Abstract
Cd1 nitrite reductase catalyzes the conversion of nitrite to NO in denitrifying bacteria. Reduction of the substrate occurs at the d1-heme site, which faces on the distal side some residues thought to be essential for substrate binding and catalysis. We report the results obtained by mutating to Ala the two invariant active site histidines, His-327 and His-369, of the enzyme from Pseudomonas aeruginosa. Both mutants have lost nitrite reductase activity but maintain the ability to reduce O2 to water. Nitrite reductase activity is impaired because of the accumulation of a catalytically inactive form, possibly because the productive displacement of NO from the ferric d1-heme iron is impaired. Moreover, the two distal His play different roles in catalysis; His-369 is absolutely essential for the stability of the Michaelis complex. The structures of both mutants show (i) the new side chain in the active site, (ii) a loss of density of Tyr-10, which slipped away with the N-terminal arm, and (iii) a large topological change in the whole c-heme domain, which is displaced 20 Å from the position occupied in the wild-type enzyme. We conclude that the two invariant His play a crucial role in the activity and the structural organization of cd1 nitrite reductase from P. aeruginosa.
The conversion of nitrite (NO2−) to nitric oxide (NO) is catalyzed in denitrifying bacteria by the periplasmic nitrite reductases (NIRs), which are either copper- or heme-containing enzymes (1, 2). Heme NIRs are homodimers of two 60-kDa subunits, each containing one covalently bound c-heme and one d1-heme. These enzymes catalyze not only the one-electron reduction of NO2− to NO but also the four-electron reduction of O2 to 2 H2O. Extensive spectroscopic and functional studies (3) have been carried out on cd1NIR from Pseudomonas aeruginosa (Pa-NIR); the c-heme domain is the electron's entry site, whereas catalysis occurs at the level of the d1-heme.
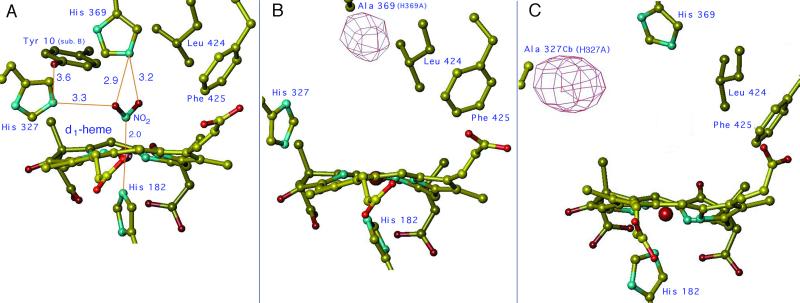
The three-dimensional (3D) structure of NIR from P. aeruginosa has been solved (by x-ray diffraction) for the oxidized, reduced, and reduced NO-bound forms of the enzyme (4, 5). The overall structure of this enzyme is similar to that published by Fulop et al. (6) for cd1 NIR from Paracoccus pantotrophus (formerly called Thiosphaera pantotropha; ref. 7). In both enzymes each subunit is organized in two structurally distinct domains: an N-terminal α-helical domain containing the c-heme and a C-terminal eight-blade β-propeller domain with the d1-heme binding site. The distal side of the d1-heme pocket is lined up with several important residues that include Tyr-10 (coming from the N-terminal arm of the other monomer) and two invariant histidine residues, His-327 and His-369 (Fig. 1A), presumably involved in substrate binding and/or protonation. Reduction of Pa-NIR leads to conformational changes (5), involving motion of a loop in the c-heme domain, rotation of Tyr-10 away from the d1-heme site, and loss of the distal d1-heme iron ligand, which in oxidized Pa-NIR is a hydroxyl (4).
Figure 1.
The active site of cd1 NIR from P. aeruginosa (Pa-NIR). The model of the d1-heme pocket of the wt reduced enzyme complexed with nitrite is shown in A. The stereochemistry of NO2− was simulated starting from the coordinates of the NO adduct of reduced Pa-NiR (5). Among the key amino acid side chains shown here, notice that Tyr-10 comes from the other monomer (identified as sub. B), as a result of a domain swapping across the 2-fold axis of the homodimer (4). The 3D structure of the d1-heme pocket of the two mutants in the oxidized state is shown in the same orientation in B (for H369A) and C (for H327A). The Fo−Fc Sigma A negative electron density map is also represented at the place of the missing side chain; this map is contoured at −3σ for B and −4σ for C.
