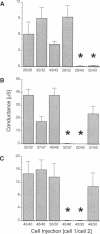
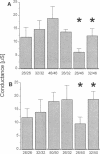
Abstract
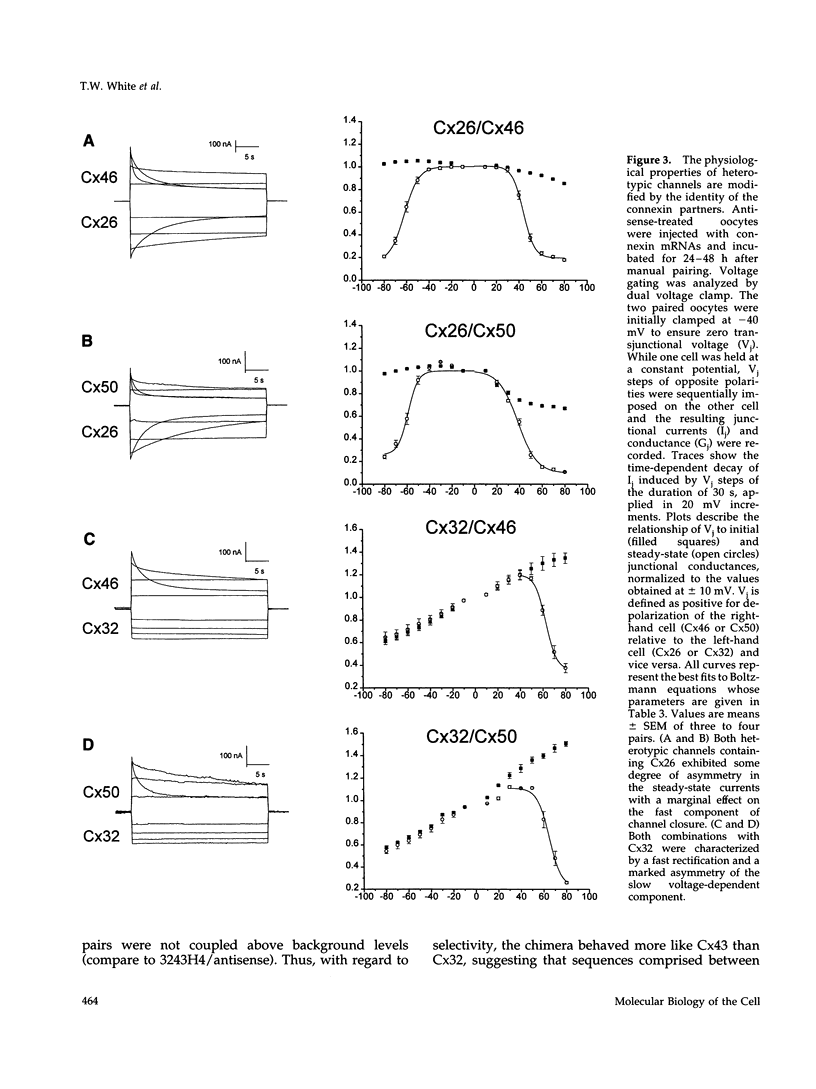
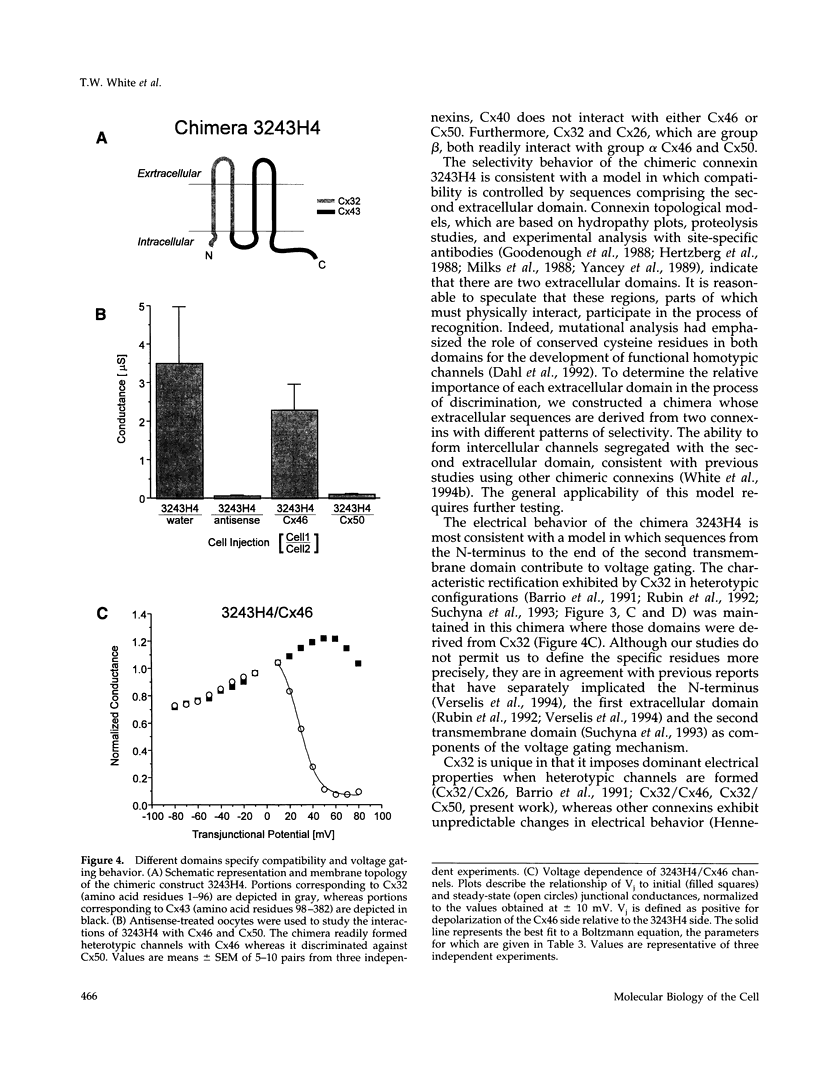
One consequence of the diversity in gap junction structural proteins is that cells expressing different connexins may come into contact and form intercellular channels that are mixed in connexin content. We have systematically examined the ability of adjacent cells expressing different connexins to communicate, and found that all connexins exhibit specificity in their interactions. Two extreme examples of selectivity were observed. Connexin40 (Cx40) was highly restricted in its ability to make heterotypic channels, functionally interacting with Cx37, but failing to do so when paired with Cx26, Cx32, Cx43, Cx46, and Cx50. In contrast, Cx46 interacted well with all connexins tested except Cx40. To explore the molecular basis of connexin compatibility and voltage gating, we utilized a chimera consisting of Cx32 from the N-terminus to the second transmembrane domain, fused to Cx43 from the middle cytoplasmic loop to the C-terminus. The chimeric connexin behaved like Cx43 with regard to selectivity and like Cx32 with regard to voltage dependence. Taken together, these results demonstrate that the second but not the first extracellular domain affects compatibility, whereas voltage gating is strongly influenced by sequences between the N-terminus and the second transmembrane domain.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., Nicholson B. J. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide B., Neyses L., Ganten D., Paul M., Willecke K., Traub O. Gap junction protein connexin40 is preferentially expressed in vascular endothelium and conductive bundles of rat myocardium and is increased under hypertensive conditions. Circ Res. 1993 Dec;73(6):1138–1149. doi: 10.1161/01.res.73.6.1138. [DOI] [PubMed] [Google Scholar]
- Bennett M. V., Barrio L. C., Bargiello T. A., Spray D. C., Hertzberg E., Sáez J. C. Gap junctions: new tools, new answers, new questions. Neuron. 1991 Mar;6(3):305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Kistler J., Paul D. L., Goodenough D. A. Antisera directed against connexin43 peptides react with a 43-kD protein localized to gap junctions in myocardium and other tissues. J Cell Biol. 1989 Feb;108(2):595–605. doi: 10.1083/jcb.108.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R., Haefliger J. A., Gimlich R. L., Paul D. L. Connexin40, a component of gap junctions in vascular endothelium, is restricted in its ability to interact with other connexins. Mol Biol Cell. 1993 Jan;4(1):7–20. doi: 10.1091/mbc.4.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R., White T. W., Paul D. L. Expression of chimeric connexins reveals new properties of the formation and gating behavior of gap junction channels. J Cell Sci. 1994 Apr;107(Pt 4):955–967. doi: 10.1242/jcs.107.4.955. [DOI] [PubMed] [Google Scholar]
- Chanson M., Orci L., Meda P. Extent and modulation of junctional communication between pancreatic acinar cells in vivo. Am J Physiol. 1991 Jul;261(1 Pt 1):G28–G36. doi: 10.1152/ajpgi.1991.261.1.G28. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., DeHaan R. L. Multiple-channel conductance states and voltage regulation of embryonic chick cardiac gap junctions. J Membr Biol. 1992 Apr;127(2):95–111. doi: 10.1007/BF00233282. [DOI] [PubMed] [Google Scholar]
