time in cocultures resulted in endothelial cells detaching as a floating sheet in the medium, while hepatocytes remained embedded in collagen as shown in the Supplementary Figure S2 (available online at www.liebertonline.com/ten). Purity of endothelial cells recovered from cocultures was >95%. Endothelial cells recovered from cocultures and monocultures were pelleted and then frozen at −80°C before further analysis. RNA was extracted from cells using nucleospin RNA II kit (Macherey-Nagel, Bethlehem, PA) according to the manufacturer's instructions. Quantitative RT-PCR was performed using the Superscript III two-Step qRT-PCR kit purchased from Invitrogen. Five hundred nanograms of cellular RNA was reverse transcribed according to the manufacturer's directions. Real-time quantitative PCR was performed using the Stratagene (La Jolla, CA) MX5000P QPCR system. Each reaction was carried out with 10 ng cDNA and 0.6 μM primers. During amplification, the cycling temperatures were 95°C for 15 s, 57°C for 1 min and 72°C for 30 s. The following primers were used for amplifying DNA—E-Selectin forward primer: CAACACATCCTGCAGTGGTC; E-Selectin reverse primer: AGCTGAAGGAGCAGGATGAA; intercellular adhesion molecule 1 (ICAM-1) forward primer: CCTCTTGCGAAGACGA GAAC; ICAM-1 reverse primer: ACTCGCTCTGGGAACGAATA; vascular cell adhesion molecule 1 (VCAM-1) forward primer: TGAAGGGGCTACATCCACAC; VCAM-1 reverse primer: GACCGTGCAGT TGACAGTGA β-actin forward primer: GTCGTACCACTGGCATTGTG; and β-actin reverse primer: CTCTCAGCTGTGGTGG TGAA. Expression levels of VCAM-1, ICAM-1, and E-Selectin were measured relative to β-actin. In monocultures and cocultures, gene expression levels for pulse stimulation were calculated as percentage of step treatment with LPS. In the case of control (without LPS), gene expression levels for monoculture were calculated relative to the coculture.
Leukocyte adhesion experiments
Peripheral blood leukocytes were purified from heparinized peripheral blood of rat using Histopaque density gradient (Sigma) according to manufacturer's instructions. Leukocytes were labeled with CM-DiI (2.5 μg/mL) at 37°C for 10 min and then suspended in the hepatocyte culture medium at a concentration of 0.8 × 106 cells/mL. At 8 h after introduction of LPS, 500 μL of leukocyte suspension was introduced in 12-well plates containing monocultures and cocultures. Leukocytes were allowed to adhere for 60 min at 37°C and then washed three times with PBS for removing nonadhered cells. Fluorescence images were captured and analyzed using Image J to estimate the degree of leukocyte adhesion to endothelial cells. In monocultures and cocultures, leukocyte adhesion (fluorescence intensity) for control and pulse treatment was expressed as percentage of step stimulation.
Statistical analysis
Results are reported as mean ± standard deviation. Statistical analysis was performed using the Student's t-test, with p < 0.05 considered significant.
Results
Real-time dynamics of NF-κB-regulated gene expression in endothelium of an organotypical model of liver
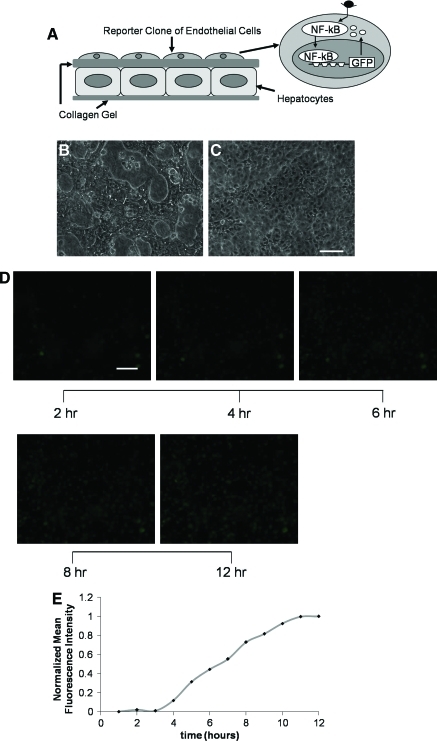
In the liver sinusoid, endothelial cells are closely associated but separated from hepatocytes by the extracellular matrix of the Space of Disse. During inflammation, intercellular communication between hepatocytes and endothelial cells can create dynamic NF-κB activation profiles in the endothelium. For real-time monitoring of NF-κB activation in endothelium, a reporter clone of endothelial cells was integrated in our previously developed organotypical model of liver. The model consists of hepatocytes embedded in collagen gel and the reporter clone of endothelial cells overlaid on top of the gel. Figure 1B and C shows phase-contrast images of hepatocytes and the reporter clone of endothelial cells in different planes of the layered coculture structure. Hepatocytes display characteristic polygonal morphology,19–21 whereas endothelial cells exhibit typical cobblestone morphology.22
FIG. 1.