To elucidate the role of the two invariant distal residues His-327 and His-369 (Fig. 1A) in the catalytic mechanism of Pa-NIR, we have prepared and characterized two site-directed mutants, H369A and H327A. The kinetic data show that these two histidines are essential for NIR activity (NIRV) but have no effect on the oxygen reductase activity. The 3D structures of both mutants shows that (i) Ala replaces His in the distal d1-heme pocket of both mutants; (ii) Tyr-10 slips away together with the N-terminal arm; and (iii) the c-heme domain experiences a large topological change relative to the d1-heme domain, which is unmodified. Our results allow us to propose a mechanism for catalysis of nitrite reduction, based largely on the essential role of the electrostatic potential imposed by the invariant His in assisting the dissociation of the product NO from the ferric d1-heme iron. The proposed mechanism may be of significance for heme proteins involved in the metabolism and transport of NO, all of which have to avoid being trapped in a “dead-end” complex with the reduced heme iron.
Materials and Methods
Mutagenesis and Protein Purification.
Mutagenesis of His-327 to Ala was carried out as in ref. 8; His-369 was mutated to Ala with the use of a U.S.E. Mutagenesis Kit (Amersham Pharmacia). Subcloning, expression in Pseudomonas putida, and purification were obtained as described (9, 10). In the P. putida expression system, the protein is synthesized with the c-heme, but no d1-heme; this semiapo-NIR is then reconstituted in vitro with the d1-heme extracted from wild-type (wt) Pa-NIR as detailed in ref. 9.
General Characterization.
Reduced derivatives were obtained by adding anaerobically excess ascorbate to oxidized NIR. Cytochrome oxidase activity was assessed at 20°C in 50 mM sodium phosphate buffer (pH 7.0) by measuring the rate of oxidation of reduced P. aeruginosa cytochrome c551 (1–20 μM) as described in ref. 11. NIR activity was measured anaerobically at 27°C in 50 mM sodium phosphate buffer (pH 6.2), either after oxidation of reduced azurin (12) or during amperometric measurement of NO production with a NO electrode (ISO-NO; World Precision Instruments, Sarasota, FL). A typical experiment at the electrode was carried out at 25°C in the presence of ascorbate (13 mM) and N,N,N,N-tetramethyl-p-phenylenediamine (0.1 mM) as electron donors and initiated by adding different concentrations of nitrite (10–1,200 μM).
Stopped-flow experiments were carried out anaerobically with the use of a TN6500 (Tracor Northern, Madison, WI) multidiode array spectrometer coupled to a Gibson–Durrum stopped-flow apparatus. Reduced NIR (4–8 μM before mixing) in degassed 50 mM sodium phosphate buffer (pH 8.0) was mixed with nitrite (0.02–1 mM) in the presence of 1 mM ascorbate at 25°C. The time course of the reaction was followed in the wavelength range of 380 to 650 nm; analysis of the experimental data was carried out as described (13).
Crystallization and Structure Determination of the H369A and H327A Mutants.
A detailed description of the crystallization procedures and of the structures has yet to be published (K.B., V. Roig-Zamboni, F.C., E.K.W., A.B., M.A., D. Nurizzo, M.B., C.C., and M.T., unpublished observations). Briefly, crystals of the H327A mutant were obtained by the vapor diffusion technique by mixing in a 1:1 ratio the protein and a reservoir solution containing 4.0% polyethylene glycol 5000 monomethyl ether, 0.1 M sodium acetate (pH 5.5). The space group is P4322 with cell dimensions 70.5 × 70.5 × 281 Å. Crystals of the H369A mutant were obtained by mixing in a 1:1 ratio the protein and a reservoir solution containing 11.5% polyethylene glycol 6000, 0.2 M imidazole/malate (pH 6.5). The space group is P41212 with cell dimensions 94.7 × 94.7 × 159.9 Å. These crystal forms are different from those obtained with the wt protein under similar crystallization conditions (14) and at high salt concentration (4) or at low pH (4). In contrast with these wt crystal forms, the mutant crystal forms both contain only one monomer per asymmetric unit, half the functional dimeric enzyme.
Before x-ray data collection, the crystals were soaked in 30% ethylene glycol in mother liquor and flash-frozen to 100 K. A H327A mutant data set was collected at 2.7-Å resolution on ID14-EH2 [European Synchrotron Radiation Facility (ESRF), Grenoble, France], integrated with denzo (15), and reduced with scala (16). The structure was solved by molecular replacement with the use of the d1 heme domain (residues 120–543) and amore (17). The c-heme domain was rigid-body placed manually in the Fo−Fc electron density map with turbo-frodo (18). The structure was refined with cns (19) to values of R and free R of 21.7 and 28.0%, respectively.