- Dahl G., Miller T., Paul D., Voellmy R., Werner R. Expression of functional cell-cell channels from cloned rat liver gap junction complementary DNA. Science. 1987 Jun 5;236(4806):1290–1293. doi: 10.1126/science.3035715. [DOI] [PubMed] [Google Scholar]
- Dahl G., Werner R., Levine E., Rabadan-Diehl C. Mutational analysis of gap junction formation. Biophys J. 1992 Apr;62(1):172–182. doi: 10.1016/S0006-3495(92)81803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermietzel R., Spray D. C. Gap junctions in the brain: where, what type, how many and why? Trends Neurosci. 1993 May;16(5):186–192. doi: 10.1016/0166-2236(93)90151-b. [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Traub O., Hwang T. K., Beyer E., Bennett M. V., Spray D. C., Willecke K. Differential expression of three gap junction proteins in developing and mature brain tissues. Proc Natl Acad Sci U S A. 1989 Dec;86(24):10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara L., Steiner E. Properties of a nonjunctional current expressed from a rat connexin46 cDNA in Xenopus oocytes. J Gen Physiol. 1993 Jul;102(1):59–74. doi: 10.1085/jgp.102.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M. L., Gilula N. B. A study of communication specificity between cells in culture. J Cell Biol. 1977 Dec;75(3):769–787. doi: 10.1083/jcb.75.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A. Gap junctions and intercellular communications. Science. 1994 Aug 19;265(5175):1017–1020. [PubMed] [Google Scholar]
- Fishman G. I., Spray D. C., Leinwand L. A. Molecular characterization and functional expression of the human cardiac gap junction channel. J Cell Biol. 1990 Aug;111(2):589–598. doi: 10.1083/jcb.111.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaume C., Kado R. T., Korn H. Voltage-clamp analysis of a crayfish rectifying synapse. J Physiol. 1987 May;386:91–112. doi: 10.1113/jphysiol.1987.sp016524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Differential regulation of the levels of three gap junction mRNAs in Xenopus embryos. J Cell Biol. 1990 Mar;110(3):597–605. doi: 10.1083/jcb.110.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliger J. A., Paul D. L. Expression of gap junction proteins Cx26, Cx31.1, Cx37, and Cx43 in developing and mature rat epidermis. Dev Dyn. 1994 May;200(1):1–13. doi: 10.1002/aja.1002000102. [DOI] [PubMed] [Google Scholar]
- Goodenough D. A., Musil L. S. Gap junctions and tissue business: problems and strategies for developing specific functional reagents. J Cell Sci Suppl. 1993;17:133–138. doi: 10.1242/jcs.1993.supplement_17.19. [DOI] [PubMed] [Google Scholar]
- Goodenough D. A., Paul D. L., Jesaitis L. Topological distribution of two connexin32 antigenic sites in intact and split rodent hepatocyte gap junctions. J Cell Biol. 1988 Nov;107(5):1817–1824. doi: 10.1083/jcb.107.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdie R. G., Severs N. J., Green C. R., Rothery S., Germroth P., Thompson R. P. The spatial distribution and relative abundance of gap-junctional connexin40 and connexin43 correlate to functional properties of components of the cardiac atrioventricular conduction system. J Cell Sci. 1993 Aug;105(Pt 4):985–991. doi: 10.1242/jcs.105.4.985. [DOI] [PubMed] [Google Scholar]