Hepatic tissue model for real-time measurement of dynamic NF-κB-regulated fluorescence response in endothelium. (A) Schematic illustrating hepatocytes embedded in collagen gel and reporter clone of endothelial cells overlaid on top of the gel. The reporter clone synthesizes GFP in response to activation and binding of NF-κB to its response element in the promoter region of GFP. (B) Phase-contrast image of hepatocytes on the bottom layer of the construct. (C) Phase-contrast image of reporter clone of endothelial cells on the top layer of the construct. (D) Time-lapse fluorescence images of endothelium in layered tissue model acquired at 2, 4, 6, 8, and 12 h after stimulation with tumor necrosis factor α. (E) Normalized mean fluorescence intensity levels for (D) with sampling frequency of 1 h. Scale bar = 50 μm. NF-κB, nuclear factor-kappa B; GFP, green fluorescent protein. Color images available online at www.liebertonline.com/ten.
For measuring the NF-κB response under inflammatory conditions, the layered model was exposed to TNFα, a classical inducer of NF-κB.23 Figure 1D shows fluorescence images acquired at different time intervals by focusing the microscope on the endothelial cell plane in the layered system. To make sure that the images were obtained from the same population of cells, the field of view in the plate was fixed during image acquisition. Figure 1E illustrates normalized mean fluorescence intensity of images acquired at 1 h frequency. The fluorescence response starts increasing at 4 h after stimulation and reaches maxima around 11 h. These results signify the utility of this approach for monitoring the dynamics of NF-κB-mediated expression in real time in a tissue-like multicellular microenvironment.
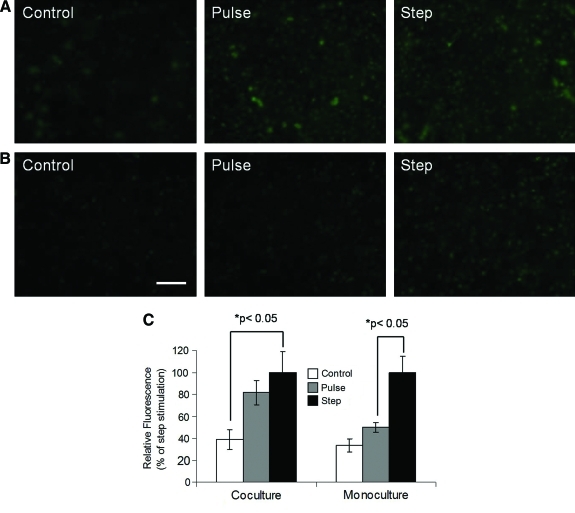
Differential GFP response of endothelial cells in coculture as compared to monoculture to bacterial endotoxin (LPS)
Even though LPS by itself is known to activate NF-κB,24 hepatocytes can influence NF-κB activation profiles in endothelial cells by secreting various soluble factors. For investigating this hepatocyte-mediated effect, endothelial cells in coculture and monoculture were exposed to LPS. Since LPS activity in vivo gets neutralized by various serum components, the cells were subjected to two types of stimulation profiles-2-h pulse and continuous (step) treatment with LPS. Pulse treatment was designed to approximate conditions where LPS remains functionally active for short period. Fluorescence images of endothelial cells were acquired at 9 h for both types of stimulation (Fig. 2A, B). Images were quantified and fluorescence was expressed as percentage of step stimulation (Fig. 2C). Figure 2A indicates that endothelial cells in coculture with hepatocytes exhibit enhanced GFP levels for both pulse and continuous treatment with LPS in comparison to control. Further, the GFP level for pulse exposure was 82% of step stimulation (difference was not statistically significant). However, as shown in the Figure 2B, for endothelial cells cultured alone enhancement in GFP levels was much more dramatic for continuous treatment than pulse exposure of LPS, as compared to the control. In this case, GFP level for pulse treatment was 50% (p < 0.05) of step stimulation. These results suggest that hepatocytes enhance NF-κB response of endothelial cells to a pulse of LPS.
