Abstract
Experimental and epidemiologic evidence indicates that variations of absolute humidity account for the onset and seasonal cycle of epidemic influenza in temperate regions. A role for absolute humidity in the transmission of pandemic influenza, such as 2009 A/H1N1, has yet to be demonstrated and, indeed, outbreaks of pandemic influenza during more humid spring, summer, and autumn months might appear to constitute evidence against an effect of humidity. However, here the authors show that variations of the basic and effective reproductive numbers for influenza, caused by seasonal changes in absolute humidity, are consistent with the general timing of pandemic influenza outbreaks observed for 2009 A/H1N1 in temperate regions, as well as wintertime transmission of epidemic influenza. Indeed, absolute humidity conditions correctly identify the region of the United States vulnerable to a third, wintertime wave of pandemic influenza. These findings suggest that the timing of pandemic influenza outbreaks is controlled by a combination of absolute humidity conditions, levels of susceptibility, and changes in population-mixing and contact rates.
Keywords: disease outbreaks; disease susceptibility; disease transmission, infectious; humidity; influenza, human
Recent studies have shown that the survival and transmission of the influenza virus (1), as well as the winter seasonality of epidemic influenza and the onset of individual wintertime influenza outbreaks (2), are strongly associated with declines in absolute humidity. This relation is nonlinear, with influenza transmission and survival most sensitive to absolute humidity variations when conditions are dry (Figure 1). In temperate regions, absolute humidity has a substantial seasonal cycle, both indoors and outdoors, which peaks in summer and reaches its nadir in winter (1). Differences in this seasonal cycle, as well as day-to-day weather, from place to place may in part explain changes in the timing of individual influenza seasons.
Figure 1.
Influenza virus survival, transmission, and the basic reproductive number, R0, plotted as a function of absolute humidity. Influenza virus survival data are from Harper (30), influenza virus transmission data are from Lowen et al. (31, 32), and R0 is based on best-fitting, absolute humidity-forced, susceptible-infected-recovered susceptible simulations from Shaman et al. (2). The solid line is R0 for the best-fitting simulation; the gray region shows the range of R0 values as a function of absolute humidity for the 10 best-fitting simulations. The measure of absolute humidity is 2 m above-ground specific humidity in kg/kg and is taken from National Center for Environmental Prediction–National Center for Atmospheric Research (NCEP-NCAR) reanalysis (23).
Sustained transmission of pandemic influenza in temperate regions, by contrast, often occurs out of season during spring, summer, and autumn. Such a transmission pattern occurred during the 2009 A/H1N1 pandemic, for example. The spread of pandemic influenza within particular populations often points to the importance of clustering of individuals in close quarters, such as military vessels (3, 4) or schools (5, 6). Such clusters of high-transmitting populations may help to sustain the outbreak in the general population, as evidenced by the influence of school opening and closing on the community-wide transmission of 2009 A/H1N1 pandemic influenza (7). Although these observations demonstrate that influenza transmission is possible in more humid conditions, the implications for the relation between pandemic influenza transmission and absolute humidity are less clear. One might imagine that the sustained transmission of pandemic influenza outside the wintertime epidemic influenza season (i.e., during periods of higher absolute humidity) argues against the importance of absolute humidity in driving the timing of influenza epidemics. Moreover, one might argue that the association of pandemic influenza outbreaks in the autumn with the resumption of school argues for a greater role for increased mixing in schools, rather than increased transmissibility from low absolute humidity, in the seasonality of epidemic influenza. These objections, if correct, would raise serious concerns about the causal role of absolute humidity in the timing of seasonal influenza.
In this article, we briefly describe the evidence underlying the absolute humidity-seasonality hypothesis for seasonal influenza, and then we consider each of these issues—nonwinter pandemics and pandemic resurgence when schools reopened—in turn. We also examine whether absolute humidity variability may have affected the geographic pattern of development, or lack thereof, of a wintertime third wave of pandemic influenza in the continental United States. We conclude that the observed patterns of 2009 A/H1N1 transmission are consistent with the hypothesis that absolute humidity modulates the survival and transmission of both epidemic and pandemic influenza viruses in temperate regions.
HUMIDITY AND THE BASIC AND EFFECTIVE REPRODUCTIVE NUMBERS
The effect of absolute humidity on epidemic influenza transmission and seasonality can be understood in terms of the basic reproductive number, R0, the number of secondary infections the average infectious person would produce in a fully susceptible population. Previous modeling work indicates that R0 varies through time as absolute humidity changes (2), that is, that transmission patterns fit a model in which
where R0(t) is the daily basic reproductive number, R0min is a constant that defines a baseline level for R0(t) at high absolute humidity, q(t) is daily specific humidity, a measure of absolute humidity, and t is time. Best-fitting parameter combinations from simulations of this model include maximal values of R0(t) between 2 and 4 at low absolute humidity, a mean infectious period of 2–4.2 days, and a duration of immunity of 3–8 years (Table 1).
