Abstract
asdA mutants of Gram-negative bacteria have an obligate requirement for diaminopimelic acid (DAP), which is an essential constituent of the peptidoglycan layer of the cell wall of these organisms. In environments deprived of DAP, i.e., animal tissues, they will undergo lysis. Deletion of the asdA gene has previously been exploited to develop antibiotic-sensitive strains of live attenuated recombinant bacterial vaccines. Introduction of an Asd+ plasmid into a ΔasdA mutant makes the bacterial strain plasmid-dependent. This dependence on the Asd+ plasmid vector creates a balanced-lethal complementation between the bacterial strain and the recombinant plasmid. E. ictaluri is an enteric Gram-negative fish pathogen that causes enteric septicemia in catfish. Because E. ictaluri is a nasal/oral invasive intracellular pathogen, this bacterium is a candidate to develop a bath/oral live recombinant attenuated Edwardsiella vaccine (RAEV) for the catfish aquaculture industry. As a first step to develop an antibiotic-sensitive RAEV strain, we characterized and deleted the E. ictaluri asdA gene. E. ictaluri ΔasdA01 mutants exhibit an absolute requirement for DAP to grow. The asdA gene of E. ictaluri was complemented by the asdA gene from Salmonella. Several Asd+ expression vectors with different origins of replication were transformed into E. ictaluri ΔasdA01. Asd+ vectors were compatible with the pEI1 and pEI2 E. ictaluri native plasmids. The balanced-lethal system was satisfactorily evaluated in vivo. Recombinant GFP, PspA, and LcrV proteins were synthesized by E. ictaluri ΔasdA01 harboring Asd+ plasmids. Here we constructed a balanced-lethal system, which is the first step to develop an antibiotic-sensitive RAEV for the aquaculture industry.
Introduction
Aspartate β-semialdehyde deshydrogenase (Asd; EC 1.2.1.11), a highly conserved homodimeric enzyme encoded by the asd gene, is involved in the conversion of β-aspartyl phosphate to aspartate β-semialdehyde. Asd is an enzyme common to the biosynthesis of the essential amino acids lysine, threonine, methionine, and isoleucine. It also performs a key step in the production of diaminopimelic acid (DAP), a required component for the peptidoglycan synthesis of Gram-negative and some Gram-positive bacterial cell walls [1], [2], [3], [4] and an immediate precursor to lysine. asd mutants have an obligate requirement for DAP, and in the absence of DAP they undergo lysis. This has been demonstrated by gene-knockout studies with Legionella pneumophila [5], Salmonella Typhimurium [6] and Streptococcus mutans [7].
The Asd enzyme is also found in plants, where lysine is synthesized via the DAP pathway [8], [9]. In contrast, mammalian cells neither synthesize nor use DAP as a substrate in any metabolic pathway, and lysine is not synthesized since it is an essential amino acid that is obtained from dietary sources [5], [10], [11]. Also lysine, threonine, methionine, and isoleucine are essential amino acids in the diet of teleostei fish [12], [13], [14], [15], [16], [17], suggesting the absence of both the DAP/lysine synthesis pathway and Asd enzyme in fish cells.
Since DAP is absent from mammalian tissues, deletion of the asd gene has been exploited to develop a balanced-lethal system for vaccine delivery vehicles using a cloned asd gene as a selective marker in place of antibiotic-resistance markers, which are totally impractical in vivo [6]. Introduction of an Asd+ plasmid into asd mutants makes the bacterial strain plasmid-dependent. This dependence on the Asd+ plasmid vector creates a balanced-lethal complementation between the bacterial strain and the recombinant plasmid [18]. Asd+ vectors introduced into live recombinant attenuated Salmonella vaccines have been used to deliver heterologous antigens [19]. The construction of live attenuated recombinant bacterial vaccines not only require the absence of antibiotic-resistance markers in their recombinant plasmid, but also in their chromosomal deletions.
Edwardsiella ictaluri, a Gram-negative bacterial pathogen, is the cause of enteric septicemia in catfish, which causes losses estimated at $50–80 million annually [20]. The current USDA licensed vaccine, live E. ictaluri AQUAVAC-ESC® (Intervet Inc.), has been selected by multiple passages in increased concentrations of the antibiotic rifampicin [21], [22], [23]. The selected spontaneous mutant strain presented an attenuated phenotype missing part of the lipopolysaccharide (LPS) [20], [24]. Although there are FDA and USDA regulations against the use of antibiotic resistance in live attenuated bacterial vaccines for birds, mammals, and humans, the catfish industry currently allows antibiotic-resistant vaccine strains. Despite the fact that the current vaccine against enteric septicemia in catfish is antibiotic resistant, by using this vaccine we have learned that E. ictaluri live attenuated vaccines can be easily delivered to young fish and stimulate both humoral and cellular immunity of long duration [25], [26]. These results provide guidance to design live attenuated antibiotic-sensitive vaccines for the catfish aquaculture.
