Abstract
The crucial role of recombination in HIV-1 biology is being increasingly recognized. In vitro studies have shown that up to 30 strand-transfer events may occur per viral replication cycle. Thus, recombination may surpass mutation as a major mechanism driving HIV-1 evolution. Currently, recombinant strains comprise 37% of the full-genome HIV-1 sequence database, including sequences representing 47 Circulating Recombinant Forms (CRFs) and more than 250 different Unique Recombinant Forms (URFs). Mapping of recombination breakpoints helps establish relationships among strains that are related by descent, such as CRF07_BC and CRF08_BC in China, and sheds light on their origin and epidemic spread. Additionally, unrelated recombinants sharing common breakpoints may reflect recombination hotspots within the viral genome. Here we present a software tool, RecDraw, for the graphical representation and efficient comparison of recombinant HIV-1 structures and breakpoints. RecDraw is a platform-flexible, Java stand-alone application available through http://www.hivresearch.org/research.php?ServiceID = 5&SubServiceID = 6.
Human immunodeficiency virus type 1 (HIV-1) is characterized by an extensive degree of genetic diversity, which constitutes a major obstacle for the design of effective therapeutic antiretroviral (ARV) drugs and preventive vaccines.1 The source of HIV-1 genetic variation lies in a crucial step of the viral replication cycle: the reverse transcription. During this step, the viral reverse transcriptase (RT) introduces substitutions in the order of one mutation per genome per cycle.2 As reverse transcription proceeds, the RT may alternate in its use of RNA template between the two copies of viral genome that were co-packaged in the viral particle in the previous cycle. For HIV-1, the advantage of recombination is two-fold. By combining pre-existing beneficial polymorphisms in different loci, recombination accelerates evolution under selective pressure. Additionally, genomic segments encoding deleterious mutations can be rapidly repaired by substitution with the cognate unaffected segment encoded in the sister genome.3 Consequently, the capacity of HIV-1 to generate recombinant genomes allows the virus to exploit the advantages of high mutation rate while decreasing the catastrophic consequences of the generation of massive amounts of defective genomes.
In vivo, 75%–80% of HIV-1-infected cells appear to harbor two or more proviruses,4 and this can lead to the generation of both homozygous and heterozygous virions.5 Recombination will occur in both cases, but it is virtually impossible to detect in homozygotes, and is technically easier to ascertain when the genomic fragments involved are rich in genetic information (i.e., when the parental strains are genetically diverse, and when the distance between recombination breakpoints is long). In defined geographic settings, where different HIV-1 subtypes co-circulate, individuals can become multiply-infected with strains from different clades. In these patients, the presence of a constellation of different recombinants has been documented, as recombinants can further recombine with parental strains or other recombinants.6 If recombination hotspots occur within the HIV-1 genome, recombinants with similar structures may be convergently generated in different individuals (i.e., identical by state).7 HIV-1 transmission to a new host is accompanied by a substantial population bottleneck; in 80% of the cases, the presence of a single founder virus can be inferred.8 If this virus happened to be a recombinant strain generated in a multiply-infected patient, it would share common breakpoints with strains from the index case. In the absence of further multiple infections, this recombinant strain can be subsequently disseminated to other hosts as a stable genetic form, named circulating recombinant form (CRF). Recombinants that have been recovered from fewer than three individuals are termed unique recombinant forms (URFs). If the carrier of a CRF or URF gets multiply-infected with other HIV-1 strains, this will lead to the generation of new recombinants that may share common recombination breakpoints with the parental recombinant strains. For instance, through the mapping of recombination breakpoints, McCutchan et al. determined that CRF07_BC and CRF08_BC, sampled in different injecting-drug user networks in China, shared a common ancestor, which is supported by other epidemiological evidence.9 Consequently, the irreversibility of HIV recombination followed by transmission allows for the use of recombination breakpoints as stable markers of relatedness by descent and can be exploited in molecular epidemiological studies.