Table 1.
Parameter Combinations for the 10 Best-Fit Susceptible-Infected-Recovered Susceptible Simulations at the Arizona, Florida, Illinois, New York, and Washington State Sitesa,b
| Ordered Model Rank | L, Years | D, Days | R0max, Persons/Person | R0min, Persons/Person |
| 1 | 5.35 | 3.24 | 3.52 | 1.12 |
| 2 | 5.40 | 2.41 | 2.89 | 1.16 |
| 3 | 3.28 | 4.18 | 3.40 | 1.22 |
| 4 | 3.70 | 2.03 | 2.05 | 1.15 |
| 5 | 7.77 | 2.59 | 3.69 | 1.30 |
| 6 | 6.23 | 2.37 | 2.71 | 1.23 |
| 7 | 6.05 | 2.56 | 3.79 | 1.06 |
| 8 | 4.61 | 2.71 | 2.61 | 1.29 |
| 9 | 7.39 | 2.85 | 3.69 | 1.27 |
| 10 | 3.58 | 3.61 | 3.19 | 1.20 |
Abbreviations: R0max, a constant that defines the maximum basic reproductive number, R0(t), at zero absolute humidity; R0min, a constant that defines a baseline level for basic reproduction number, R0(t), at high absolute humidity; RMS, root mean square; SIRS, susceptible-infected-recovered susceptible.
Adapted from Shaman et al. (2).
At each site, 5,000 simulations were performed with the parameters R0max, R0min, D (mean infection period), and L (mean duration of immunity) randomly chosen from within specified ranges. Best-fit SIRS simulations were selected for the 5 sites in aggregate based on RMS error after scaling the 31-year mean daily infection number to the 31-year mean observed daily excess pneumonia and influenza mortality rate at each site.
The basic reproductive number sets an upper bound for the possible intensity of transmission, but the actual number of secondary cases infected by a typical primary case depends on the proportion of contacts who are susceptible to infection. This quantity, the effective reproductive number, RE(t), is given (for a simple model) by
| (1) |
where N is the total population, S(t) is the number of persons susceptible to influenza infection, and S(t)/N is the population susceptibility to influenza infection.
For an outbreak of influenza to occur, RE(t) must be greater than 1. As long as RE(t) > 1, the number of influenza infections will grow; however, as an outbreak proceeds and more susceptibles are infected, the proportion of the population remaining susceptible (S(t)/N) decreases. Eventually, RE(t) falls below 1, at which point infection numbers decrease and the outbreak subsides (Figure 2).
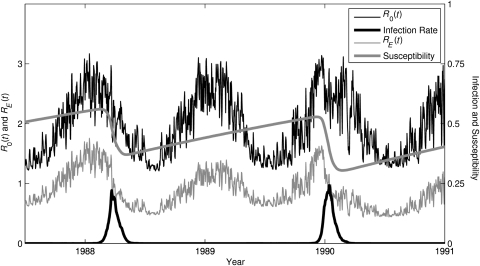
Figure 2.
Time series of simulated epidemic influenza in New York State from an absolute humidity-forced, susceptible-infected-recovered susceptible (SIRS) model. Simulation is shown from July 1987 through December 1990 for the best-fitting parameter combination (Table 1). The SIRS model simulates 2 influenza subtypes (A/H3N2 and A/H1N1), but only the time series for A/H3N2 is shown. Plotted lines indicate the steady rise of susceptibility (S(t)/N) to A/H3N2 in the time between outbreaks (thick gray line); the seasonal cycle of the basic reproductive number, R0(t) (thin black line), due to seasonal changes in absolute humidity plus shorter time-scale variability due to changes in absolute humidity due to weather variability; the time series of the effective reproductive number, RE(t) (thin gray line); and the time series of infection rate (proportion infected × 10, thick black line). During outbreaks, both S(t) and RE(t) drop precipitously as susceptibles are infected and S(t) decreases. Once RE(t) drops below 1, the outbreak begins to abate. Of note, A/H3N2 was not present in the simulation from April 1988 to March 1989; hence, no outbreak was possible during this winter (A/H1N1 was present and is not shown).