- Gros D., Jarry-Guichard T., Ten Velde I., de Maziere A., van Kempen M. J., Davoust J., Briand J. P., Moorman A. F., Jongsma H. J. Restricted distribution of connexin40, a gap junctional protein, in mammalian heart. Circ Res. 1994 May;74(5):839–851. doi: 10.1161/01.res.74.5.839. [DOI] [PubMed] [Google Scholar]
- Haefliger J. A., Bruzzone R., Jenkins N. A., Gilbert D. J., Copeland N. G., Paul D. L. Four novel members of the connexin family of gap junction proteins. Molecular cloning, expression, and chromosome mapping. J Biol Chem. 1992 Jan 25;267(3):2057–2064. [PubMed] [Google Scholar]
- Hennemann H., Dahl E., White J. B., Schwarz H. J., Lalley P. A., Chang S., Nicholson B. J., Willecke K. Two gap junction genes, connexin 31.1 and 30.3, are closely linked on mouse chromosome 4 and preferentially expressed in skin. J Biol Chem. 1992 Aug 25;267(24):17225–17233. [PubMed] [Google Scholar]
- Hennemann H., Suchyna T., Lichtenberg-Fraté H., Jungbluth S., Dahl E., Schwarz J., Nicholson B. J., Willecke K. Molecular cloning and functional expression of mouse connexin40, a second gap junction gene preferentially expressed in lung. J Cell Biol. 1992 Jun;117(6):1299–1310. doi: 10.1083/jcb.117.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg E. L., Disher R. M., Tiller A. A., Zhou Y., Cook R. G. Topology of the Mr 27,000 liver gap junction protein. Cytoplasmic localization of amino- and carboxyl termini and a hydrophilic domain which is protease-hypersensitive. J Biol Chem. 1988 Dec 15;263(35):19105–19111. [PubMed] [Google Scholar]
- Kam E., Watt F. M., Pitts J. D. Patterns of junctional communication in skin: studies on cultured keratinocytes. Exp Cell Res. 1987 Dec;173(2):431–438. doi: 10.1016/0014-4827(87)90283-7. [DOI] [PubMed] [Google Scholar]
- Kanemitsu M. Y., Lau A. F. Epidermal growth factor stimulates the disruption of gap junctional communication and connexin43 phosphorylation independent of 12-0-tetradecanoylphorbol 13-acetate-sensitive protein kinase C: the possible involvement of mitogen-activated protein kinase. Mol Biol Cell. 1993 Aug;4(8):837–848. doi: 10.1091/mbc.4.8.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter H. L., Saffitz J. E., Beyer E. C. Cardiac myocytes express multiple gap junction proteins. Circ Res. 1992 Feb;70(2):438–444. doi: 10.1161/01.res.70.2.438. [DOI] [PubMed] [Google Scholar]
- Kettenmann H., Orkand R. K., Schachner M. Coupling among identified cells in mammalian nervous system cultures. J Neurosci. 1983 Mar;3(3):506–516. doi: 10.1523/JNEUROSCI.03-03-00506.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Molecular biology and genetics of gap junction channels. Semin Cell Biol. 1992 Feb;3(1):3–16. doi: 10.1016/s1043-4682(10)80003-0. [DOI] [PubMed] [Google Scholar]
- Lo Turco J. J., Kriegstein A. R. Clusters of coupled neuroblasts in embryonic neocortex. Science. 1991 Apr 26;252(5005):563–566. doi: 10.1126/science.1850552. [DOI] [PubMed] [Google Scholar]
