Abstract
Chloroplast sensor kinase (CSK) is a bacterial-type sensor histidine kinase found in chloroplasts—photosynthetic plastids—in eukaryotic plants and algae. Using a yeast two-hybrid screen, we demonstrate recognition and interactions between: CSK, plastid transcription kinase (PTK), and a bacterial-type RNA polymerase sigma factor-1 (SIG-1). CSK interacts with itself, with SIG-1, and with PTK. PTK also interacts directly with SIG-1. PTK has previously been shown to catalyze phosphorylation of plastid-encoded RNA polymerase (PEP), suppressing plastid transcription nonspecifically. Phospho-PTK is inactive as a PEP kinase. Here, we propose that phospho-CSK acts as a PTK kinase, releasing PTK repression of chloroplast transcription, while CSK also acts as a SIG-1 kinase, blocking transcription specifically at the gene promoter of chloroplast photosystem I. Oxidation of the photosynthetic electron carrier plastoquinone triggers phosphorylation of CSK, inducing chloroplast photosystem II while suppressing photosystem I. CSK places photosystem gene transcription under the control of photosynthetic electron transport. This redox signaling pathway has its origin in cyanobacteria, photosynthetic prokaryotes from which chloroplasts evolved. The persistence of this mechanism in cytoplasmic organelles of photosynthetic eukaryotes is in precise agreement with the CoRR hypothesis for the function of organellar genomes: the plastid genome and its primary gene products are Co-located for Redox Regulation. Genes are retained in plastids primarily in order for their expression to be subject to this rapid and robust redox regulatory transcriptional control mechanism, whereas plastid genes also encode genetic system components, such as some ribosomal proteins and RNAs, that exist in order to support this primary, redox regulatory control of photosynthesis genes. Plastid genome function permits adaptation of the photosynthetic apparatus to changing environmental conditions of light quantity and quality.
Keywords: chloroplast sensor kinase, plastid transcription kinase, sigma factor, cytoplasmic inheritance, protein phosphorylation, Co-location for Redox Regulation (CoRR)
Plastids are cytoplasmic organelles of plant and algal cells (Kirk and Tilney-Bassett 1978). They contain DNA, RNA, ribosomes, and a complete apparatus of gene expression. Chloroplasts are green, chlorophyll-containing plastids (Link 1996) that contain this cytoplasmic genetic system in close association with a complete photosynthetic apparatus that uses light energy to release oxygen from water, makes adenosine triphosphate, and assimilates atmospheric carbon dioxide. Photosynthesis in chloroplasts involves an electron transport chain containing two photosystems, I and II (Hill and Bendall 1960). Each photosystem contains a photochemical reaction center together with its associated light-harvesting pigments (Blankenship 2002). Chlorophyll and prosthetic groups of the two reaction centers are located in the chloroplast thylakoid membrane where they are bound by membrane intrinsic apoproteins. Reaction center apoproteins are products of genes in chloroplast DNA (Ohyama et al. 1986; Shinozaki et al. 1986). Photosystem I and II are connected in series for noncyclic electron transport, and so their rates of light energy conversion must be equal. Efficiency in utilization of absorbed light energy is maintained by adjustment of light-harvesting antenna size (Bonaventura and Myers 1969; Murata 1969; Allen et al. 1981) and also by adjustment of the stoichiometry of the two photochemical reaction centers (Melis et al. 1989; Chow et al. 1990; Murakami et al. 1997). These adjustments occur in response to changes in spectral composition of incident light that would otherwise favor one photosystem at the expense of the other.
Both of these types of adjustment involve a sensor of the reduction–oxidation—“redox”—state of an electron carrier, plastoquinone, linking the two photosystems, and maintenance of a null position whereby electrons enter the plastoquinone pool from photosystem II and leave for photosystem I at the same rate (Allen 1995). Any transient departure from this steady state initiates changes that restore it. In the case of photosystem stoichiometry adjustment, reduced plastoquinone induces photosystem I and/or represses photosystem II, whereas oxidized plastoquinone induces photosystem II and/or represses photosystem I (Pfannschmidt, Nilsson, and Allen 1999; Pfannschmidt, Nilsson, Tullberg, et al. 1999; Allen and Pfannschmidt 2000).