EPIDEMIC VERSUS PANDEMIC INFLUENZA
The timing of outbreaks of both epidemic and pandemic influenza can be understood with reference to the effective reproductive number and its relation to the basic reproductive number and absolute humidity. To examine these issues, we further analyze results from model simulations of epidemic influenza, as presented by Shaman et al. (2), for New York State, which experienced considerable spring and late summer/early autumn 2009 A/H1N1 transmission (5, 8). Although the real world is much more complex than these idealized simulations, these model representations of transmission provide insight into the dynamics underlying outbreak events.
The parameter combination used in this representative example is the 1972–2002 (31-year) best-fit simulation of epidemic influenza (Table 1). Given these parameter values and observed absolute humidity levels for New York State, R0(t) ranges seasonally on average from a summertime low of 1.24 to a wintertime high of 3.14. Susceptibility ranges from an average postoutbreak minimum of 0.34 to an average preoutbreak maximum of 0.52. In summer, when humidity is high, these susceptibilities imply an RE(t) ranging from 0.42 to 0.64. Thus, even at the highest population susceptibility level of 0.52, summertime RE(t) remains well below 1 and substantial outbreaks of influenza are not possible. However, during winter, when R0(t) is high, RE(t) rises well above 1 (Figure 2) and epidemics do occur. Thus, the seasonality of R0(t), which varies with absolute humidity, strongly favors wintertime epidemics in temperate regions. This finding, in which absolute humidity and susceptibility preclude epidemic transmission during summer, is also evident for other best-fitting model parameter combinations (Table 1) and simulated susceptibilities (not shown).
For pandemic influenza, preoutbreak susceptibility is much higher than for epidemic influenza, particularly among younger individuals, including school-aged populations. Prior to the 2009 A/H1N1 pandemic, little immunity to this virus was measurable in individuals under 30 years of age, who are thought to be the main drivers of transmission of influenza and most other respiratory infections (9, 10). We can use absolute humidity conditions in New York City during 2009 to examine both R0(t) and RE(t) with respect to 2009 A/H1N1 (Figure 3). The city was slightly more humid during late April and early May 2009 than normal (the 1948–2008 average) when the first pandemic wave developed (5). RE(t) is shown for several population susceptibility levels and reaches its nadir during August. In this simplified model, susceptibility above 80% permits epidemic growth even at the nadir of transmissibility in August, while transmissibility above about 60% permits epidemic growth in May–June, when the main epidemic occurred in New York City. After August, RE(t) rises as humidity levels fall, and transmission of influenza becomes possible for even lower levels of susceptibility.
Figure 3.
Time series of New York City observed specific humidity, estimated basic reproductive number, and estimated effective reproductive number for 1948–2008 and 2009. Top, plots of observed specific humidity, q(t), and estimated basic reproductive number, R0(t), from the susceptible-infected-recovered susceptible (SIRS) model best-fitting parameter combination. Bottom, plots of estimated effective reproductive number, RE(t), for various population susceptibility levels; the dash-dot line shows RE(t) = 1.
These numbers are not intended as precise estimates of R0(t) or RE(t) for New York City; moreover, RE(t) is likely to increase after school opening (7). Nonetheless, our model demonstrates the potential patterns of pandemic and epidemic influenza transmission facilitated by susceptibility and absolute humidity in temperate regions. High susceptibility (>60%–80% in this example) to a novel strain of influenza, such as 2009 A/H1N1, can support transmission even in the presence of high spring or summer absolute humidity, even in places where seasonal influenza could not spread. Sustained transmission of 2009 A/H1N1 did occur in many temperate locations, including New York City, during spring and summer (11, 12).
In temperate regions, typically, absolute humidity declines and R0(t) rises beginning in September. At the same time, increased close contact, particularly between schoolchildren in classrooms and among college students in group residences, begins to occur. Both trends may have contributed to the autumn outbreaks of 2009 A/H1N1 observed in the United States (3, 4, 7) and elsewhere.
In contrast, susceptibility is typically well below 60% for epidemic influenza that, in conjunction with high absolute humidity, precludes significant influenza transmission during summer and early fall (in this perfectly mixed model example). Only during late fall and winter, when absolute humidity is at its lowest, does RE(t) rise above 1 for epidemic influenza (Figure 3). Population structure in human populations leads to more variability than this simple model would suggest. In temperate zones, epidemic influenza transmission does typically peak during winter; however, localized outbreaks during spring, summer, and autumn do occur. These out-of-season epidemic outbreaks occur where locally RE(t) has risen above 1. Real-world populations are clustered. This heterogeneity can group susceptible individuals together and create subpopulations in which the increased susceptibility to epidemic influenza, combined with high enough R0(t), as dictated by absolute humidity conditions, is sufficient to push RE(t) above 1.