FIG. 2.
NF-κB-regulated fluorescence response of endothelial cells stimulated with pulse or step of LPS under different culture conditions. (A) Fluorescence images of endothelial cells on top of collagen-embedded hepatocytes (coculture). (B) Fluorescence images of endothelial cells on top of the collagen gel (monoculture). (C) Relative fluorescence intensity levels for (A) and (B) with control and pulse expressed as percentage of step stimulation, respectively. Scale bar = 50 μm. LPS, lipopolysaccharide. Color images available online at www.liebertonline.com/ten.
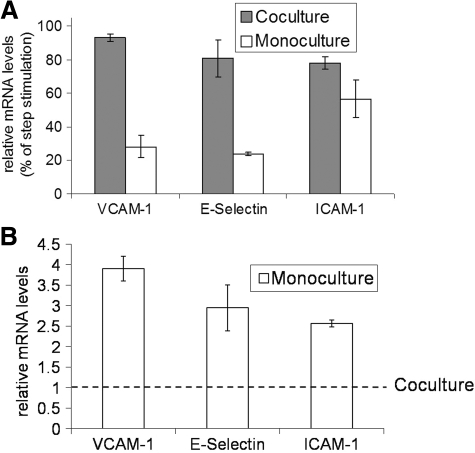
Relating differential GFP profiles in coculture and monoculture to expression of adhesion molecules in endothelial cells
Past studies have identified involvement of NF-κB in regulating expression of adhesion molecules ICAM-1, VCAM-1, and E-selectin in endothelial cells.25 We measured expression of these adhesion molecules for probing the efficacy of GFP profiles as an indicator of NF-κB-regulated gene expression in endothelial cells. Figure 3A shows gene expression levels (6 h) of VCAM-1, E-selectin, and ICAM-1 in endothelial cells exposed to pulse of LPS relative to step stimulation. It indicates that VCAM-1 gene expression level for pulse exposure was 28% of step treatment in the case of endothelial cells cultured alone. By contrast, in response to pulse treatment, endothelial cells in co-culture with hepatocytes exhibited VCAM-1 gene expression levels (93%) that approached those for step stimulation. Similarly, gene expression levels of E-selectin and ICAM-1 for pulse exposure relative to step stimulation was lower in monocultures (24% and 57%) of endothelial cells in comparison to those in coculture (80% and 78%) with hepatocytes. This is in agreement with GFP profiles observed in Figure 2 where endothelial cells in coculture showed similar level of activation for pulse and step exposure of LPS, while endothelial cells in monoculture demonstrated higher level of activation for step in comparison to pulse treatment of LPS. For estimating if coculture with hepatocytes devoid of any LPS stimulation has any effect on the activation state of endothelial cells, we compared gene expression levels of adhesion molecules in the control condition of cocultures and monocultures. Figure 3B indicates that expression of all three adhesion molecules in endothelial cells is elevated for monocultures relative to cocultures. These results suggest that hepatocytes alter the activation state of endothelial cells both in the presence and absence of any LPS stimulation.
FIG. 3.
Relative changes in gene expression of VCAM-1, ICAM-1, and E-Selectin in endothelial cells cultured under different conditions. (A) Gene expression levels were measured for pulse and step treatment of LPS in both monocultures and cocultures. Relative mRNA levels for pulse are expressed as percentage of step treatment. (B) Gene expression levels were measured in the control (no LPS stimulation) cocultures and monocultures. mRNA levels for monocultures expressed relative to cocultures. VCAM-1, vascular cell adhesion molecule 1; ICAM-1, intercellular adhesion molecule 1.