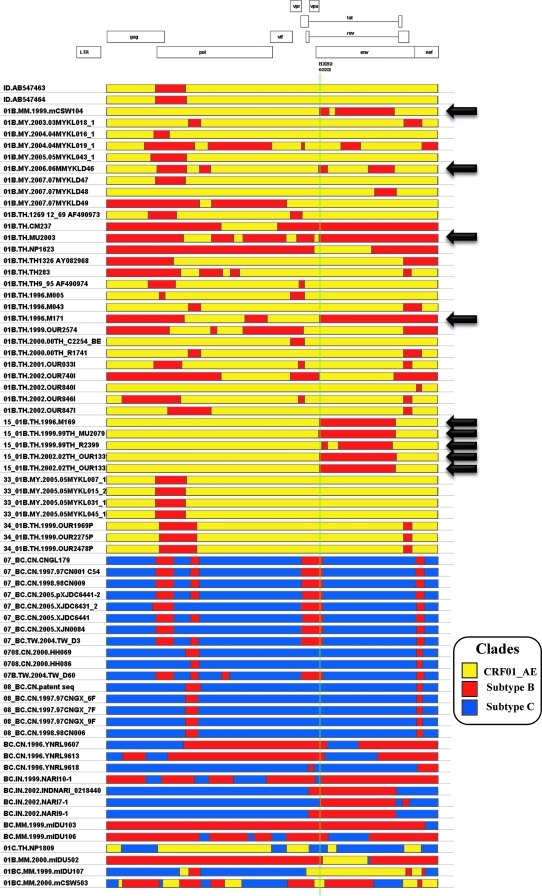
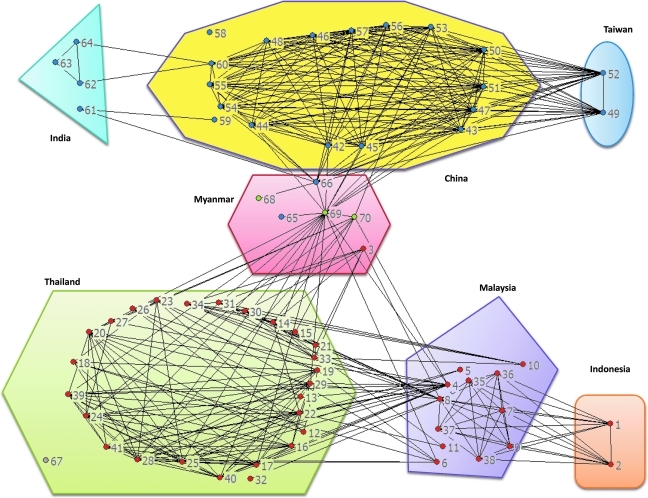
The availability of several bioinformatic tools for the detection of recombination and breakpoint mapping, each based on different phylogenetic and statistical models, has been instrumental for the advancement of the field.10–13 The Recombinant HIV-1 Drawing Tool allows for the graphical representation of HIV-1 recombinant structures, but only one strain at a time (http://www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html; last accessed on May 3, 2010). A major obstacle for analyzing commonalities among these strains is that no tool has been developed to date for the management and visualization of multiple recombinant structures. To support the study of HIV-1 recombination, we have developed a software package, RecDraw. The input file consists of a user-generated list of genetic structures of recombinant strains that includes the number of segments, their subtype, and their boundaries. To allow for comparison among different sequences, the coordinates of the recombination breakpoints need to be referred to the HXB2 strain. Through an intuitive interface, the user can select the order in which the strains are depicted, and the colors to represent the different subtypes. Positions of interest can be highlighted with a fine vertical line. Figure 1 illustrates the use of RecDraw to represent recombinants among CRF01_AE, and subtypes B and C sampled in Asia. The HXB2 location 6332 is highlighted, showing a shared breakpoint among 01B.MM.1999.mCSW104, 01B.MY.2006.06MMYKLD46, 01B.TH.1996.M171, 01B.TH.-.MU2003, and CRF15_01B strains.The obtained diagrams can be saved as PNG files. An additional feature is the generation of a table of genomic segments (i.e., boundaries and subtypes), which can be queried for positions of interest. As the mapping of recombination breakpoints is usually accompanied with a level of imprecision,9,11 the query can accommodate user-defined tolerance for what is to be considered the same breakpoint. An included module performs the pair-wise comparison between strains, recording the number of shared breakpoints (i.e., the same location ± imprecision, and the same directionality of subtype transition). The resulting matrix of shared breakpoints can be imported into commonly used programs for network analysis (e.g., UCINET, Pajek,14 etc). Figure 2 shows that 68/70 recombinants sampled in Asia appear to be linked in the same network. A high degree of interconnectivity is evidenced among recombinants from Thailand, even as these strains were sampled from individuals with different risk factors for HIV-1 and over a period of time that exceeds 10 years.15–18 Strains sampled in Malaysia bridge connections between 01B recombinants from Thailand and Indonesia.19 Recombinants from Burma/Myanmar play a central role in this extended network, as complex recombinants combining CRF01_AE, and subtypes B and C serve as bridges between 01B recombinants from Southeast Asia and BC recombinants from China. Overall, this analysis further supports previous observations on geographical hotspots in western parts of Yunnan Province in China and Central Myanmar,20 where routes of drugs and human trafficking converge (www.unodc.org; last accessed on May 3, 2010).