Thus, although absolute humidity conditions determine the general phase organization of epidemic influenza transmission, such that the majority of temperate region infections occur during winter, absolute humidity conditions alone do not preclude out-of-season epidemic influenza transmission in select subpopulations. Rather, absolute humidity conditions must be evaluated in conjunction with local levels of susceptibility to determine whether RE(t) is >1 and transmission can be supported. The school environment is one such location where susceptible subpopulations cluster, and RE(t) may rise above 1 prior to winter.
THE TIMING OF PANDEMIC AND EPIDEMIC INFLUENZA
The 2009 A/H1N1 pandemic showed that, when conditions are conducive for influenza transmission, that is, RE(t) > 1, the transmission response to population mixing and increased contact in schools is rapid (5, 7). Because of the clustering of susceptible children during academic terms, it is likely that the opening of schools in late summer/early autumn also contributes to the spread of epidemic influenza, even though the peak of epidemic influenza is much later, typically between December and February. However, by itself, the school calendar does not explain the seasonality of epidemic influenza. In particular, the seasonal cycle of epidemic influenza has a greater amplitude and a more consistent phase in temperate regions (2, 13, 14) than in subtropical and tropical regions (13, 15–17) despite the existence of school calendars in some countries in the latter regions (e.g., Hong Kong, Thailand) that are similar to those in temperate Western countries. This circumstance indicates that factors other than the school calendar must be contributing to the seasonality of epidemic influenza.
Furthermore, we have previously described 2 findings that indicate that changes in absolute humidity affect the transmission of epidemic influenza (2): 1) An absolute humidity-driven model of smoothly varying influenza transmissibility, peaking in midwinter, fits the seasonal cycle of influenza transmission better than one in which transmissibility increases as a step function when schools are in session; and 2) negative absolute humidity anomalies are associated with the onset of sustained wintertime influenza transmission in the United States.
Here, we argue that the timing of pandemic influenza waves provides further support for the importance of absolute humidity in determining influenza transmissibility. Specifically, if the potential for influenza transmission (R0(t)) were determined primarily by schools—as a step function increasing around September in the United States—then the effective transmissibility, RE(t), would remain nearly fixed or decline during the course of the school year, as susceptibles are depleted. On the other hand, evidence that RE(t) increases within the course of a school year would suggest that some factor that varies with season—beyond school terms themselves—is contributing to RE(t). Two waves of sustained transmission occurred during the school years in the pandemics of 1918, 1957, and 2009. For each of these pandemics, there was a wave of influenza soon after schools opened in the United States during September and October. This wave subsided by November, but in some parts of the country there was a subsequent resurgent wave of sustained transmission during the winter months of December–February (8, 18, 19).
In 1918, the resumption of transmission was due at least in part to the relaxation of intense control measures in certain cities (20–22), but no such explanation is available for 1957 or 2009. These resurgences imply an increase of R0(t), as there is no other simple mechanism by which a declining epidemic could turn into a growing one. A reasonable explanation for the resurgence of pandemic influenza during the winters of 1957 and 2009, and possibly 1918, is that absolute humidity conditions became more favorable as the winter set in. In 2009, the winter wave in the United States was limited to the southeastern part of the country. In the next section, we assess whether the geographic pattern of the winter part of the 2009 pandemic is consistent with predictions made using our absolute humidity-influenza model (2).
A THIRD WAVE OF PANDEMIC 2009 A/H1N1
Given the historical precedents of 1918 and 1957, health officials and researchers were aware that a wintertime wave of 2009 A/H1N1 transmission might follow the August–November outbreak. We explored this possibility of a third wave in terms of the effective reproductive number, the basic reproductive number, and absolute humidity.
We used regional data on influenza-like illness and viral positivity that are publicly available from the Centers for Disease Control and Prevention (CDC) (8) to make estimates of the upper bounds on RE(t) during weeks 44–49 of 2009, following the fall outbreak (Table 2). Specifically, the weekly effective reproductive number for week tw was estimated as
| (2) |
where is a relative measure of weekly influenza incidence for week tw, estimated as the percentage of influenza-like illness among physician visits × the percentage of collected specimens testing positive for influenza during that week, and μ is the mean serial interval for influenza, that is, the mean time in days between the infection of an individual and the infection of others by that individual. A further description of this derivation is provided in the Appendix.
Table 2.