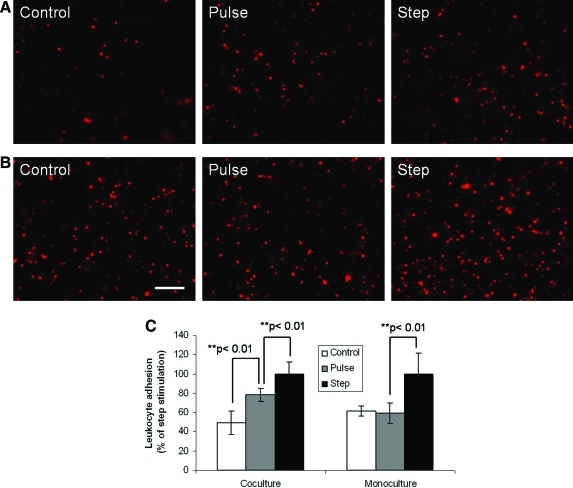
Leukocyte adhesion to endothelial cells in monoculture and coculture with hepatocytes
Leukocytes recruited during endotoxemia have been implicated in hepatic damage. Leukocytes adhesion to endothelial cells is an important step in leukocytes recruitment. We evaluated if differences observed in fluorescence response of our reporter were sufficient in inducing disparity in leukocytes adhesion to endothelial cells. Figure 4 shows leukocytes adhesion to endothelial cells in monoculture and coculture with hepatocytes subjected to control, pulse, and step stimulation of LPS. Figure 4A indicates that endothelial cells in coculture with hepatocytes show enhanced binding of leukocytes both for pulse and step treatment of LPS in comparison to control. Additionally, leukocyte binding response for pulse treatment was more than control but less than step stimulation of LPS (Fig. 4C). However, in the case of endothelial cells cultured alone (Fig. 4B) enhanced leukocyte adhesion was only observed for step treatment of LPS, while pulse exposure exhibited similar leukocyte binding as control (Fig. 4C). Further, in the absence of LPS stimulation, endothelial cells cultured alone displayed a higher level of leukocyte binding than those in coculture with hepatocytes (controls of Fig. 4A, B).
FIG. 4.
Leukocyte (labeled with a fluorescent dye CM-DiI) adhesion to endothelial cells stimulated with pulse or step of LPS under different culture conditions. (A) Fluorescence images of leukocytes adhering to endothelial cells on top of collagen embedded hepatocytes (coculture). (B) Fluorescence images of leukocytes adhering to endothelial cells on top of the collagen gel (monoculture). (C) Relative leukocyte adhesion (or fluorescence intensity) levels for (A) and (B) with control and pulse expressed as percentage of step stimulation. Scale bar = 50 μm. Color images available online at www.liebertonline.com/ten.
Discussion
In this report, we describe the development of an in vitro model of hepatic tissue that facilitates real-time monitoring of NF-κB activation in the endothelium in a complex tissue-like microenvironment. This was achieved by integrating a GFP reporter clone of endothelial cells in our previously developed organotypical model of liver tissue. We demonstrate the efficacy of our approach by identifying a differential NF-κB-regulated fluorescence response of endothelial cells to pulse and step of LPS, depending on whether they were in coculture with hepatocytes or in monoculture. Endothelial cells in coculture with hepatocytes exhibited similar fluorescence response for both pulse and step stimulation of LPS. By contrast, endothelial cells in monoculture displayed enhanced fluorescence response for step in comparison to pulse of LPS. We were able to correlate the disparity in the NF-κB-mediated fluorescence response to the expression level of several NF-κB-regulated genes such as ICAM-1, VCAM-1, and E-Selectin, which ultimately lead to differences in leukocyte adhesion to endothelial cells.
In the liver sinusoid, endothelial cells and hepatocytes are arranged in layers with the intervening space occupied by the extracellular matrix of the space of Disse. Our three-dimensional model mimics this arrangement by overlaying endothelial cells on top of collagen-embedded hepatocytes. In a previous report, we demonstrated the efficacy of this three-dimensional layering in maintaining hepatocyte differentiated function. This work focuses on utilizing this model for investigating hepatic inflammation. In the context of inflammation, our three-dimensional layered tissue model facilitates the creation of more in vivo-like conditions where endothelial cells are the first cell type to come in contact with leukocytes, while at the same time their adhesion is influenced by a signaling environment affected by hepatocytes.