FIG. 1.
Genomic structure of 70 HIV-1 inter-subtype recombinants sampled in Southeast Asia, China, and India, represented using RecDraw. Full-genome sequences were retrieved from the Los Alamos HIV database and recombination breakpoints were mapped using jpHMM.11 The vertical line highlights HXB2 position 6332, and strains sharing a common CRF01_AE ->subtype B recombination breakpoint are indicated by black arrows. See text for details.
FIG. 2.
Extended network of inter-subtype recombinant HIV strains in Southeast Asia, China, and India. 01B, BC, 01C, and 01BC inter-subtype recombinants are represented as red, blue, gray, and green nodes, respectively. Lines connecting the nodes link strains that share common recombination breakpoints, allowing for 27 bp of mapping imprecision (i.e., the average distance between the jpHMM-predicted and previously published breakpoints, as determined by Schultz et al.11). The network diagram was built using UCINET version 6.275/ NetDraw version 2.091 (Analytic Technologies, Lexington, KY). Strains are grouped by country of sampling. Depicted strains include: 1: ID.AB547463, 2: ID.AB547464, 3: 01B.MM.1999.mCSW104, 4: 01B.MY.2003.03MYKL018_1, 5: 01B.MY.2004.04MYKL016_1, 6: 01B.MY.2004.04MYKL019_1, 7: 01B.MY.2005.05MYKL043_1, 8: 01B.MY.2006.06MMYKLD46, 9: 01B.MY.2007.07MYKLD47, 10: 01B.MY.2007.07MYKLD48, 11: 01B.MY.2007.07MYKLD49, 12: 01B.TH.1269 12_69 AF490973, 13: 01B.TH.CM237, 14: 01B.TH.MU2003, 15: 01B.TH.NP1623, 16: 01B.TH.TH1326 AY082968, 17: 01B.TH.TH283, 18: 01B.TH.TH9_95 AF490974, 19: 01B.TH.1996.M005, 20: 01B.TH.1996.M043, 21: 01B.TH.1996.M171, 22: 01B.TH.1999.OUR2574, 23: 01B.TH.2000.00TH_C2254_BE, 24: 01B.TH.2000.00TH_R1741, 25: 01B.TH.2001.OUR033I, 26: 01B.TH.2002.OUR740I, 27: 01B.TH.2002.OUR840I, 28: 01B.TH.2002.OUR846I, 29: 01B.TH.2002.OUR847I, 30: 15_01B.TH.1996.M169, 31: 15_01B.TH.1999.99TH_MU2079, 32: 15_01B.TH.1999.99TH_R2399, 33: 15_01B.TH.2002.02TH_OUR1331, 34: 15_01B.TH.2002.02TH_OUR1332, 35: 33_01B.MY.2005.05MYKL007_1, 36: 33_01B.MY.2005.05MYKL015_2, 37: 33_01B.MY.2005.05MYKL031_1, 38: 33_01B.MY.2005.05MYKL045_1, 39: 34_01B.TH.1999.OUR1969P, 40: 34_01B.TH.1999.OUR2275P, 41: 34_01B.TH.1999.OUR2478P, 42: 07_BC.CN.CNGL179, 43: 07_BC.CN.1997.97CN001 C54, 44: 07_BC.CN.1998.98CN009, 45: 07_BC.CN.2005.pXJDC6441-2, 46: 07_BC.CN.2005.XJDC6431_2, 47: 07_BC.CN.2005.XJDC6441, 48: 07_BC.CN.2005.XJN0084, 49: 07_BC.TW.2004.TW_D3, 50: 0708.CN.2000.HH069, 51: 0708.CN.2000.HH086, 52: 07B.TW.2004.TW_D60, 53: 08_BC.CN.patent seq, 54: 08_BC.CN.1997.97CNGX_6F, 55: 08_BC.CN.1997.97CNGX_7F, 56: 08_BC.CN.1997.97CNGX_9F, 57: 08_BC.CN.1998.98CN006, 58: BC.CN.1996.YNRL9607, 59: BC.CN.1996.YNRL9613, 60: BC.CN.1996.YNRL9618, 61: BC.IN.1999.NARI10-1, 62: BC.IN.2002.INDNARI_0218440, 63: BC.IN.2002.NARI7-1, 64: BC.IN.2002.NARI9-1, 65: BC.MM.1999.mIDU103, 66: BC.MM.1999.mIDU106, 67: 01C.TH.NP1809, 68: 01B.MM.2000.mIDU502, 69: 01BC.MM.1999.mIDU107, 70: 01BC.MM.2000.mCSW503. See text for details.