Upper Bounds of RE(t) for Different Weeks in Different Geographic Regions of the United States During the Fall of 2009a
| Region | Weeks 44–46 | Weeks 45–47 | Weeks 46–48 | Weeks 47–49 |
| 1 | 0.862 | 0.788 | 0.767 | 0.732 |
| 2 | 0.879 | 0.827 | 0.842 | 0.845 |
| 3 | 0.815 | 0.714 | 0.745 | 0.730 |
| 4 | 0.874 | 0.868 | 0.938 | 0.936 |
| 5 | 0.814 | 0.801 | 0.782 | 0.773 |
| 6 | 0.853 | 0.790 | 0.823 | 0.913 |
| 7 | 0.753 | 0.752 | 0.838 | 0.780 |
| 8 | 0.767 | 0.843 | 0.870 | 0.839 |
| 9 | 0.902 | 0.998 | 0.886 | 0.786 |
| 10 | 0.847 | 0.793 | 0.804 | 0.823 |
Abbreviation: RE(t), effective reproduction number.
The regions are defined as follows: region 1—Connecticut, Maine, Massachusetts, New Hampshire, Rhode Island, and Vermont; region 2—New Jersey, New York, Puerto Rico, and US Virgin Islands; region 3—Delaware, District of Columbia, Maryland, Pennsylvania, Virginia, and West Virginia; region 4—Alabama, Florida, Georgia, Kentucky, Mississippi, North Carolina, South Carolina, and Tennessee; region 5—Illinois, Indiana, Michigan, Minnesota, Ohio, and Wisconsin; region 6—Arkansas, Louisiana, New Mexico, Oklahoma, and Texas; region 7—Iowa, Kansas, Missouri, and Nebraska; region 8—Colorado, Montana, North Dakota, South Dakota, Utah, and Wyoming; region 9—Arizona, California, Guam, Hawaii, and Nevada; and region 10—Alaska, Idaho, Oregon, and Washington.
We then used 2009–2010 wintertime 2 m above-ground specific humidity conditions (23) in conjunction with parameters derived from the best-fitting model simulation (Table 1, model 1) to estimate R0(t) during the fall and winter throughout the United States. These R0(t) values were then used to project changes to upper-bound RE(t) during the 2009–2010 winter by using the expression:
| (3) |
where t1 is weeks 47–49, t2 is a subsequent time period, and RE(t1) is the estimates of RE(t) for weeks 47–49 (Table 2; Figure 4A).
Figure 4.
Distributed maps of estimated and projected upper-bound effective reproductive number, RE(t), for 2009 A/H1N1 in the United States during the winter of 2009–2010. A, 2009 week 47–49 estimates of RE(t) (from Table 2); B, the ratio of projected 2010 week 1–3 RE(t) to 2009 week 47–49 estimates of RE(t) showing the proportional change of RE(t); C, as for B, but for projected 2010 week 4–6 RE(t); D–F, 3-week projections of upper-bound RE(t) made by using the 2009 week 47–49 estimates of susceptibility and estimates of 3-week average basic reproductive number, R0(t). Both R0(t) and the upper-bound estimates of RE(t) were made by using 2-m above-ground specific humidity, q(t), from National Center for Environmental Prediction–National Center for Atmospheric Research (NCEP-NCAR) reanalysis (2). R0(t) was calculated by using equation 4 of Shaman et al. (2) and the best-fit susceptible-infected-recovered susceptible (SIRS) parameter estimates of maximum and minimum basic reproductive number. RE(t) was calculated per equation 3. D, 2009 week 50–52 projections of RE(t); E, 2010 week 1–3 projections of RE(t); F, 2010 week 4–6 projections of RE(t).
These absolute humidity-based projections indicate that RE(t) (and R0(t)) rose considerably in the eastern United States during the first weeks of 2010 (Figure 4, B and C). In the southeastern United States, this increase was sufficient to drive RE(t) above 1 from mid-December through mid-February (Figure 4, D–F). These high levels were due to, in part, the lingering high level of RE(t) in this region following the fall wave (Table 2, region 4; Figure 4A) and, in part, the decreased absolute humidity levels during January and February, relative to late November/early December, that led to an increase of R0(t) and hence RE(t) in the southeastern United States (Figure 4, B and C). We also performed similar projections using other best-fitting model parameter combinations with a mean infectious period of greater than 2.5 days (Table 1). These projections produced similar results (Web Figures 1–3; these supplementary figures are posted on the Journal’s Web site (http://aje.oupjournals.org/)). Indeed, the southeastern United States did experience a third wave of A/H1N1 (8, 24), while other regions did not.
DISCUSSION
Variations of absolute humidity provide a framework that helps to explain the timing of both epidemic and pandemic influenza in temperate regions. As a key modulator of R0(t), absolute humidity facilitates influenza transmission should the virus be present and susceptibility within subpopulations be appropriate. Increased contact within schools provides a further boost to transmission, but it does not explain the entire seasonal variation in transmission of pandemic or epidemic influenza.