There are several advantages of integrating reporter cells in tissue models for investigating gene expression patterns. In a tissue model that employs multiple cell types, conventional methods for gene expression analysis require separation of individual cell types before ascertaining gene expression levels in the cell type of interest. Additionally, each measurement requires destruction of cells, which is not optimal for obtaining dynamic information. In contrast, utilization of reporter cells not only obviates the need for separating cells, but temporal expression patterns can be obtained from the same population of cells. In our layered tissue model, dynamic NF-κB-regulated fluorescence response was measured in real time from the endothelium plane under inflammatory conditions. Since hepatocytes and endothelial cells were in different planes, the background autofluorescence from hepatocytes was minimized when the microscope was focused on the endothelium plane. The temporal fluorescence profile was similar to that observed in other reporter clones with a characteristic delay associated with transcription, translation, and maturation of GFP after activation of NF-κB.11,12,14
Although, in vivo, the liver sinusoid is lined by the sinusoidal endothelial cells,26 we used a GFP reporter clone of primary RHMEC in our model. Liver sinusoidal endothelial cells have limited proliferation capacity, which renders them unsuitable for creating a GFP reporter clone. We employed primary rat heart endothelial cells for creating a GFP reporter clone due to their capacity to proliferate and ease of maintaining them in culture. Use of heart endothelial cells is a limitation of our model as endothelial cells from different organs are known to exhibit heterogeneous phenotype. Nevertheless, we reasoned that primary rat heart endothelial cells will be a compromise in terms of creating a GFP reporter clone that enables real-time monitoring of NF-κB activation at the tissue level, while at the same time useful studies in the context of hepatic inflammation can be conducted as intercellular communication by various growth factors and cytokines are not omitted, since they belong to the same species as the hepatocytes. In fact, a wide variety of cell types, including cells belonging to other species, have shown remarkable ability to interact with hepatocytes and influence their function.27
LPS released from the outer cell membrane of gram-negative bacteria is a potent inducer of the inflammatory response.28 We used our model for investigating the role played by intercellular communication between hepatocytes and endothelial cells during LPS-induced inflammatory response. In vivo, LPS present in circulation can get neutralized by various proteins present in plasma.29 This can lead to conditions where LPS remains functionally active only transiently. To mimic such conditions, we subjected our system to a pulse of LPS, where LPS was introduced for a brief period and then cells were exposed to the medium for the remainder of the experiment. Pulse response was compared to step stimulation where LPS was not externally removed. Our results revealed that in the presence of hepatocytes, endothelial cells get activated similarly to both pulse and step stimulation. However, when endothelial cells were cultured alone, the level of activation was much stronger for step than pulse. Additionally, in the absence of any stimulation, hepatocytes reduce the activated state of endothelial cells. This suggests that hepatocytes play an important role in modulating the endothelial response whereby under noninflammatory conditions they contribute to maintaining the endothelium in a nonactivated state, while under inflammatory conditions they enhance the activation of endothelial cells upon encountering pathogen-associated products even for brief periods.
Tissues respond to injury or infection by recruiting leukocytes as part of the host defense strategy to ward off source of injury. However, in liver leukocytes recruited during endotoxemia play an important role in causing hepatic damage.30–31 There are several reports where blocking of leukocyte recruitment to liver results in diminished damage.31–33 Since NF-κB is believed to play a critical role in leukocyte recruitment,34 we evaluated the applicability of our model in applying the NF-κB-mediated fluorescence response to the functional output of leukocyte adhesion to endothelium, which is a key step in their recruitment. Our experiments showed that the disparity observed in the level of fluorescence response was sufficient in eliciting differential leukocyte adhesion to endothelial cells cultured under different conditions. Hepatocytes seemed to reduce adhesion of leukocytes to endothelial cells under noninflammatory (control) condition, while stimulation with pulse of LPS resulted in reversal of trend. This could be a useful strategy whereby hepatocytes communicate with the endothelium for ascertaining if hepatic tissue needs to recruit leukocytes—presence of mediators of inflammation induces leukocyte recruitment, whereas somewhat healthy condition is not favorable for leukocyte recruitment.