Photosystem stoichiometry refers to the quantity of photosystem I (PS I) relative to that of photosystem II (PS II) in the photosynthetic, thylakoid membrane of chloroplasts and cyanobacteria. Changes in photosystem stoichiometry occur as acclimatory responses to changes in light quality (Melis et al. 1989; Chow et al. 1990; Fujita 1997; Murakami et al. 1997) and are initiated by changes in the redox state of the interphotosystem electron carrier, plastoquinone (PQ) (Pfannschmidt, Nilsson, and Allen 1999; Pfannschmidt, Nilsson, Tullberg, et al. 1999; Allen and Pfannschmidt 2000). In mustard chloroplasts, photosystem stoichiometry adjustments involve regulation of both photosystem I and photosystem II reaction center genes (Pfannschmidt, Nilsson, and Allen 1999; Pfannschmidt, Nilsson, Tullberg, et al. 1999). However, in Arabidopsis and pea (Pisum sativum) chloroplasts, only photosystem I genes seem to be regulated at the level of transcription (Tullberg et al. 2000; Fey et al. 2005; Puthiyaveetil and Allen 2008; Shimizu et al. 2010), consistent with the model proposed for cyanobacteria by Fujita (1997) (Murakami et al. 1997).
Chloroplast sensor kinase (CSK) is a sensor histidine kinase that communicates the redox state of plastoquinone to the chloroplast transcriptional apparatus, initiating the appropriate change in photosystem stoichiometry (Puthiyaveetil et al. 2008). The precise upstream mechanism of redox sensing by CSK is under investigation but not yet known. Here, we report on downstream events involving interactions between CSK and candidates for its cognate transcriptional regulator.
Plastid transcription kinase (PTK) is one candidate. PTK is a protein kinase that catalyzes phosphorylation of one or more proteins acting as regulatory cofactors of the chloroplast RNA polymerase (Baginsky et al. 1999; Ogrzewalla et al. 2002). Another candidate for the interaction partner of CSK is chloroplast sigma factor-1 (SIG-1) (Shimizu et al. 2010). SIG-1 has been shown, like CSK, to be required for repression of the psaA gene, which encodes a reaction center apoprotein of photosystem I (Shimizu et al. 2010). Like CSK, chloroplast sigma factors (Schweer et al. 2009, 2010) indicate that chloroplasts retain a prokaryotic type of transcriptional regulatory system (Tiller et al. 1991) acting on a chloroplast-encoded bacterial-type RNA polymerase (Suzuki et al. 2004). A third proposal for a chloroplast redox response regulator is a 34 kDa protein abbreviated as TCP34 (tetratricopeptide-containing chloroplast protein of 34 kDa) (Weber et al. 2006). TCP34 is a DNA-binding protein containing a tetratricopeptide motif and exhibiting sequence homology with bacterial response regulators (Weber et al. 2006).
Conservation of CSK throughout the Evolutionary Transition from Endosymbiont to Subcellular Organelle
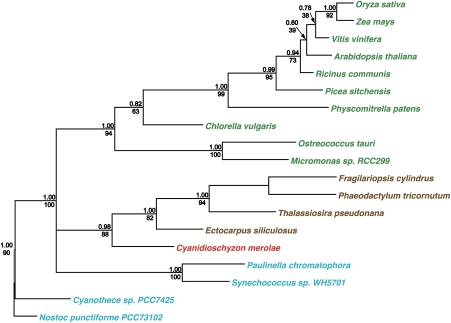
Figure 1 shows a phylogenetic tree of CSK. Phylogenetic analysis of CSK reveals CSK orthologues in all major plant and algal lineages. Cyanobacterial lineages also contain a recognizable CSK homolog, confirming the evolutionary origin of this chloroplast protein from cyanobacteria. CSK orthologues in diatoms and phaeophytes cluster with the red algal CSK (fig. 1), consistent with the secondary symbiotic origin of diatom and phaeophycean plastids from red algae. Interestingly, a CSK ortholog is also found in “chromatophores”—in this context, cyanobacterial endosymbionts—of the amoeboid eukaryote Paulinella chromatophora. The symbiotic origin of Paulinella chromatophores has an independent evolutionary history from the symbiotic event that gave rise to chloroplasts (Nowack et al. 2008).
FIG. 1.—
Phylogeny of CSK. CSK is present in all major plant and algal lineages and evolved from a cyanobacterial sensor histidine kinase. Bayesian posterior probabilities are shown above nodes, PHYML 3.0 bootstrap values are shown below nodes. The name of each taxon is colored according to the major photosynthetic pigment characteristic of that group.
The phylogenetic analysis presented in figure 1 and table 1 also reveal an interesting feature of molecular evolution in CSK. CSK occurs as a canonical sensor histidine kinase in cyanobacteria, red algae, diatoms, and phaeophytes (table 1), whereas in green algae and plants, CSK is a modified histidine kinase as the conserved histidine autophosphorylation site in CSK has been lost in these lineages. Studies in cyanobacteria suggest that the NarL-type response regulator ycf29 is the cognate partner of CSK (Sato et al. 2007). CSK seems to retain its cognate response regulator partner in cyanobacteria and in nongreen algae (table 1) but not in green algae and plants—lineages in which CSK occurs as a modified histidine kinase (table 1). The evolutionary loss of a bacterial-type response regulator from chloroplasts may therefore correlate with a modified kinase activity of CSK (Puthiyaveetil and Allen 2009). CSK nevertheless regulates chloroplast transcription in plants (Puthiyaveetil et al. 2008). Nonresponse regulator partners of CSK were thus specifically sought in our study.