The same procedure was used with the H369A mutant, which diffracted to 2.8-Å resolution on ID14-EH2 (ESRF). However, after molecular replacement with the use of the d1 heme domain, no electron density for the c-heme domain was visible in the Fo − Fc maps. Furthermore, a two-body molecular replacement procedure failed to yield the position of the c-heme domain. We therefore decided to apply multiple wavelength anomalous diffraction (MAD) techniques to the anomalous signal of the 2 Fe ions. Three MAD data sets were collected on BM14 (ESRF) at 3.8-Å resolution. Finally, the complete structure was determined by a combination of MAD techniques and phase combination, with the use of the molecular replacement results with the d1-heme domain. The resolution was extended to 2.8 Å with the use of a data set already collected on ID14-EH2 (ESRF) at a single wavelength. The refined model has a Rwork of 22.9% and a Rfree of 28.5%.
Results
Spectral and Steady-State Kinetic Properties.
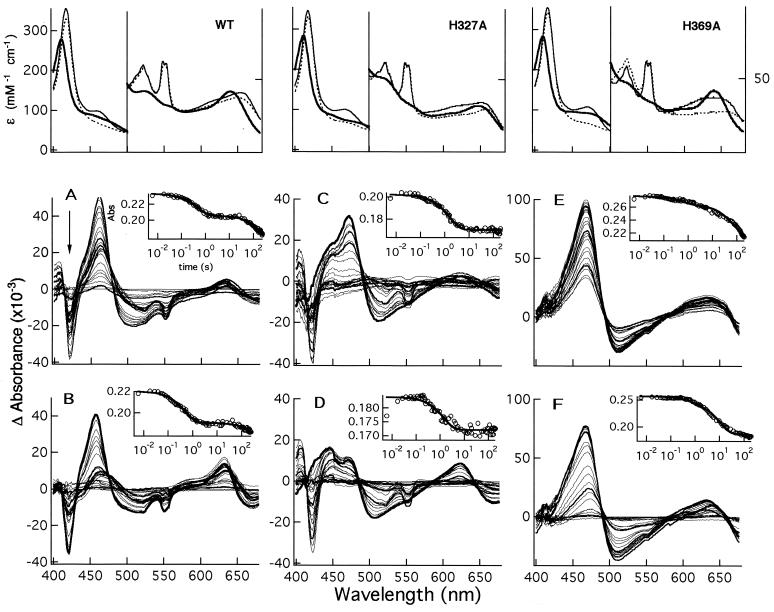
The optical spectra of the two mutants, H327A and H369A, are different from that of wt NIR (Fig. 2), mainly where the d1-heme absorbs (450–500 nm and 600–700 nm). In the oxidized state, the largest changes are seen for the H327A mutant, the 641-nm peak being red-shifted by 8 nm; in the reduced state changes are observed for both mutants with a 10-nm red-shift (from 462 to 472 nm) of the d1-heme Soret band. The spectrum of the reduced NO-bound derivative was essentially unchanged.
Figure 2.
Static and transient optical spectra of wt and mutants Pa-NIR. (Upper) Absolute spectra of the oxidized (bold line), reduced (thin line), and reduced NO (dotted line) derivatives for the wt NIR (Left), mutant H327A (Center), and mutant H369A (Right). (Lower) Time evolution of the kinetic difference spectra observed for the same proteins, after the reduced enzyme is mixed with nitrite anaerobically. The two sets of difference spectra for each protein refer to experiments carried out at the lowest (10 μM; A, C, and E) and the highest (0.15 or 0.5 mM; B, D, and F) nitrite concentrations. The arrow indicates the direction of the time course, from 6 ms to 245 s. To better follow the spectral evolution with time, the difference spectra at selected times (6 ms and 1, 25, and 180 s) are drawn as thick lines. The insets show the time course as followed at the maximum of reduced d1-heme (462 nm for the wt and 472 nm for the two mutants), fitted to two or three exponentials (continuous lines). Kinetic experiments were carried out in 50 mM phosphate buffer (pH 8.0) and 25°C.
The most interesting feature of the two mutants relates to catalysis. As shown in Table 1, both mutants catalyze the reduction of oxygen to water as efficiently as the wt NIR; in contrast, reduction of nitrite to NO is severely compromised for both mutants (the activity is ≈1% of wt). The same effect on the nitrite reductase activity was observed for both mutants between pH 5 and 7 (not shown).
Table 1.
Enzymatic activities of wt and mutant cd1 NIR
| NIR | Nitrite
reductase*
|
Oxygen
reductase†
|
||
|---|---|---|---|---|
| Km, μM | Turnover number, s−1 | Km, μM | Kcat, μM min−1 | |
| wt | 6 | 8 | 2.0 (±0.5) | 1.5 (±0.5) |
| H327A | — | 0.08 | 1.8 (±0.8) | 2.6 (±0.5) |
| H369A | — | 0.08 | 7.5 (±2) | 1.9 (±0.6) |
pH 6.2 and 25°C. Error is ±20%.
pH 7.0 and 20°C. Km values were obtained with cytochrome c551 as a substrate.
Stopped-Flow Experiments on the Nitrite Reaction.
Wt NIR.