In summary, bioinformatic software for the study of recombination structures and breakpoints recombination markers are powerful tools for molecular epidemiology and virology, and can add another dimension to our understanding of HIV-1 evolution. RecDraw runs as a stand-alone application and it was written in JAVA, to lend it cross-platform executability. The software is distributed through http://www.hivresearch.org/research.php?ServiceID = 5&SubServiceID = 6; last accessed on May 3, 2010).
Acknowledgment
This work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD), and by the National Institute for Allergy and Infectious Diseases, National Institutes of Health (YD1 AI2642-12, NIAID/MRMC HIV/AIDS Vaccine Research and Development Program). The views and opinions expressed herein do not necessarily reflect those of the U.S. Army, the Department of Defense, or the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kijak GH. McCutchan FE. HIV diversity, molecular epidemiology, and the role of recombination. Curr Infect Dis Rep. 2005;7:480–488. doi: 10.1007/s11908-005-0051-8. [DOI] [PubMed] [Google Scholar]
- 2.Mansky LM. Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenstein J. Yokoyama S. The evolutionary advantage of recombination. II. Individual selection for recombination. Genetics. 1976;83:845–859. doi: 10.1093/genetics/83.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung A. Maier R. Vartanian JP, et al. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 5.Hu WS. Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 6.McCutchan FE. Hoelscher M. Tovanabutra S, et al. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: Evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol. 2005;79:11693–11704. doi: 10.1128/JVI.79.18.11693-11704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan J. Negroni M. Robertson DL. The distribution of HIV-1 recombination breakpoints. Infect Genet Evol. 2007;7:717–723. doi: 10.1016/j.meegid.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Keele BF. Giorgi EE. Salazar–Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCutchan FE. Carr JK. Murphy D, et al. Precise mapping of recombination breakpoints suggests a common parent of two BC recombinant HIV type 1 strains circulating in China. AIDS Res Hum Retroviruses. 2002;18:1135–1140. doi: 10.1089/088922202320567879. [DOI] [PubMed] [Google Scholar]
- 10.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz AK. Zhang M. Bulla I, et al. jpHMM: Improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37(Web Server issue):W647–651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salminen MO. Carr JK. Burke DS. McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 13.de Oliveira T. Deforche K. Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 14.deNooy W. Mrvar A. Batagelj V. Exploratory Social Network Analysis with Pajek. Cambridge; United Kingdom: 2005. [Google Scholar]
- 15.Tovanabutra S. Sanders EJ. Graham SM, et al. Evaluation of HIV type 1 strains in men having sex with men and in female sex workers in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2010;26:123–131. doi: 10.1089/aid.2009.0115. [DOI] [PubMed] [Google Scholar]
- 16.Tovanabutra S. Polonis V. De Souza M, et al. First CRF01_AE/B recombinant of HIV-1 is found in Thailand. AIDS. 2001;15:1063–1065. doi: 10.1097/00002030-200105250-00018. [DOI] [PubMed] [Google Scholar]
- 17.Tovanabutra S. Kijak GH. Beyrer C, et al. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses. 2007;23:829–833. doi: 10.1089/aid.2006.0300. [DOI] [PubMed] [Google Scholar]
- 18.Kijak GH. Tovanabutra S. Sanders–Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v.2. Virology. 2007;358:178–191. doi: 10.1016/j.virol.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 19.Tee KK. Li XJ. Nohtomi K. Ng KP. Kamarulzaman A. Takebe Y. Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr. 2006;43:523–529. doi: 10.1097/01.qai.0000242451.74779.a7. [DOI] [PubMed] [Google Scholar]
- 20.Takebe Y. Motomura K. Tatsumi M. Lwin HH. Zaw M. Kusagawa S. High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: Geographical hot spot of extensive recombination. AIDS. 2003;17:2077–2087. doi: 10.1097/00002030-200309260-00009. [DOI] [PubMed] [Google Scholar]