Table 1.
Distribution of CSK, ycf29, and PTK in Photosynthetic Organisms
| Taxonomic Group/Organism | CSK | Ycf29 | PTK | |||
| Cyanobacteria | ||||||
| Nostoc punctiforme PCC 73102 | + | + | − | |||
| Cyanothecesp. PCC 7425 | + | + | − | |||
| Synechococcussp. WH 5701 | + | + | − | |||
| Paulinella chromatophora | + | + | nk | |||
| N | P | N | P | N | P | |
| Rhodophytes | ||||||
| Cyanidioschyzon merolae | + | − | − | + | − | − |
| Phaeophytes | ||||||
| Ectocarpus siliculosus | + | − | + | − | − | − |
| Bacillariophytes | ||||||
| Fragilariopsis cylindrus | + | − | + | − | − | − |
| Phaeodactylum tricornutum | + | − | + | − | − | − |
| Thalassiosira pseudonana | + | − | + | − | − | − |
| Viridiplantae | ||||||
| Ostreococcus tauri | + | − | − | − | + | − |
| Ostreococcus lucimarinus | + | − | − | − | + | − |
| Micromonas pusilla | + | − | − | − | + | − |
| Chlorellasp. NC64A | + | − | − | − | + | − |
| Chlamydomonas reinhardtii | − | − | − | − | + | − |
| Physcomitrella patens | + | − | − | − | + | − |
| Picea sitchensis | + | − | nk | − | + | − |
| Oryza sativa | + | − | − | − | + | − |
| Zea mays | + | − | − | − | + | − |
| Lotus japonicus | + | − | − | − | + | − |
| Vitis vinifera | + | − | − | − | + | − |
| Populus trichocarpa | + | − | − | − | + | − |
| Arabidopsis thaliana | + | − | − | − | + | − |
NOTE.—The plus (+) indicates the presence and the minus (−) indicates the absence of CSK or ycf29 or PTK. The abbreviation “nk” indicates that the complete genome sequence for that taxon is not available, so the presence or absence of CSK or ycf29 or PTK is not known. The subcategories “N” and “P” in the CSK, ycf29, and PTK occurrence column stand for “nuclear” and “plastid,” respectively, and indicate the genetic compartment in which these proteins are encoded. For P. chromatophora, both CSK and ycf29 genes are found in the chromatophore genome. The accession numbers of C. punctiforme PCC 73102 CSK and ycf29 homologs are ACC82407 and ACC80206, respectively; for C .sp. PCC 7425, ACK71103 and ACK71358; for S.sp. WH 5701, EAQ75489, and EAQ76691. The taxonomic group “Viridiplantae” means “Green Plants,” and includes green algae, lower, and higher plants.
Interactions of CSK
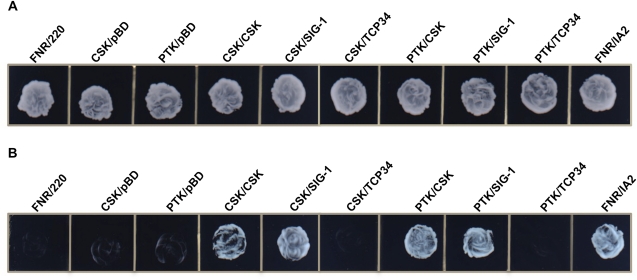
In order to investigate protein–protein interactions between CSK and its putative functional partners, we undertook a yeast two-hybrid analysis. Figure 2 shows the results of the yeast two-hybrid screen. Figure 2A shows growth of all bait–prey combinations in a medium lacking leucine and tryptophan, confirming the successful transformation of yeast cells with both bait and prey plasmids. Figure 2B shows growth of yeast cells in a medium lacking leucine, tryptophan, and histidine. Growth on this latter medium reports on interactions of bait and prey proteins, which together activate the HIS reporter gene, enabling yeast cells to grow in a medium lacking histidine. Figure 2B shows that functional interactions occurred between the following pairs of bait and prey proteins: CSK with CSK; CSK with SIG-1; PTK with CSK; PTK with SIG-1; ferredoxin-NADP+ reductase (FNR) with IA2. FNR and IA2 are two chloroplast proteins that are known to interact (Kuchler et al. 2002) and are therefore used as a positive control as in Juric et al. (2009). The bait–prey combination FNR/220 is used as a negative control as these chloroplast proteins do not interact (Juric et al. 2009) and are thus unable to permit growth in a medium lacking histidine (fig. 2B). The combinations CSK/pBD and PTK/pBD are additional negative controls designed to reveal self-activation of the bait proteins—CSK and PTK. The sparse growth of these negative controls (fig. 2B) rules out self-activation of the bait proteins.