Hepatocytes can influence the microenvironment of endothelial cells dynamically by both secretion and/or uptake of factors from the medium. It is plausible that endothelial cells in coculture were subjected to different combination of factors under control condition and pulse stimulation of LPS, which resulted in reversal of trend from reduced to enhanced activation as the medium was supplemented with LPS for short duration. The factor(s) responsible for enhanced activation of endothelial cells in monoculture as compared to coculture under control condition are unknown. It remains unclear if somewhat higher activation of endothelial cells in monoculture (control condition) before LPS stimulation played any role in reduced responsiveness to LPS pulse. Although, step stimulation of LPS was able to further enhance the activation of endothelial cells in monoculture demonstrating that the cells had not reached maximum activation under control condition and retained ability to respond to LPS. These results suggest that the differential response of endothelial cells in monoculture and coculture to LPS pulse was hepatocytes mediated. The mechanism responsible for this differential response was not investigated in this study. Nevertheless, based on literature reports, involvement of certain factors secreted by hepatocytes can be speculated. Hepatocytes are known to secrete various factors such as vascular endothelial growth factor (VEGF)35 and LPS binding protein (LBP),36 which can influence the coculture response of endothelial cells to LPS. In a recent report, LPS administered to mice resulted in increased levels of VEGF in liver.37 VEGF is known to activate expression of adhesion molecules on endothelial cells.25 LBP is an acute-phase protein that binds to LPS and is thought to enhance LPS signaling.38,39 LBP in conjunction with other unidentified factors has also been implicated in LPS-induced higher cytokine production by nonparenchymal cells of liver in coculture with hepatocytes.40
In summary, our model provides a powerful platform for evaluating hepatic endothelium activation in real time under inflammatory conditions. Since endothelium activation and leukocyte recruitment play critical role in a number of disease states associated with inflammation, it can potentially be used as a high-throughput tool for identifying drug targets and gaining mechanistic insight into the role played by intercellular mediators in regulating hepatic inflammation.
Supplementary Material
Acknowledgments
We thank Candice Calhoun, and Carley Shulman for the isolation of cells from rat livers. This work was supported by NIH BioMEMS Resource Center Grant P41 EB-002503 and NIH Grant RO1AI063795. Microscopic imaging studies were made possible by a Core Morphology Facility, and gene expression measurements were carried out at the Genomic and Proteomic Facility at Boston's Shriners Burns Hospital.
Disclosure Statement
No competing financial interests exist.
References
- 1.Schmid-Schonbein G.W. Analysis of inflammation. Ann Rev Biomed Eng. 2006;8:93. doi: 10.1146/annurev.bioeng.8.061505.095708. [DOI] [PubMed] [Google Scholar]
- 2.Szabo G. Romics L., Jr. Frendl G. Liver in sepsis and systemic inflammatory response syndrome. Clin Liver Dis. 2002;6:1045. doi: 10.1016/s1089-3261(02)00058-2. [DOI] [PubMed] [Google Scholar]
- 3.Jaeschke H. Molecular mechanisms of hepatic ischemia-reperfusion injury and preconditioning. Am J Physiol. 2003;284:G15. doi: 10.1152/ajpgi.00342.2002. [DOI] [PubMed] [Google Scholar]
- 4.Wieckowska A. Mccullough A.J. Feldstein A.E. Noninvasive diagnosis and monitoring of nonalcoholic steatohepatitis: present and future. Hepatology (Baltimore, MD) 2007;46:582. doi: 10.1002/hep.21768. [DOI] [PubMed] [Google Scholar]
- 5.Brenner D.A. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361. [PMC free article] [PubMed] [Google Scholar]