- Lo C. W., Gilula N. B. Gap junctional communication in the post-implantation mouse embryo. Cell. 1979 Oct;18(2):411–422. doi: 10.1016/0092-8674(79)90060-6. [DOI] [PubMed] [Google Scholar]
- Meda P., Michaels R. L., Halban P. A., Orci L., Sheridan J. D. In vivo modulation of gap junctions and dye coupling between B-cells of the intact pancreatic islet. Diabetes. 1983 Sep;32(9):858–868. doi: 10.2337/diab.32.9.858. [DOI] [PubMed] [Google Scholar]
- Mege R. M., Matsuzaki F., Gallin W. J., Goldberg J. I., Cunningham B. A., Edelman G. M. Construction of epithelioid sheets by transfection of mouse sarcoma cells with cDNAs for chicken cell adhesion molecules. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7274–7278. doi: 10.1073/pnas.85.19.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnil M., Fraslin J. M., Piccoli C., Yamasaki H., Guguen-Guillouzo C. Cell contact but not junctional communication (dye coupling) with biliary epithelial cells is required for hepatocytes to maintain differentiated functions. Exp Cell Res. 1987 Dec;173(2):524–533. doi: 10.1016/0014-4827(87)90292-8. [DOI] [PubMed] [Google Scholar]
- Meyer R. A., Laird D. W., Revel J. P., Johnson R. G. Inhibition of gap junction and adherens junction assembly by connexin and A-CAM antibodies. J Cell Biol. 1992 Oct;119(1):179–189. doi: 10.1083/jcb.119.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalke W., Loewenstein W. R. Communication between cells of different type. Nature. 1971 Jul 9;232(5306):121–122. doi: 10.1038/232121b0. [DOI] [PubMed] [Google Scholar]
- Milks L. C., Kumar N. M., Houghten R., Unwin N., Gilula N. B. Topology of the 32-kd liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988 Oct;7(10):2967–2975. doi: 10.1002/j.1460-2075.1988.tb03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A. P., Eghbali B., Spray D. C. Connexin32 gap junction channels in stably transfected cells: unitary conductance. Biophys J. 1991 Nov;60(5):1254–1266. doi: 10.1016/S0006-3495(91)82159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A. P., Sáez J. C., Fishman G. I., Spray D. C. Human connexin43 gap junction channels. Regulation of unitary conductances by phosphorylation. Circ Res. 1994 Jun;74(6):1050–1057. doi: 10.1161/01.res.74.6.1050. [DOI] [PubMed] [Google Scholar]
- Nedergaard M. Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science. 1994 Mar 25;263(5154):1768–1771. doi: 10.1126/science.8134839. [DOI] [PubMed] [Google Scholar]
- Nicholson B., Dermietzel R., Teplow D., Traub O., Willecke K., Revel J. P. Two homologous protein components of hepatic gap junctions. Nature. 1987 Oct 22;329(6141):732–734. doi: 10.1038/329732a0. [DOI] [PubMed] [Google Scholar]
- Nishi M., Kumar N. M., Gilula N. B. Developmental regulation of gap junction gene expression during mouse embryonic development. Dev Biol. 1991 Jul;146(1):117–130. doi: 10.1016/0012-1606(91)90452-9. [DOI] [PubMed] [Google Scholar]
- Paul D. L., Ebihara L., Takemoto L. J., Swenson K. I., Goodenough D. A. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J Cell Biol. 1991 Nov;115(4):1077–1089. doi: 10.1083/jcb.115.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risek B., Gilula N. B. Spatiotemporal expression of three gap junction gene products involved in fetomaternal communication during rat pregnancy. Development. 1991 Sep;113(1):165–181. doi: 10.1242/dev.113.1.165. [DOI] [PubMed] [Google Scholar]