Differences between pandemic and epidemic influenza transmission dynamics appear primarily to be due to differences in population susceptibility to these pathogens, particularly among school-aged children. With little immunity, population mixing and increased person-to-person contact at the start of the school year can trigger transmission during late summer and fall; however, with extensive immunity, as with epidemic influenza, the start of the school term will not typically initiate an influenza outbreak. Rather, epidemic influenza typically peaks in the winter when low absolute humidity maximizes R0(t).
School closure reduces RE(t), in part, by reducing opportunities for transmission among susceptible school-aged individuals. For a new pandemic, if school closures drive RE(t) below 1 in a given area, outbreaks may temporarily abate or be averted. The efficacy of school closure will depend on immunity levels to the pandemic strain in the broader population and when the pandemic arrives. As shown in Figure 3, the modulation of RE(t) by absolute humidity suggests that a pandemic virus that arrives in a temperate region during winter will be harder to control, through school closure or other measures, than a pandemic arriving during more humid months.
Observed absolute humidity changes during the 2009–2010 winter, in conjunction with upper-bound estimates of RE(t) following the autumn wave of pandemic 2009 A/H1N1, correctly identify the southeastern United States as the region within this country most vulnerable to a subsequent winter resurgence of pandemic influenza. Had these projections been made in real time. they could have utilized weather forecasts in the short term (1–5 days) and historical conditions for that area and time of year for longer time scales (>5 days). In the future, such a framework could be used in real time to assess influenza outbreak risk in temperate regions.
The simulations and projections presented here are highly idealized; the model used (2) simulates a perfectly mixed, unstructured population and utilizes simplified influenza transmission dynamics. However, in spite of this simple framework, model behavior is consistent with the observed transmission patterns of both epidemic and pandemic influenza in temperate regions, and the model correctly simulates the third winter wave of 2009 A/H1N1 in the United States. Future study, however, might use a more detailed, structured model of influenza transmission and provide more precise estimates of absolute humidity-modulated effects.
The findings presented here are for temperate regions, where the relation between absolute humidity and influenza is best established. In the tropics, influenza often peaks during more humid and rainy seasons (25, 26). The relation presented in Figure 1 indicates that, in areas of high year-round absolute humidity, such as the tropics, seasonal absolute humidity-based modulations of influenza virus survival and transmission would be very reduced. In such an environment, another factor might control the seasonal timing of influenza. Alternately, the relation presented in Figure 1 might be incomplete; a few laboratory studies found influenza survival minimal at moderate humidity but increased at both low and high levels (27, 28), and some recent theoretical work suggests that virus desiccation may be reduced at high absolute humidity (29). These studies suggest a bimodal relation between absolute humidity and influenza transmission and that, in the humid tropics, higher absolute humidity would favor influenza transmission. Further investigation of this issue is needed.
Overall, the hypothesis that absolute humidity modulates influenza virus survival and transmission provides a framework for understanding outbreaks of both epidemic and pandemic influenza in temperate regions. Further, more detailed study of the effects of absolute humidity on pandemic influenza transmission is needed.
Supplementary Material
Acknowledgments
Author affiliations: College of Oceanic and Atmospheric Sciences, Oregon State University, Corvallis, Oregon (Jeffrey Shaman); Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard School of Public Health, Harvard University, Boston, Massachusetts (Edward Goldstein, Marc Lipsitch); and Department of Immunology and Infectious Diseases, Harvard School of Public Health, Harvard University, Boston, Massachusetts (Marc Lipsitch).
This work was supported by the US National Institutes of Health Models of Infectious Disease Agent Study program (cooperative agreement 1U54GM088558).
Dr. Marc Lipsitch discloses consulting or honorarium income from the Avian/Pandemic Flu Registry (Outcome Sciences, funded in part by Roche) and from Pfizer and Novartis.
Glossary
Abbreviation
- CDC
Centers for Disease Control and Prevention
APPENDIX
Deriving an Upper Bound on RE(t)
To estimate an upper bound on RE(tw) for week tw, we define w(t) to be the serial interval distribution for influenza, with mean μ; namely, it is the distribution of time (in days, t) between an individual's infection and the infection of his infectees. We let r be the daily growth (or decline) rate, such that the weekly change of influenza incidence is given by
| (A1) |
In addition, let I(t) be the daily incidence t days prior to week tw; thus, 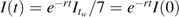 . We assume that the serial interval is no longer than a week and that the effective reproductive number does not grow between weeks tw−1 and tw. Thus, the effective reproductive number is bounded by RE(tw). By the Euler-Lotka equation,
. We assume that the serial interval is no longer than a week and that the effective reproductive number does not grow between weeks tw−1 and tw. Thus, the effective reproductive number is bounded by RE(tw). By the Euler-Lotka equation,
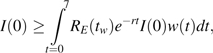 |
where the inequality stems from a lower bound on RE(tw). Thus, by Jensen's inequality,
 |
that in view of equation A1 is equivalent to equation 2 (33).