- 6.Ganey P.E. Roth R.A. Concurrent inflammation as a determinant of susceptibility to toxicity from xenobiotic agents. Toxicology. 2001;169:195. doi: 10.1016/s0300-483x(01)00523-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu S.F. Malik A.B. NF-kappa B activation as a pathological mechanism of septic shock and inflammation. Am J Physiol Lung Cell Mol Physiol. 2006;290:L622. doi: 10.1152/ajplung.00477.2005. [DOI] [PubMed] [Google Scholar]
- 8.Essani N.A. Mcguire G.M. Manning A.M. Jaeschke H. Endotoxin-induced activation of the nuclear transcription factor kappa B and expression of E-selectin messenger RNA in hepatocytes, Kupffer cells, and endothelial cells in vivo. J Immunol. 1996;156:2956. [PubMed] [Google Scholar]
- 9.Kuhnel F. Zender L. Paul Y. Tietze M.K. Trautwein C. Manns M., et al. NFkappaB mediates apoptosis through transcriptional activation of Fas (CD95) in adenoviral hepatitis. J Biol Chem. 2000;275:6421. doi: 10.1074/jbc.275.9.6421. [DOI] [PubMed] [Google Scholar]
- 10.Barnes P.J. Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 11.King K.R. Wang S. Irimia D. Jayaraman A. Toner M. Yarmush M.L. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieder K.J. King K.R. Thompson D.M. Zia C. Yarmush M.L. Jayaraman A. Optimization of reporter cells for expression profiling in a microfluidic device. Biomed Microdevices. 2005;7:213. doi: 10.1007/s10544-005-3028-3. [DOI] [PubMed] [Google Scholar]
- 13.Canton I. Sarwar U. Kemp E.H. Ryan A.J. Macneil S. Haycock J.W. Real-time detection of stress in 3D tissue-engineered constructs using NF-kappaB activation in transiently transfected human dermal fibroblast cells. Tissue Eng. 2007;13:1013. doi: 10.1089/ten.2006.0357. [DOI] [PubMed] [Google Scholar]
- 14.Thompson D.M. King K.R. Wieder K.J. Toner M. Yarmush M.L. Jayaraman A. Dynamic gene expression profiling using a microfabricated living cell array. Anal Chem. 2004;76:4098. doi: 10.1021/ac0354241. [DOI] [PubMed] [Google Scholar]
- 15.Jindal R. Nahmias Y. Tilles A.W. Berthiaume F. Yarmush M.L. Amino acid-mediated heterotypic interaction governs performance of a hepatic tissue model. Faseb J. 2009;23:2288. doi: 10.1096/fj.08-114934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seglen P.O. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 17.Dunn J.C. Tompkins R.G. Yarmush M.L. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog. 1991;7:237. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- 18.Moghe P.V. Coger R.N. Toner M. Yarmush M.L. Cell-cell interactions are essential for maintenance of hepatocyte function in collagen gel but not on Matrigel. Biotechnol Bioeng. 1997;56:706. doi: 10.1002/(SICI)1097-0290(19971220)56:6<706::AID-BIT14>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Dunn J.C. Yarmush M.L. Koebe H.G. Tompkins R.G. Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. Faseb J. 1989;3:174. doi: 10.1096/fasebj.3.2.2914628. [DOI] [PubMed] [Google Scholar]
- 20.Park J. Berthiaume F. Toner M. Yarmush M.L. Tilles A.W. Microfabricated grooved substrates as platforms for bioartificial liver reactors. Biotechnol Bioeng. 2005;90:632. doi: 10.1002/bit.20463. [DOI] [PubMed] [Google Scholar]
- 21.Ryan C.M. Carter E.A. Jenkins R.L. Sterling L.M. Yarmush M.L. Malt R.A., et al. Isolation and long-term culture of human hepatocytes. Surgery. 1993;113:48. [PubMed] [Google Scholar]
- 22.Deroanne C.F. Lapiere C.M. Nusgens B.V. In vitro tubulogenesis of endothelial cells by relaxation of the coupling extracellular matrix-cytoskeleton. Cardiovasc Res. 2001;49:647. doi: 10.1016/s0008-6363(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 23.Wallach D. Varfolomeev E.E. Malinin N.L. Goltsev Y.V. Kovalenko A.V. Boldin M.P. Tumor necrosis factor receptor and Fas signaling mechanisms. Ann Rev Immunol. 1999;17:331. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 24.Faure E. Equils O. Sieling P.A. Thomas L. Zhang F.X. Kirschning C.J., et al. Bacterial lipopolysaccharide activates NF-kappaB through toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J Biol Chem. 2000;275:11058. doi: 10.1074/jbc.275.15.11058. [DOI] [PubMed] [Google Scholar]