This reaction was followed at several nitrite concentrations and at pH 8.0, which is higher than that of the steady-state experiments (pH 6.2), to better resolve the kinetics. The spectra recorded at different times are shown in Fig. 2 as the difference between each spectrum and that of the fully reduced NO bound enzyme, the final product of the reaction. The redox state of the two hemes is followed by looking at the time evolution of the absorption peaks characteristic of each chromophore (420 and 550 nm for the c-heme; 460 and 640 nm for the d1-heme). As an example, in Fig. 2 a negative peak at 550 nm is diagnostic of oxidation of the c-heme, because the reference spectrum is that of a fully reduced c-heme.
The first spectrum recorded after reduced NIR was mixed with 10 μM nitrite (at 6 ms) still contains a significant fraction of the starting reduced species (Fig. 2A). Because only minor oxidation of the c-heme is observed (≤15%), it is likely that the Michaelis complex (c+2d1+2 NO2−) is one of the species populated at this time. At higher nitrite concentrations (150 μM, Fig. 2B) more oxidized c-heme is observed at 6 ms. The initial mixture evolves through a nitrite-independent process and populates other species. After absorbance changes in the spectral regions characteristic of both the c- and d1-hemes, it is observed that more c-heme becomes oxidized (up to 20–25%). This process (see Insets in Fig. 2) occurs between 0.1 and 10 s, with the optical changes assigned to the d1-heme preceding those of the c-heme (with rate constants of 2 and 0.3 s−1, respectively). Finally, at times between 10 s and 4 min the mixture slowly drifts toward the c+2d1+2 NO species, and the c-heme returns to the fully reduced state.
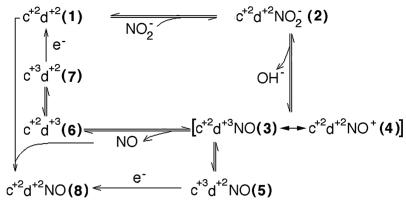
At least four spectra thus can be assigned during the overall time course (but only two of them refer to pure species): (i) that of the fully reduced c+2d1+2 derivative, present in the driving syringe; (ii) that of the pre-steady-state mixture, clearly evident in the first spectrum after mixing (6 ms); (iii) that corresponding to a more complex mixture, containing a clearly detectable amount of the mixed valence complex c+3d1+2 NO; and (iv) that of the final species c+2d1+2 NO (dead-end state). The time course at 460 nm (Insets of Fig. 2 A and B) is thus representative of two processes, namely evolution of the initial mixture to the steady state and formation of the dead-end species. An overall scheme showing the various possible intermediates, as well as information obtained by George et al. (20), is depicted in Fig. 3 and detailed in the figure legend.
Figure 3.
A proposed scheme for the reaction mechanism of cd1 NIR with NO2−. When nitrite is mixed with the reduced enzyme (species 1), the formation of the Michaelis complex (species 2) is followed by very rapid formation of NO, involving bond breaking and loss of a hydroxide ion; yet this process is assumed to be reversible. The resulting mixed-valence species 3 or 4 (c+2d1+3 NO or c+2d1+2 NO+) may dissociate NO and be reduced to species 1 (via 6 and 7) to enter a new productive cycle; however, in vitro it seems to be progressively inhibited to a dead-end state with NO bound to the ferrous d1-heme (species 8). Species 1–5 equilibrate rapidly [see also George et al. (20)], accounting for a fraction of oxidized c-heme formed during the dead time of the stopped flow. The relative population of each species depends on experimental conditions, such as pH and concentration of substrate and reductant, and it may even differ in NIR from different species. The Michaelis complex, formed rapidly even at low nitrite concentrations (e.g., 100 μM), accounts for considerably less than 100% of the enzyme; therefore the bimolecular rate constant is fast, but the affinity is lower than previously suggested (24). Nevertheless, our kinetic data are consistent with the value of Km = 6 μM NO2−, which was independently determined (not shown). Species 6 builds up slowly (k = 2 s−1) and incompletely under our experimental conditions; the internal redox equilibrium between species 6 and 7 is assigned a rate constant of 0.3 s−1. Because it is well established that the electron accepting site is the c-heme, only species 5 and 7 can be reduced with the use of ascorbate, azurin, or cytochrome c551. In the scheme, there are two paths leading to c+2d1+2 NO, the dead-end species 8: either by reaction of the reduced enzyme with NO or by reduction of species 5 by external reductants. In P. pantotrotrophus NiR, species 5 forms completely and instantaneously at [NO2−] = 0.2–5.0 mM; thereafter it decays to species 4 at 38 s−1, as shown by George et al. (20); it is possible that a similar reequilibration also occurs in Pa-NIR, but if so, it is lost in the dead time.