FIG. 2.—
Yeast two-hybrid analysis based on the activation of the reporter gene HIS. Different combinations of bait and prey proteins are indicated above the growth plates. FNR/220, CSK/pBD, and PTK/pBD are negative controls. Test combination of bait and prey proteins are CSK/CSK, CSK/SIG-1, CSK/TCP34, PTK/CSK, PTK/SIG-1, and PTK/TCP34. The combination FNR/IA2 is the positive control. (A) Yeast cells growing on synthetic SD media plates lacking leucine and tryptophan. The growth of all bait and prey combinations in this medium confirms successful transformation of yeast cells with both bait and prey plasmids. (B) Growth of yeast cells in SD media plates without leucine, tryptophan, and histidine. The growth in –His plates require the activation of the histidine biosynthetic gene (His reporter gene), which in turn requires the functional interaction between bait and prey proteins.
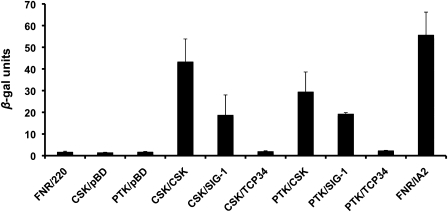
Figure 3 shows validation of the protein–protein interactions inferred from the results in figure 2 by use of a second reporter gene, β-galactosidase. This gene is under the control of a separate promoter from that of the HIS reporter gene, and its activation is therefore an independent measure of interaction. The results from the β-galactosidase assay shown in figure 3 are also indicative of the relative strength of the different protein–protein interactions. Among the test combinations, CSK/CSK shows the highest degree of interaction; followed by CSK/PTK, with CSK/SIG-1 and PTK/SIG-1 showing weaker and more or less equal interactions (fig. 3). The results in figures 2 and 3 suggest clearly that TCP34 does not interact with CSK or PTK. Our results therefore do not support the response regulator function of TCP34 in plant chloroplasts.
FIG. 3.—
Yeast two-hybrid analysis as a function of the β-galactosidase assay. Activation of the β-galactosidase reporter gene (lacZ gene), which is driven by a separate promoter from the HIS reporter gene, requires functional interaction between bait and prey proteins. β-galactosidase activity is also a measure of the strength of interaction, with higher activity suggesting stronger interaction. One β-gal unit hydrolyses 1 μmol of o-nitrophenyl-β-D-galactopyranoside (ONPG) to o-nitrophenol and galactose per minute at pH 7.5 and 37 °C. Error bars indicate the standard error of the mean of 3 individual measurements, with each measurement representing a different sample.
CSK Acts on Transcriptional Regulators in Control of Photosynthesis Gene Expression
Arabidopsis knockout mutants of the CSK gene are unable to repress photosystem I genes in photosystem I light and therefore cannot regulate the relative quantities of photosystem I and photosystem II (Puthiyaveetil et al. 2008). Sigma factors assist RNA polymerase in promoter recognition and DNA-melting, two processes that determine faithful and efficient transcription (Wosten 1998). Chloroplast sigma factors are cyanobacterial in origin (Tiller et al. 1991), as is CSK (Puthiyaveetil et al. 2008). As many as six sigma factors are found in Arabidopsis chloroplasts (Lysenko 2007). SIG-1 has been shown to be phosphorylated under PQ oxidizing conditions, when incident light favors photosystem I (light 1) (Shimizu et al. 2010). Phosphorylated SIG-1 represses transcription at psa (photosystem I reaction center) promoters while efficiently transcribing psb (photosystem II) promoters (Shimizu et al. 2010). SIG-1 phosphorylation is part of the control of gene expression involved in photosystem stoichiometry adjustments, but the identity of the SIG-1 kinase is yet to be revealed (Shimizu et al. 2010).
Here, we propose that CSK is the SIG-1 kinase based on the following observations. First, CSK function and SIG-1 phosphorylation have the same gene expression phenotype, which is suppression of psa genes (Puthiyaveetil et al. 2008; Shimizu et al. 2010). This identical gene-regulatory property of these two proteins suggests they are part of the same signal transduction pathway. Secondly, CSK and SIG-1 interact in vivo in yeast (figs. 2 and 3). Thirdly, oxidized PQ, the signal for sigma factor phosphorylation, is also the signal that promotes the kinase activity of CSK (Ibrahim IM, Puthiyaveetil S, Allen JF, unpublished data). The high degree of interaction between CSK monomers, as noted in our yeast two-hybrid assay (figs. 2 and 3), suggests that CSK exists as a homodimer in its functional form. This property of CSK is consistent with the proposed signal sensing function, as dimerization is well known in bacterial sensor kinases (Wolanin et al. 2002).