We first used CDC data (8) to give a bound on the RE(tw) for the whole of the United States (Appendix Table 1). Following numerous studies (34–38), we assumed that μ ≥ 2.5 days. Consequently, the inequality shown in equation 2 applies with μ = 2.5 days for a declining epidemic. Table 3 shows the upper bounds on RE(tw) for the last weeks of the fall 2009 A/H1N1 outbreak in the United States. During weeks 45–49, the national upper bound on RE(tw) is 0.833.
We then used regional CDC data (8) based on influenza-like illness and specimen testing to estimate triweekly upper bounds on RE(tw) in each region for the same last weeks of the fall 2009 A/H1N1 outbreak (Table 2). Because the weekly counts for positive influenza tests descended below 100 for some regions toward weeks 48–49, we used a biweekly average for the estimates in equation 2. Thus, for instance, the estimate for weeks 44–46 draws on I(44), I(45), and I(46).
Appendix Table 1.
Upper-Bound Estimates of RE(tw) for the Last Weeks of the Fall 2009 A/H1N1 Outbreak in the United States
| Weeks 45–46 | Weeks 46–47 | Weeks 47–48 | Weeks 48–49 | |
| RE(tw) bound | 0.810 | 0.857 | 0.814 | 0.851 |
Abbreviation: RE(tw), effective reproductive number for weeks tw.
References
- 1.Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A. 2009;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaman J, Pitzer VE, Viboud C, et al. Absolute humidity and the seasonal onset of influenza in the continental United States [electronic article] PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000316. 8(2):e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crum-Cianflone NF, Blair PJ, Faix D, et al. Clinical and epidemiologic characteristics of an outbreak of novel H1N1 (swine origin) influenza A virus among United States military beneficiaries. Clin Infect Dis. 2009;49(12):1801–1810. doi: 10.1086/648508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dill CE, Favata MA. Novel influenza A (H1N1) outbreak on board a US Navy vessel. Disaster Med Public Health Prep. 2009;3(suppl 2):S117–S120. doi: 10.1097/DMP.0b013e3181bf249b. [DOI] [PubMed] [Google Scholar]
- 5.Paterson B, Durrheim DN, Tuyl F. Influenza: H1N1 goes to school. Science. 2009;325(5944):1071–1072. doi: 10.1126/science.325_1071b. author reply 1072–1073. [DOI] [PubMed] [Google Scholar]
- 6.Witkop CT, Duff MR, Macias EA, et al. Novel influenza A (H1N1) outbreak at the U.S. Air Force Academy: epidemiology and viral shedding duration. Am J Prev Med. 2010;38(2):121–126. doi: 10.1016/j.amepre.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Chao DL, Halloran ME, Longini IM., Jr School opening dates predict pandemic influenza A (H1N1) outbreaks in the United States. J Infect Dis. 2010;202(6):877–880. doi: 10.1086/655810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flu activity & surveillance. Atlanta, GA: Centers for Disease Control and Prevention; 2010. ( http://www.cdc.gov/flu/weekly/fluactivity.htm). (Accessed April 2010) [Google Scholar]
- 9.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 10.Miller E, Hoschler K, Hardelid P, et al. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet. 2010;375(9720):1100–1108. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 11.Update: influenza activity—United States, April–August 2009. MMWR Morb Mortal Wkly Rep. 2009;58(36):1009–1012. [PubMed] [Google Scholar]
- 12.Scriven J, McEwen R, Mistry S, et al. Swine flu: a Birmingham experience. Clin Med. 2009;9(6):534–538. doi: 10.7861/clinmedicine.9-6-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions [electronic article] PLoS Med. 2006;3(4):e89. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelman BS, Viboud C, Koelle K, et al. Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: viral coexistence and latitudinal gradients [electronic article] PLoS One. 2007;2(12):e1296. doi: 10.1371/journal.pone.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong CM, Yang L, Chan KP, et al. Influenza-associated hospitalization in a subtropical city [electronic article] PLoS Med. 2006;3(4):e121. doi: 10.1371/journal.pmed.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee VJ, Yap J, Ong JB, et al. Influenza excess mortality from 1950–2000 in tropical Singapore [electronic article] PLoS One. 2009;4(12):e8096. doi: 10.1371/journal.pone.0008096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmerman JM, Chittaganpitch M, Levy J, et al. Incidence, seasonality and mortality associated with influenza pneumonia in Thailand: 2005–2008 [electronic article] PLoS One. 2009;4(11):e7776. doi: 10.1371/journal.pone.0007776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins SD, Frost WH, Gover M, et al. Mortality from influenza and pneumonia in 50 large cities of the United States 1910–1929. Public Health Rep. 1930;45(39):2277–2328. [Google Scholar]