Additional information on this reaction has been obtained with the following experiments: (i) if the steady-state mixture obtained as described above is quickly prepared and then mixed with NO (100 μM) within 20 s, a species with the spectrum of c+2d1+2 NO is obtained with an apparent rate constant of 3 s−1, whereas mixing with degassed buffer has no effect (not shown). This experiment shows that the initial mixture of species must transiently allow formation of the unliganded reduced derivative, either by cycling or by dissociation of the weakly bound substrate. (ii) If reduced NIR is mixed with NO and nitrite, only the NO complex is obtained within the dead time of the instrument (2 ms), showing that the affinity and rate constants for combination with NO must be high. (iii) Reduction of fully oxidized wt NIR by ascorbate populates a mixed-valence intermediate and displays a complex time course: the faster phase has a rate constant of 0.1–0.2 s−1, dependent on the concentration of ascorbate, and is followed by much slower process(es) (not shown). This rate constant is compatible with the hypothesis that irreversible inhibition of the enzyme in vitro may occur by reduction of the mixed valence state with formation of c+2d1+2 NO in the presence of NO.
H369A and H327A Mutants.
For the mutant H327A, the first spectrum after mixing depends on nitrite concentration (Fig. 2 C and D). At [NO2−] = 10 μM (Fig. 2C) this spectrum corresponds to a mixture containing a significant percentage of the fully reduced unliganded enzyme (see, for example, the band at 472 nm); at [NO2−] = 0.5 mM (Fig. 2D) the more populated component is not one that can be obtained at equilibrium and recalls the spectrum of the first intermediate observed in the experiment carried out on wt NIR. The final spectrum, corresponding to the reduced NO complex, is reached in ≈1 min, at least at the highest NO2− concentration (0.5 mM). We conclude that substrate binding to the mutant H327A is somewhat similar to wt, but its conversion to the dead-end species occurs faster.
In the case of mutant H369A, the first spectrum corresponds largely to reduced unliganded NIR, which then evolves in a multiphasic process to yield the reduced, NO-bound species, more clearly so at high nitrite concentration, i.e., 0.5 mM (Fig. 2F). The observed time course (Fig. 2F, Inset) can be fitted to two exponentials; the faster process has the same rate (k = 0.3 s−1) at high and low nitrite, whereas the slower one is concentration dependent (k = 0.045 s−1 at 0.5 mM NO2− and 0.004 s−1 at 10 μM NO2−). Despite the complexity of the time course (which frustrated attempts to describe quantitatively the kinetics), it seems obvious from examination of the difference spectra (Fig. 2) that, compared with wt NIR, H369A has a considerably reduced affinity for NO2−. The dead-end state is reached more rapidly than wt (≈3 min), and the reaction scarcely requires cycling; thus compared with wt, either the mixed-valence NO complex is reduced faster or dissociation of NO occurs more slowly, or both. The faster evolution to the dead-end species at higher substrate concentration may simply reflect the increase of the catalytic rate with substrate concentration, given that in this mutant all backward rate constants are probably higher than in wt NIR.
Reduction of the c-heme by ascorbate starting from the fully oxidized His mutants corresponds to a single kinetic process, with a rate constant of ≈0.1 s−1 at 75 μM ascorbate, compared with heterogeneous kinetics with values of 0.1–0.01 s−1 for the wt enzyme. When azurin was used as the reductant in the presence of excess CO (21), reduction of the c-heme in the mutants occurs 2-fold more rapidly than for wt, whereas no significant difference in the c-to-d1 electron transfer rate could be detected (W. Sun, M.A., A.B., F.C., and M.B., unpublished observations).
X-Ray Structure of the H327A and H369A Mutants.
In both mutants, no 2Fo−Fc electron density is visible at the place of the mutated His, whereas a strong negative density was observed in the Fo−fc map, as expected (see Fig. 1 B and C). The whole N-terminal segment (residues 1–25) has no detectable electron density in either mutant, likely because of molecular flexibility. Consequently, Tyr-10 is not visible in the distal pocket of the d1-heme, contrary to what is seen in the oxidized wt protein (4, 5). For the H369A mutant, the other residues in the active site have conformations that are almost unchanged compared with the wt or differ as different data sets at this resolution usually do (rms deviation ≅ 0.5 Å). The active site crevice of H327A is clearly more open (on average by 2 Å), with the distances between the Fe and the Cβ of residues His-369, Ala-327, and Phe-425 increasing by about 3.8, 1.5, and 1.8 Å, as compared with the wt enzyme and the H369A mutant.
Both mutants H327A and H369A are dimers in solution, as determined by gel filtration chromatography (data not shown). Although a single monomer is contained in the crystal asymmetric unit, a dimer identical to the wt enzyme can be reconstructed by taking into account a 2-fold symmetry-related molecule (Fig. 4). The crystallographic monomer is composed of the β-propeller domain carrying the d1-heme (residues 149–543) and of the c-heme domain (residues 26–115), linked by a 34-residue segment. Similar to the wt enzyme (oxidized or reduced), the B factors of the c-heme domains of both mutants are higher than those of the d1-heme domain, to such an extent that for the mutant H369A, segments of the polypeptide chain were difficult to follow.