PTK is a eukaryotic serine/threonine protein kinase of the casein kinase II family (Baginsky et al. 1999). PTK is usually found associated with the plastid-encoded RNA polymerase (PEP), acting as a global regulator of chloroplast transcription (Link 2003). In low light conditions, PTK keeps chloroplast transcription at a low level by phosphorylating PEP—phosphorylated PEP transcribes chloroplast genes less effectively than unphosphorylated PEP (Baginsky et al. 1999; Baena-Gonzalez et al. 2001). A single subunit of PEP—the 72 kDa β' subunit—is usually found in its phosphorylated form in low light (Baginsky et al. 1999; Baena-Gonzalez et al. 2001). PTK from mustard (Sinapis alba L.) can also phosphorylate SIG-1 in vitro (Ogrzewalla et al. 2002). These observations, taken together with our yeast two-hybrid results (figs. 2 and 3), suggest that PTK nonspecifically suppresses chloroplast transcription in low light by phosphorylating PEP structural (β' subunit) and regulatory (sigma factor) subunits.
Specificity in Transcriptional Regulation of Plastid Genes for Reaction Center Proteins of Chloroplast Photosystem I and Photosystem II
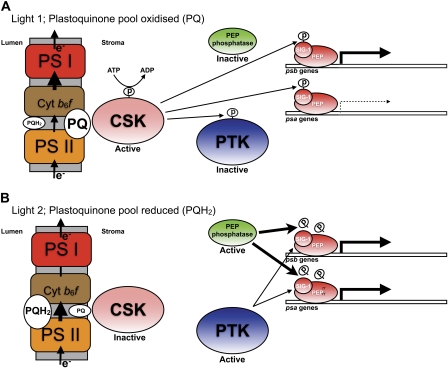
Light 1 (photosystem I light) and light 2 (photosystem II light) are selective for electron transport through photosystem I or photosystem II only when light intensity is rate limiting for photosynthesis, and other factors, such as CO2 concentration or temperature, are not. For selective transcriptional control of reaction center gene transcription in Arabidopsis thaliana, these “low light” conditions correspond to photon flux densities of the order of 12 μE m−2 s−1 (Puthiyaveetil and Allen 2008). Because PTK-mediated suppression of chloroplast transcription occurs at low incident light intensity (Baena-Gonzalez et al. 2001), it is interesting to ask how might specific reaction center gene transcription be achieved in light 1 and 2? We suggest that, under these conditions, the activity of PTK is overridden by two regulatory proteins—CSK and an as yet unidentified PEP phosphatase (fig. 4A and B).
FIG. 4.—
The proposed chloroplast signal transduction pathway coupling the redox state of the photosynthetic electron carrier plastoquinone in the chloroplast thylakoid membrane with initiation of transcription of chloroplast DNA at the promoter regions of the genes psa (encoding reaction center proteins of photosystem I) and psb (encoding reaction center proteins of photosystem II). (A) Under light 1, a rate-limiting low light selective for photosystem I, electron flow through photosystem I (PS I) has a greater potential than that through photosystem II (PS II), and so PQ pool is maintained in its oxidized form. CSK is autophosphorylated and active as a protein kinase using both SIG-1 and PTK as substrates: SIG-1 and PTK are thus maintained in their phosphorylated forms. Phospho-SIG-1 represses transcription at the psa promoter while allowing transcription of psb genes. Phospho-PTK is inactive; therefore, it cannot suppress chloroplast transcription nonspecifically, and under this circumstance, only CSK-mediated—via phospho-SIG-1—specific repression of psa genes occurs. This differential reaction center gene transcription increases the stoichiometry of photosystem II relative to photosystem I. (B) Under light 2, a rate-limiting low light selective for photosystem II, electron flow through photosystem I (PS I) has a lower potential than that through photosystem II (PS II), and so PQ pool is maintained in its reduced form (PQH2). CSK is inactive as a protein kinase. The repression of psa genes, occurred during light 1, is now relieved by the action of a PEP phosphatase that catalyzes dephosphorylation of phospho-SIG-1. As a result, PS I transcription increases. Under this light condition, PTK is active as a protein kinase acting on subunits of PEP. However, the action of PEP phosphatase overrides PTK activity by dephosphorylating both SIG-1 as well as PEP subunits so that nonspecific repression of reaction center genes is counteracted. The increase in photosystem I transcription in light 2, therefore, leads to an increase in photosystem I units relative to photosystem II.