- Risek B., Guthrie S., Kumar N., Gilula N. B. Modulation of gap junction transcript and protein expression during pregnancy in the rat. J Cell Biol. 1990 Feb;110(2):269–282. doi: 10.1083/jcb.110.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risek B., Klier F. G., Gilula N. B. Developmental regulation and structural organization of connexins in epidermal gap junctions. Dev Biol. 1994 Jul;164(1):183–196. doi: 10.1006/dbio.1994.1190. [DOI] [PubMed] [Google Scholar]
- Robinson S. R., Hampson E. C., Munro M. N., Vaney D. I. Unidirectional coupling of gap junctions between neuroglia. Science. 1993 Nov 12;262(5136):1072–1074. doi: 10.1126/science.8093125. [DOI] [PubMed] [Google Scholar]
- Rubin J. B., Verselis V. K., Bennett M. V., Bargiello T. A. Molecular analysis of voltage dependence of heterotypic gap junctions formed by connexins 26 and 32. Biophys J. 1992 Apr;62(1):183–195. doi: 10.1016/S0006-3495(92)81804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray D. C., Harris A. L., Bennett M. V. Equilibrium properties of a voltage-dependent junctional conductance. J Gen Physiol. 1981 Jan;77(1):77–93. doi: 10.1085/jgp.77.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg T. H., Civitelli R., Geist S. T., Robertson A. J., Hick E., Veenstra R. D., Wang H. Z., Warlow P. M., Westphale E. M., Laing J. G. Connexin43 and connexin45 form gap junctions with different molecular permeabilities in osteoblastic cells. EMBO J. 1994 Feb 15;13(4):744–750. doi: 10.1002/j.1460-2075.1994.tb06316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchyna T. M., Xu L. X., Gao F., Fourtner C. R., Nicholson B. J. Identification of a proline residue as a transduction element involved in voltage gating of gap junctions. Nature. 1993 Oct 28;365(6449):847–849. doi: 10.1038/365847a0. [DOI] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Swenson K. I., Piwnica-Worms H., McNamee H., Paul D. L. Tyrosine phosphorylation of the gap junction protein connexin43 is required for the pp60v-src-induced inhibition of communication. Cell Regul. 1990 Dec;1(13):989–1002. doi: 10.1091/mbc.1.13.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takens-Kwak B. R., Jongsma H. J. Cardiac gap junctions: three distinct single channel conductances and their modulation by phosphorylating treatments. Pflugers Arch. 1992 Nov;422(2):198–200. doi: 10.1007/BF00370421. [DOI] [PubMed] [Google Scholar]
- Tomasetto C., Neveu M. J., Daley J., Horan P. K., Sager R. Specificity of gap junction communication among human mammary cells and connexin transfectants in culture. J Cell Biol. 1993 Jul;122(1):157–167. doi: 10.1083/jcb.122.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub O., Look J., Dermietzel R., Brümmer F., Hülser D., Willecke K. Comparative characterization of the 21-kD and 26-kD gap junction proteins in murine liver and cultured hepatocytes. J Cell Biol. 1989 Mar;108(3):1039–1051. doi: 10.1083/jcb.108.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdimarsson G., De Sousa P. A., Beyer E. C., Paul D. L., Kidder G. M. Zygotic expression of the connexin43 gene supplies subunits for gap junction assembly during mouse preimplantation development. Mol Reprod Dev. 1991 Sep;30(1):18–26. doi: 10.1002/mrd.1080300103. [DOI] [PubMed] [Google Scholar]