Figure 4.
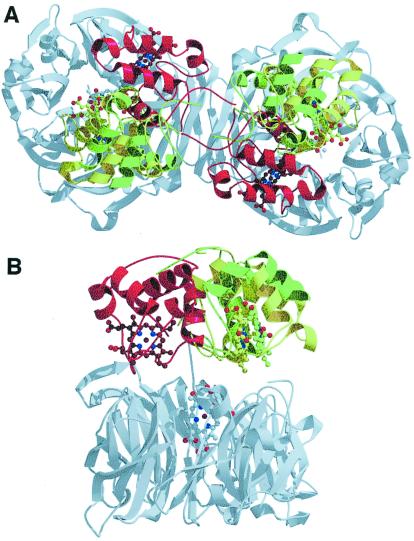
Crystallographic structure of the two mutants of Pa-NiR. (A) Top view of the mutant proteins superimposed on the wt enzyme. Color code for the c-heme domains: dark green, H327A; light green, H369A; red, wt. It is evident that the c-heme domains glide away in a new configuration, which is almost the same for the two mutants. Notice also that the 3D structure of the d1-heme domain (gray) is identical for the three proteins. (B) Side view of the three structures (same color code); only one monomer is shown for the sake of clarity.
In both mutants, the d1-heme domain is superimposable on that of the wt protein, whereas the c-heme domain's orientation is completely different (Fig. 4). The latter has been subjected to a rigid-body gliding motion on top of the d1-heme domain surface, resulting in a rotation of about 60° around an axis parallel to the 2-fold molecular axis and passing through Gln-115. As a result, the c-heme domains have moved about 20 Å, on average, from the original position in the wt (Fig. 4). The distance between the c- and d1-heme iron (21 Å) is almost unchanged, whereas the edge-to-edge distance (11.4 Å in the wt) has increased by 2 and 4 Å for H327A and H369A, respectively. In the mutants, the two c-heme domains in the dimer are closer to each other (3.8 Å), and the angle between them is about 13°, perpendicular to the 2-fold molecular axis. The c-heme domains, in their new positions, are involved in contacts with symmetry-related molecules: in H327A, the loop 92–95 interacts with the bottom of a symmetry-related d1-heme domain (Lys-431), the residues 61 and 70 interact with the Asn-502 of a symmetry-related molecule, and the loop 71–85 interacts with the same loop of a symmetry-related molecule. In H369A, the segment 78–85 interacts with a symmetry-related loop (residues 522–524) of the d1-heme domain.
The contact area between the c- and d1-heme domains in the wt enzyme has a surface of about 350 Å2. This area is almost maintained in the mutant H369A) (≈300 Å2) and is increased to ≈500 Å2 in the mutant H327A. Despite some similarities in the overall interdomain surface area, the high B values observed with the H369A mutant may indicate a mobility of the c-heme domain higher than those in the wt and H327A structures.
Discussion
Periplasmic cd1NIR catalyzes in denitrifying bacteria the conversion of nitrite to NO. This reaction poses a problem because the product NO is known to bind with very high affinity to reduced hemes and thus must be quickly released in the bulk to avoid inhibition. Structural and spectroscopic information on the enzyme from two different species is now available (3, 6, 8, 22–24), and very recently, the reaction of reduced P. pantotrophus NIR with nitrite at very high concentration (e.g., 5 mM) and in the absence of excess reductant has been reported (20). Our study provides insight into the enzyme mechanism and allows us to propose the scheme shown in Fig. 3, which is an extension of that initially proposed by Averill (1). It implicitly assumes that functional interactions between the two monomers can be neglected, which may not necessarily be the case.
Insight into the molecular mechanism of catalysis comes from the results on the two His mutants. Substitution of either of the two invariant His with Ala has a dramatic effect on nitrite reduction, but absolutely no effect on the oxygen reductase activity. In addition, it was somewhat surprising to discover that the two His are not equally important in the reaction with nitrite and to find out that His-369 is essential in controlling the affinity for this ligand. This disparity in importance is shown by the stopped-flow experiment carried out with the H369A mutant (see Fig. 2 E and F), in which no significant amounts of the Michaelis complex can be detected; the reaction with NO2− proceeds from the reduced state (species 1, Fig. 3) to the dead-end product (species 8) without detectable intermediates. In the mutant H327A, on the other hand, a significant fraction of the pre-steady-state mixture of species was detected, as shown in Fig. 2 C and D.