We thus propose that in light 1, in order to bring about specific down-regulation of psa genes, PTK becomes inhibited (fig. 4A). Phosphorylation inhibits PTK (Link 2003), and yet no PTK kinase has previously been identified. In the light of the strong interaction between CSK and PTK found in our yeast two-hybrid assay (figs. 2 and 3), we here propose that CSK acts as the PTK kinase, catalyzing phosphorylation and inactivation of PTK. CSK itself then catalyzes phosphorylation of SIG-1, which has the effect of repressing only psa transcription. As discussed earlier, psb transcription is unaffected by SIG-1 phosphorylation, thus the quantity of photosystem II increases in light 1 relative to photosystem I (fig. 4A) (Shimizu et al. 2010). In light 2, however, when photosystem I is rate limiting, sigma factors are predicted to become dephosphorylated to relieve the repression on psa genes (fig. 4B). The nonspecific suppression of chloroplast genes by PTK must also be countered by some means. The phospho-PEP phosphatase can perform both of these functions by dephosphorylating SIG-1 as well as the PEP structural subunits (fig. 4B). Whether such a phospho-PEP phosphatase is constitutively active or regulated by redox signals remains to be seen.
We suggest that PTK, a eukaryotic-type protein kinase, displaced an analogous prokaryotic-type response regulator as the functional partner of CSK in a two-component regulatory system. This proposal is supported by the distribution of CSK, Ycf29, and PTK between different eukaryotic lineages (table 1). PTK seems to be present in lineages that have lost ycf29, thus supporting the notion of PTK replacing ycf29 as the functional partner of CSK.
We conclude that regulation of photosystem stoichiometry in chloroplasts (Allen 1995, 2005; Pfannschmidt, Nilsson, and Allen 1999) results from effects of the redox state of the plastoquinone pool on reversible phosphorylation of CSK, PTK, and SIG-1. These proteins interact with each other and together comprise the redox signal transduction pathway that acts to regulate transcription of chloroplast reaction center genes in response to imbalance in rates of energy conversion in photosystems I and II (Pfannschmidt, Nilsson, and Allen 1999; Pfannschmidt, Nilsson, Tullberg, et al. 1999; Allen and Pfannschmidt 2000; Puthiyaveetil and Allen 2008). This genetic switch has been retained from the cyanobacterial ancestor of chloroplasts (Puthiyaveetil and Allen 2009) and is a transcriptional analog of posttranslational modification of the activity of photosystem I and II by phosphorylation of chloroplast light-harvesting complex II (Allen et al. 1981; Allen 1992). Redox control of transcription (Allen 1993c) has been proposed as a necessary condition for the retention, in evolution, of the genetic systems of chloroplasts and mitochondria as extranuclear, cytoplasmic elements that produce non-Mendelian inheritance of characters associated with photosynthesis and respiration in eukaryotic cells (Allen 1993a, 1993b; Race et al. 1999)—the “CoRR” hypothesis (Allen 2003a, 2003b; Allen et al. 2005). The results described here are in agreement with CoRR and further resolve the mechanism by which a modified bacterial two-component system continues to operate in chloroplasts in order to secure a functionally intelligible adjustment of the stoichiometry of the chloroplast photosystems in response to changing light regimes.
Materials and Methods
Phylogenetic Analysis
Multiple alignment of the amino acid sequence corresponding to the catalytic domain of CSK and its homologs was generated across a representative selection of photosynthetic eukaryotes and cyanobacteria using ClustalX and adjusted manually using Jalview (Clamp et al. 2004). CSK tree was reconstructed from 91 characters. Bayesian phylogeny was computed using Mr. Bayes 3.1 (Ronquist and Huelsenbeck 2003) from 2,000,000 generations divided between two parallel runs of 1,000,000, each with sampling every 1,000 generations. The substitution model was inferred using a mixed model of amino acid substitution, and rate across sites variation was modeled on a discrete gamma distribution approximated using 4 gamma categories and 1 category of invariable sites. Bootstraps were generated using PHYML 3.0 (Guindon and Gascuel 2003) using the Whelan and Goldman substitution model and rate across sites variation modeled on an approximate gamma distribution using 4 gamma categories and one category of invariable sites.