- Veenstra R. D., Wang H. Z., Beyer E. C., Ramanan S. V., Brink P. R. Connexin37 forms high conductance gap junction channels with subconductance state activity and selective dye and ionic permeabilities. Biophys J. 1994 Jun;66(6):1915–1928. doi: 10.1016/S0006-3495(94)80985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra R. D., Wang H. Z., Westphale E. M., Beyer E. C. Multiple connexins confer distinct regulatory and conductance properties of gap junctions in developing heart. Circ Res. 1992 Nov;71(5):1277–1283. doi: 10.1161/01.res.71.5.1277. [DOI] [PubMed] [Google Scholar]
- Verselis V. K., Ginter C. S., Bargiello T. A. Opposite voltage gating polarities of two closely related connexins. Nature. 1994 Mar 24;368(6469):348–351. doi: 10.1038/368348a0. [DOI] [PubMed] [Google Scholar]
- Warner A. E., Lawrence P. A. Permeability of gap junctions at the segmental border in insect epidermis. Cell. 1982 Feb;28(2):243–252. doi: 10.1016/0092-8674(82)90342-7. [DOI] [PubMed] [Google Scholar]
- Weir M. P., Lo C. W. Gap junctional communication compartments in the Drosophila wing disk. Proc Natl Acad Sci U S A. 1982 May;79(10):3232–3235. doi: 10.1073/pnas.79.10.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R., Levine E., Rabadan-Diehl C., Dahl G. Formation of hybrid cell-cell channels. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5380–5384. doi: 10.1073/pnas.86.14.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. W., Bruzzone R., Goodenough D. A., Paul D. L. Mouse Cx50, a functional member of the connexin family of gap junction proteins, is the lens fiber protein MP70. Mol Biol Cell. 1992 Jul;3(7):711–720. doi: 10.1091/mbc.3.7.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. W., Bruzzone R., Goodenough D. A., Paul D. L. Voltage gating of connexins. Nature. 1994 Sep 15;371(6494):208–209. doi: 10.1038/371208a0. [DOI] [PubMed] [Google Scholar]
- White T. W., Bruzzone R., Wolfram S., Paul D. L., Goodenough D. A. Selective interactions among the multiple connexin proteins expressed in the vertebrate lens: the second extracellular domain is a determinant of compatibility between connexins. J Cell Biol. 1994 May;125(4):879–892. doi: 10.1083/jcb.125.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey S. B., John S. A., Lal R., Austin B. J., Revel J. P. The 43-kD polypeptide of heart gap junctions: immunolocalization, topology, and functional domains. J Cell Biol. 1989 Jun;108(6):2241–2254. doi: 10.1083/jcb.108.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R., Peinado A., Katz L. C. Neuronal domains in developing neocortex. Science. 1992 Jul 31;257(5070):665–669. doi: 10.1126/science.1496379. [DOI] [PubMed] [Google Scholar]
- de Laat S. W., Tertoolen L. G., Dorresteijn A. W., van den Biggelaar J. A. Intercellular communication patterns are involved in cell determination in early molluscan development. Nature. 1980 Oct 9;287(5782):546–548. doi: 10.1038/287546a0. [DOI] [PubMed] [Google Scholar]