- 25.Kim I. Moon S.O. Kim S.H. Kim H.J. Koh Y.S. Koh G.Y. Vascular endothelial growth factor expression of intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin through nuclear factor-kappa B activation in endothelial cells. J Biol Chem. 2001;276:7614. doi: 10.1074/jbc.M009705200. [DOI] [PubMed] [Google Scholar]
- 26.Tilles A.W. Baskaran H. Roy P. Yarmush M.L. Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol Bioeng. 2001;73:379. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- 27.Chan C. Berthiaume F. Nath B.D. Tilles A.W. Toner M. Yarmush M.L. Hepatic tissue engineering for adjunct and temporary liver support: critical technologies. Liver Transplant. 2004;10:1331. doi: 10.1002/lt.20229. [DOI] [PubMed] [Google Scholar]
- 28.Moulin F. Copple B.L. Ganey P.E. Roth R.A. Hepatic and extrahepatic factors critical for liver injury during lipopolysaccharide exposure. Am J Physiol. 2001;281:G1423. doi: 10.1152/ajpgi.2001.281.6.G1423. [DOI] [PubMed] [Google Scholar]
- 29.Dedrick R.L. Conlon P.J. Prolonged expression of lipopolysaccharide (LPS)-induced inflammatory genes in whole blood requires continual exposure to LPS. Infect Immun. 1995;63:1362. doi: 10.1128/iai.63.4.1362-1368.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewett J.A. Schultze A.E. Vancise S. Roth R.A. Neutrophil depletion protects against liver injury from bacterial endotoxin. Lab Invest J Tech Methods Pathol. 1992;66:347. [PubMed] [Google Scholar]
- 31.Jaeschke H. Farhood A. Smith C.W. Neutrophil-induced liver cell injury in endotoxin shock is a CD11b/CD18-dependent mechanism. Am J Physiol. 1991;261:G1051. doi: 10.1152/ajpgi.1991.261.6.G1051. [DOI] [PubMed] [Google Scholar]
- 32.Li X. Klintman D. Liu Q. Sato T. Jeppsson B. Thorlacius H. Critical role of CXC chemokines in endotoxemic liver injury in mice. J Leukoc Biol. 2004;75:443. doi: 10.1189/jlb.0603297. [DOI] [PubMed] [Google Scholar]
- 33.Gujral J.S. Liu J. Farhood A. Hinson J.A. Jaeschke H. Functional importance of ICAM-1 in the mechanism of neutrophil-induced liver injury in bile duct-ligated mice. Am J Physiol. 2004;286:G499. doi: 10.1152/ajpgi.00318.2003. [DOI] [PubMed] [Google Scholar]
- 34.Alcamo E. Mizgerd J.P. Horwitz B.H. Bronson R. Beg A.A. Scott M., et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J Immunol. 2001;167:1592. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu H. Miyazaki M. Wakabayashi Y. Mitsuhashi N. Kato A. Ito H., et al. Vascular endothelial growth factor secreted by replicating hepatocytes induces sinusoidal endothelial cell proliferation during regeneration after partial hepatectomy in rats. J Hepatol. 2001;34:683. doi: 10.1016/s0168-8278(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 36.Grube B.J. Cochane C.G. Ye R.D. Green C.E. Mcphail M.E. Ulevitch R.J., et al. Lipopolysaccharide binding protein expression in primary human hepatocytes and HepG2 hepatoma cells. J Biol Chem. 1994;269:8477. [PubMed] [Google Scholar]
- 37.Yano K. Liaw P.C. Mullington J.M. Shih S.C. Okada H. Bodyak N., et al. Vascular endothelial growth factor is an important determinant of sepsis morbidity and mortality. J Exp Med. 2006;203:1447. doi: 10.1084/jem.20060375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright S.D. Ramos R.A. Tobias P.S. Ulevitch R.J. Mathison J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 39.Jack R.S. Fan X. Bernheiden M. Rune G. Ehlers M. Weber A., et al. Lipopolysaccharide-binding protein is required to combat a murine gram-negative bacterial infection. Nature. 1997;389:742. doi: 10.1038/39622. [DOI] [PubMed] [Google Scholar]
- 40.Scott M.J. Liu S. Su G.L. Vodovotz Y. Billiar T.R. Hepatocytes enhance effects of lipopolysaccharide on liver nonparenchymal cells through close cell interactions. Shock (Augusta, GA) 2005;23:453. doi: 10.1097/01.shk.0000160939.08385.f1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.