Yeast Two-Hybrid Analysis
The yeast two-hybrid assay employed in our study is based on the GAL4 system (Stratagene). Bait proteins were fused to the activation domain (AD) and prey proteins to the binding domain (BD). cDNAs encoding bait proteins are therefore cloned into the pAD-Gal4-2.1 vector and prey cDNAs into the pBD-GAL4 vector. A cDNA fragment encoding the kinase domain (Q301-A611) of Arabidopsis CSK (product of gene At1g67840) was amplified (for primers, see supplementary table 1, Supplementary Material online) from a full-length CSK cDNA clone used in an earlier study (Puthiyaveetil et al. 2008). This was then used as a bait protein. In order to study whether CSK forms dimers, the same cDNA region of CSK was also used as a prey protein. The other prey proteins tested as candidates of CSK’s partner were SIG-1 (product of gene At1g64860) and TCP34 (product of gene At3g26580). For SIG-1, the cDNA region encoding the mature Arabidopsis protein (A81-N502) was amplified (primers, supplementary table 1, Supplementary Material online) from the full-length Arabidopsis Biological Resource Centre (ABRC) clone U16526. For TCP34, the cDNA region encoding the mature Arabidopsis protein—without the C-terminal transmembrane anchor—(L28-G324) was amplified (primers, supplementary table 1, Supplementary Material online) from the ABRC clone U24715. The mature Arabidopsis PTK protein (A56-Q432) was also used as a bait. The cDNA region encoding the mature PTK protein (product of gene At2g23070) was amplified (primers, supplementary table 1, Supplementary Material online) from the ABRC clone U15758. The prey proteins tested against the PTK bait were CSK; SIG-1; and TCP34. The cDNA regions cloned for these prey proteins were the same as those described above. In a negative control, the complete FNR protein was used as the bait against the “220” domain of the TROL protein, essentially as described (Juric et al. 2009). In additional negative controls, CSK and PTK bait proteins were tested for self-activation by using BD—encoded by the pBD-GAL4 vector—as prey proteins. The positive control uses the complete FNR protein as bait and the “IA2” domain of Tic62 protein as prey, as described previously (Juric et al. 2009). Successful cloning of bait and prey plasmids were confirmed by DNA sequencing. All bait and prey plasmid combinations were transformed using LiAc method into the yeast strain Saccharomyces cerevisiae YF53 (Mata ura3–52 his3–200 ade2–101 lys2–801 trp1–901 leu2–3,112 gal4–542 gal80–538, with HIS3 and lacZ reporter gene constructs LYS2::UASGAL1–TATAGAL1–HIS3 and URA3::UASGAL4 17 mers (x3)—TATACYC1–lacZ). Yeast transformants were selected on synthetic dropout (SD) media without leucine and tryptophan. Protein–protein interactions were detected by growth on SD media lacking leucine, tryptophan, and histidine and by the β-galactosidase assay. β-galactosidase activity was determined from yeast cultures by the method of Adams et al. (1997). All assays were performed in duplicates from three independent colonies in 0.5 ml of reaction buffer, and β-galactosidase units were calculated as described (Feilotter et al. 1994).
Supplementary Material
Supplementary table 1 is available at Genome Biology and Evolutiononline (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a Leverhulme Trust Early Career Fellowship to S.P., a Leverhulme Trust research grant to J.F.A., and by the Croatian Ministry of Science, Education and Sports as grant 098-0982193-2838 to H.F.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in yeast genetics: a laboratory course manual. In methods in yeast genetics: a laboratory course manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Allen JF. Protein phosphorylation in regulation of rhotosynthesis. BiochimBiophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Allen JF. Control of gene-expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol. 1993a;165:609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- Allen JF. Redox control of gene expression and the function of chloroplast genomes—an hypothesis. Photosynth Res. 1993b;36:95–102. doi: 10.1007/BF00016274. [DOI] [PubMed] [Google Scholar]
- Allen JF. Redox control of transcription—sensors, response regulators, activators and repressors. FEBS Lett. 1993c;332:203–207. doi: 10.1016/0014-5793(93)80631-4. [DOI] [PubMed] [Google Scholar]
- Allen JF. Thylakoid protein-phosphorylation, state-1-state-2 transitions, and photosystem stoichiometry adjustment—redox control at multiple levels of gene expression. Physiol Plant. 1995;93:196–205. [Google Scholar]
- Allen JF. The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci. 2003a;358:19–37. doi: 10.1098/rstb.2002.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Why chloroplasts and mitochondria contain genomes. Comp Funct Genomics. 2003b;4:31–36. doi: 10.1002/cfg.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Photosynthesis: the processing of redox signals in chloroplasts. Curr Biol. 2005;15:R929–R932. doi: 10.1016/j.cub.2005.10.061. [DOI] [PubMed] [Google Scholar]
- Allen JF, Bennett J, Steinback KE, Arntzen CJ. Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature. 1981;291:25–29. [Google Scholar]