It is known that the affinity of ferrous heme proteins for anions is usually very low; as an example, ferrous hemoglobin binds cyanide with a Kd ≈ 1 M (25). On the other hand, in the case of NIR, the ferrous d1-heme displays high affinity for both nitrite and cyanide (Kd ≈ 10−6 M). The 3D structure of the cyanide derivative of reduced P. pantotrophus NIR (23) shows that the two His and the Tyr in the active site are within H-bonding distance of cyanide, suggesting that a positive electrostatic potential in the pocket is essential for stabilization of the bound anion. An unusually high positive charge density has also been found in the heme pocket of the Escherichia coli sulfite reductase, an enzyme capable of binding both sulfite and nitrite with high affinity, catalyzing their reduction to sulfide and ammonia, respectively (26). The unique role of His-369 in stabilizing the enzyme–substrate complex, although unexpected, seems to be in agreement with the structural data shown in Fig. 1. In fact, although both His are fairly close to the bound nitrite, His-369 is a shorter distance from both oxygens of the substrate, thus accounting for a dominant effect on the stability of the Michaelis complex, via the formation of (possibly) two H-bonds.
Over and above the visible loss of density due to substitution of His with Ala in the active site (Fig. 1 B and C), the most striking structural feature of both mutants is a large displacement of the whole domain containing the c-heme relative to the d1-heme, which stays put (Fig. 4). An important issue is to understand whether and how this conformational change is related to the dramatic inhibition of NIR activity reported above for the two mutants. The overall distance between the c- and d1-hemes and the surface of contact between these two domains is similar in the His mutants compared with wt; therefore the rate of intramolecular electron transfer should not be drastically modified, a fact that has been experimentally verified (W. Sun, M.A., A.B., F.C., and M.B., unpublished observations). Furthermore, the large area of contact between the c- and d1-heme domains and the fact that the c-domain position is very similar for the two mutants (and therefore unlikely to be opportunistic) suggest that the structure is a rather stable one.
These results illustrate the plasticity of the redox enzymes, a phenomenon often postulated but seldom substantiated (27, 28). In our case the structural change shown in Fig. 4 may be used to gauge the decreased interactions of Tyr-10 with the two invariant His-327 and His-369 in the active site and the associated motion of the N-terminal arm away from the pocket. This structural change is accompanied by a relocation of the c-heme domain in a minimum of lower free energy. A similar relocation of the c-heme domain may take place also in the mutant Y10F, which, despite being functionally and spectroscopically identical to wt (9), has been very difficult to crystallize. In a nutshell, we suggest that the c-heme domain relocation and the loss of favorable interactions of Tyr-10 in the active site of the two His mutants may be due to reduction of the positive electrostatic potential in the pocket. Insofar as this hypothesis is correct, a structural interpretation of the inhibition of the NIR activity observed with either one of the two His mutants may focus on the observation that they are trapped in the reduced NO-bound dead-end species faster than is wt NIR. According to the scheme shown in Fig. 3, dissociation of NO from the mixed-valence species 3 (c+2d1+3 NO) should be faster than electron transfer to the d1-heme to yield a catalytically competent enzyme. Given that the intramolecular electron transfer rate is unchanged in the His mutants (see above), the increased probability of trapping the dead-end species may originate in a faster reduction of the mutants by ascorbate (as observed; see Results) or a decrease in the rate of NO dissociation from the ferric d1-heme of species 3, or both. We thus propose that substitution of either one of the two invariant active-site His with Ala and reduction of the positive potential in the pocket is associated with loss of the hydroxyl, which was found to be coordinated with the ferric d1-heme iron in the wt enzyme (4) and should assist NO dissociation. This hypothesis is in agreement with quantum mechanical calculations (29), indicating that interaction with the active-site His can stabilize and orient nucleophilic ligands of the d1-heme, the presence of which imposes an unfavorable orientation on NO and thereby facilitates its release. It is worth noting that it has been recently shown that a buried carboxylate in the active site of nitrophorins (a class of heme proteins with vasodilatory activity associated with NO release) promotes dissociation of NO from the ferric heme iron (30). Finally, the interpretation proposed above is consistent with our finding that mutation of either one of the two invariant active-site His with Ala has no effect on the O2 reductase activity of NIR. In this case the substrate is uncharged, and its protonation may be assisted by the remaining His and bulk water, now accessible to the d1-heme distal pocket, which is wide open.
Acknowledgments
We thank R. Dagai and L. Nicolini (Istituto Superiore di Sanità, Rome, Italy) for fermentation of bacterial strains and V. Zamboni (Marseille, France) for crystallizing the mutants. Grants from the Consiglio Nazionale delle Ricerche of Italy (Target Project on Biotechnology) and the Ministero dell'Università e della Ricerca Scientifica e Tecnologica of Italy (Progetto di Ricerca di Interesse Nazionale 1999, “Dinamica Strutturale di Emoproteine”) are gratefully acknowledged.