- 19.Housworth J, Langmuir AD. Excess mortality from epidemic influenza, 1957–1966. Am J Epidemiol. 1974;100(1):40–48. doi: 10.1093/oxfordjournals.aje.a112007. [DOI] [PubMed] [Google Scholar]
- 20.Bootsma MC, Ferguson NM. The effect of public health measures on the 1918 influenza pandemic in U.S. cities. Proc Natl Acad Sci U S A. 2007;104(18):7588–7593. doi: 10.1073/pnas.0611071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatchett RJ, Mecher CE, Lipsitch M. Public health interventions and epidemic intensity during the 1918 influenza pandemic. Proc Natl Acad Sci U S A. 2007;104(18):7582–7587. doi: 10.1073/pnas.0610941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markel H, Lipman HB, Navarro JA, et al. Nonpharmaceutical interventions implemented by US cities during the 1918–1919 influenza pandemic. JAMA. 2007;298(6):644–654. doi: 10.1001/jama.298.6.644. [DOI] [PubMed] [Google Scholar]
- 23.Kalnay E, Kanamitsu M, Kistler R, et al. NCEP/NCAR 40-year reanalysis project. Bull Am Meteorol Soc. 1996;77(3):437–471. [Google Scholar]
- 24.All influenza hospitalizations and deaths in Georgia. Atlanta, GA: Georgia Department of Community Health; 2010. ( http://dch.georgia.gov/00/channel_title/0, 2094, 31446711_148304655,00.html). (Accessed April 2010) [Google Scholar]
- 25.Hampson AW. Epidemiological data on influenza in Asian countries. Vaccine. 1999;17(suppl 1):S19–S23. doi: 10.1016/s0264-410x(99)00100-0. [DOI] [PubMed] [Google Scholar]
- 26.Beckett CG, Kosasih H, Ma'roef C, et al. Influenza surveillance in Indonesia: 1999–2003. Clin Infect Dis. 2004;39(4):443–449. doi: 10.1086/422314. [DOI] [PubMed] [Google Scholar]
- 27.Shechmeister IL. Studies on the experimental epidemiology of respiratory infections. III. Certain aspects of the behavior of type A influenza virus as an air-borne cloud. J Infect Dis. 1950;87(2):128–132. doi: 10.1093/infdis/87.2.128. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer FL, Soergel ME, Straube DC. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch Virol. 1976;51(4):263–273. doi: 10.1007/BF01317930. [DOI] [PubMed] [Google Scholar]
- 29.Minhaz Ud-Dean SM. Structural explanation for the effect of humidity on persistence of airborne virus: seasonality of influenza. J Theor Biol. 2010;264(3):822–829. doi: 10.1016/j.jtbi.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Harper GJ. Airborne micro-organisms: survival tests with four viruses. J Hyg (Lond) 1961;59(4):479–486. doi: 10.1017/s0022172400039176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowen AC, Mubareka S, Steel J, et al. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;3(10):1470–1476. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowen AC, Steel J, Mubareka S, et al. High temperature (30 degrees C) blocks aerosol but not contact transmission of influenza virus. J Virol. 2008;82(11):5650–5652. doi: 10.1128/JVI.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274(1609):599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cauchemez S, Donnelly CA, Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med. 2009;361(27):2619–2627. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowling BJ, Fang VJ, Riley S, et al. Estimation of the serial interval of influenza. Epidemiology. 2009;20(3):344–347. doi: 10.1097/EDE.0b013e31819d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghani AC, Baguelin M, Griffin J, et al. The early transmission dynamics of H1N1pdm influenza in the United Kingdom [electronic article] PLoS Curr Influenza. 2009;16 doi: 10.1371/currents.RRN1130. RRN1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White LF, Wallinga J, Finelli L, et al. Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza Other Respi Viruses. 2009;3(6):267–276. doi: 10.1111/j.1750-2659.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Sugimoto JD, Halloran ME, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326(5953):729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.