- Allen JF, Pfannschmidt T. Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 2000;355:1351–1357. doi: 10.1098/rstb.2000.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Puthiyaveetil S, Strom J, Allen CA. Energy transduction anchors genes in organelles. Bioessays. 2005;27:426–435. doi: 10.1002/bies.20194. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, et al. Chloroplast transcription at different light intensities. Glutathione-mediated phosphorylation of the major RNA polymerase involved in redox-regulated organellar gene expression. Plant Physiol. 2001;127:1044–1052. doi: 10.1104/pp.010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol Biol. 1999;39:1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Oxford: Blackwell Science Ltd; 2002. Molecular mechanisms of photosynthesis. [Google Scholar]
- Bonaventura C, Myers J. Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim Biophys Acta. 1969;189:366–383. doi: 10.1016/0005-2728(69)90168-6. [DOI] [PubMed] [Google Scholar]
- Chow WS, Melis A, Anderson JM. Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci U S A. 1990;87:7502–7506. doi: 10.1073/pnas.87.19.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- Fujita Y. A study on the dynamic features of photosystem stoichiometry: accomplishments and problems for future studies. Photosynth Res. 1997;53:83–93. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hill R, Bendall F. Function of the two cytochrome components in chloroplasts—a working hypothesis. Nature. 1960;186:136–137. [Google Scholar]
- Juric S, et al. Tethering of ferredoxin:NADP+ oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J. 2009;60:783–794. doi: 10.1111/j.1365-313X.2009.03999.x. [DOI] [PubMed] [Google Scholar]
- Kirk JTO, Tilney-Bassett RAE. Amsterdam (The Netherlands): Elsevier/North Holland; 1978. The plastids. Their chemistry, structure, growth and inheritance. [Google Scholar]
- Kuchler M, Decker S, Hormann F, Soll J, Heins L. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 2002;21:6136–6145. doi: 10.1093/emboj/cdf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. Green life: control of chloroplast gene transcription. Bioessays. 1996;18:465–471. [Google Scholar]
- Link G. Redox regulation of chloroplast transcription. Antioxid Redox Signal. 2003;5:79–87. doi: 10.1089/152308603321223568. [DOI] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007;26:845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- Melis A, Mullineaux CW, Allen JF. Acclimation of the photosynthetic apparatus to photosystem-I or photosystem-II light—evidence from quantum yield measurements and fluorescence spectroscopy of cyanobacterial cells. Z Naturforsch C. 1989;44:109–118. [Google Scholar]
- Murakami A, Kim SJ, Fujita Y. Changes in photosystem stoichiometry in response to environmental conditions for cell growth observed with the cyanophyte Synechocystis PCC 6714. Plant Cell Physiol. 1997;38:392–397. doi: 10.1093/oxfordjournals.pcp.a029181. [DOI] [PubMed] [Google Scholar]
- Murata N. Control of excitation transfer in photosynthesis. I. Light-induced changes of chlorophyll a fluorescence in Porphyridium cruentum. Biochim Biophys Acta. 1969;172:242–251. doi: 10.1016/0005-2728(69)90067-x. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem. 2002;269:3329–3337. [PubMed] [Google Scholar]
- Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF. Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life. 1999;48:271–276. doi: 10.1080/713803507. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Allen JF. Transients in chloroplast gene transcription. Biochem Biophys Res Commun. 2008;368:871–874. doi: 10.1016/j.bbrc.2008.01.167. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Allen JF. Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression. Philos Trans R Soc Lond B Biol Sci. 2009;276:2133–2145. doi: 10.1098/rspb.2008.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, et al. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci U S A. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race HL, Herrmann RG, Martin W. Why have organelles retained genomes? Trends Genet. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sato S, et al. A large-scale protein–protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Geimer S, Meurer J, Link G. Arabidopsis mutants carrying chimeric sigma factor genes reveal regulatory determinants for plastid gene expression. Plant Cell Physiol. 2009;50:1382–1386. doi: 10.1093/pcp/pcp069. [DOI] [PubMed] [Google Scholar]
- Schweer J, Turkeri H, Link B, Link G. AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J. 2010;62:192–202. doi: 10.1111/j.1365-313X.2010.04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, et al. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc Natl Acad Sci U S A. 2010;107:10760–10764. doi: 10.1073/pnas.0911692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JY, et al. Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 2004;40:164–172. doi: 10.1111/j.1365-313X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Eisermann A, Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.): evidence for three different transcription factors that resemble bacterial sigma factors. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- Tullberg A, Alexciev A, Pfannschmidt T, Allen JF. Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 2000;41:1045–1054. doi: 10.1093/pcp/pcd031. [DOI] [PubMed] [Google Scholar]
- Weber P, et al. TCP34, a nuclear-encoded response regulator-like TPR protein of higher plant chloroplasts. J Mol Biol. 2006;357:535–549. doi: 10.1016/j.jmb.2005.12.079. [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]