Abbreviations
- NIR
nitrite reductase
- Pa-NIR
Pseudomonas aeruginosa NIR
- wt
wild type
- 3D
three-dimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1HZU and 1HZV).
References
- 1.Averill B A. Chem Rev. 1996;96:2951–2964. doi: 10.1021/cr950056p. [DOI] [PubMed] [Google Scholar]
- 2.Zumft W G. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutruzzolà F. Biochim Biophys Acta. 1999;4730:1–19. doi: 10.1016/s0005-2728(99)00017-1. [DOI] [PubMed] [Google Scholar]
- 4.Nurizzo D, Silvestrini M C, Mathieu M, Cutruzzolà F, Bourgeois D, Fülop V, Hajdu J, Brunori M, Tegoni M, Cambillau C. Structure (London) 1997;5:1157–1171. doi: 10.1016/s0969-2126(97)00267-0. [DOI] [PubMed] [Google Scholar]
- 5.Nurizzo D, Cutruzzolà F, Arese M, Bourgeois D, Brunori M, Tegoni M, Cambillau C. Biochemistry. 1998;37:13987–13996. doi: 10.1021/bi981348y. [DOI] [PubMed] [Google Scholar]
- 6.Fülöp V, Moir J W B, Ferguson S J, Hajdu J. Cell. 1995;81:369–377. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- 7.Rainey F A, Kelly D P, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, Wood A P. Int J Syst Bacteriol. 1999;49:645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 8.Wilson E K, Bellelli A, Liberti S, Arese M, Grasso S, Cutruzzolà F, Brunori M, Brzezinski P. Biochemistry. 1999;38:7556–7564. doi: 10.1021/bi990179u. [DOI] [PubMed] [Google Scholar]
- 9.Silvestrini M C, Cutruzzolà F, D'Alessandro R, Brunori M, Fochesato N, Zennaro E. Biochem J. 1992;285:661–666. doi: 10.1042/bj2850661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutruzzolà F, Arese M, Grasso S, Bellelli A, Brunori M. FEBS Lett. 1997;412:365–369. doi: 10.1016/s0014-5793(97)00583-8. [DOI] [PubMed] [Google Scholar]
- 11.Tordi M G, Silvestrini M C, Colosimo A, Tuttobello L, Brunori M. Biochem J. 1985;230:797–805. doi: 10.1042/bj2300797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silvestrini M C, Colosimo A, Brunori M, Walsh T A, Barber D, Greenwood C. Biochem J. 1979;183:701–709. doi: 10.1042/bj1830701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonini G, Bellelli A, Brunori M, Falcioni G. Biochem J. 1996;314:533–540. doi: 10.1042/bj3140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tegoni M, Silvestrini M C, Lamzin V S, Brunori M, Cambillau C. J Mol Biol. 1994;243:347–350. doi: 10.1006/jmbi.1994.1659. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 17.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 18.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1991. p. 81. [Google Scholar]
- 19.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.George S J, Allen J W, Ferguson S J, Thorneley R N. J Biol Chem. 2000;275:33231–33237. doi: 10.1074/jbc.M005033200. [DOI] [PubMed] [Google Scholar]
- 21.Parr S R, Barber D, Greenwood C, Brunori M. Biochem J. 1977;167:447–455. doi: 10.1042/bj1670447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams P A, Fülöp V, Garman E F, Saunders N F W, Ferguson S J, Hajdu J. Nature (London) 1997;389:406–412. doi: 10.1038/38775. [DOI] [PubMed] [Google Scholar]
- 23.Jafferji A, Allen J W A, Ferguson S J, Fulop V. J Biol Chem. 2000;275:25089–25094. doi: 10.1074/jbc.M001377200. [DOI] [PubMed] [Google Scholar]
- 24.Silvestrini M C, Tordi M G, Musci G, Brunori M. J Biol Chem. 1990;265:11783–11787. [PubMed] [Google Scholar]
- 25.Stitt F, Coryell C D. J Am Chem Soc. 1939;61:1263–1266. [Google Scholar]
- 26.Barber D, Parr S R, Greenwood C. Biochem J. 1978;175:239–249. doi: 10.1042/bj1750239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H, Xia D, Yu C A, Xia J Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Proc Natl Acad Sci USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 29.Ranghino G, Scorza E, Sjogren T, Williams P, Ricci M, Hajdu J. Biochemistry. 2000;39:10958–10966. doi: 10.1021/bi000178y. [DOI] [PubMed] [Google Scholar]
- 30.Andersen J F, Montfort W R. J Biol Chem. 2000;275:30496–30503. doi: 10.1074/jbc.M002857200. [DOI] [PubMed] [Google Scholar]