As a first step in developing an antibiotic-sensitive live recombinant E. ictaluri vaccine strain (RAEV), we adapted suicide vector technology [27] to E. ictaluri to construct defined unmarked chromosomal deletion mutations, for instance the asd deletion. Two E. ictaluri asd genes were identified, a functional asdA and a non-functional asdB pseudogene. The asdA gene was deleted by using the described suicide vector technology. Using Asd+ expression vectors [19], we developed a balance-lethal system compatible with E. ictaluri native plasmids, to express and secrete heterologous proteins through the type II secretion system. The virulence of the E. ictaluri ΔasdA mutant, harboring an AsdA+ expression vector, was evaluated in vivo in the catfish (Ictalurus punctatus) and in the zebrafish (Danio rerio) host models. Here we report the first balanced-lethal vector-host system in E. ictaluri, a key in constructing antibiotic-sensitive live RAEV for the catfish industry.
Results
Sequence analysis
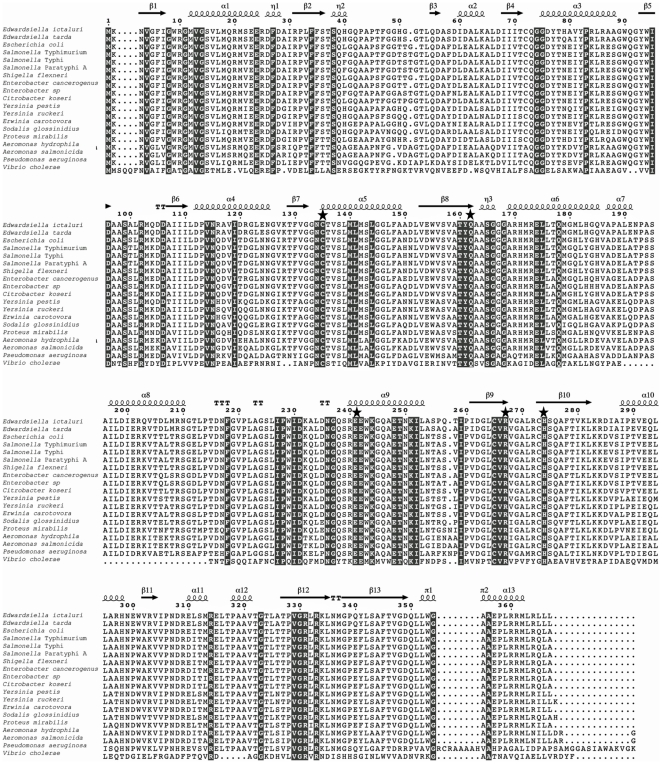
To develop a balanced-lethal system we first characterized the asd genes present in E. ictaluri. The genome of E. ictaluri has two asd gene sequences, asdA (gene ID 7960734) and asdB (gene ID 7959931). Sequence and structural alignment between functional representative bacterial Asd proteins reveals that 22 amino acid residues (∼6%) are strictly conserved out of 367 residues in E. ictaluri AsdA (Fig. 1). E. ictaluri AsdA has 28%, 81%, 82%, 84%, and 97% amino acid similarity to the Asd of Streptococcus mutans, Salmonella enterica, Escherichia coli, Yersinia (Y. pestis and Y. ruckeri), and E. tarda, respectively. The overall domain organization of E. ictaluri AsdA is similar to other Gram-negative Asd-family members, presenting an N-terminal domain comprising the NAD binding site and a C-terminal catalytic domain (Fig. 1). The same set of key functional groups in the active sites (Cys-135, Gln-162, Glu-241, Arg-267, and His-274) are conserved in E. ictaluri AsdA and likely have the same catalytic mechanism as other Asd enzymes (Fig. 1).
Figure 1. Sequence alignment among representative members of the AsdA family.
The secondary structure at the top of the alignment corresponds to the E. ictaluri AsdA enzyme (spirals represent α-helix; arrows represent β-sheet). Conserved amino acids residues are indicated in grey. The stars indicated the key catalytic active site residues (Cys-135, Gln-162, Glu-241, Arg-267, and His-274). The AsdA sequences were obtained from NCBI's Entrez Protein database for Edwardsiella ictaluri YP_002935083.1; Edwardsiella tarda YP_003297386.1; Escherichia coli AP_004358.1; Salmonella Typhi NP_807591.1; Salmonella Paratyphi A YP_152515.1; Salmonella Typhimurium AAB69392.1; Shigella flexnieri YP_690789.1; Shigella sonnei YP_312455.1; Citrobacter koseri YP_001456333.1; Enterobacter cancerogenus ZP_05969786.1; Enterobacter sp. YP_001178547.1; Yersinia pestis NP_671174.1; Yersinia ruckeri ZP_04615435.1; Proteus mirabilis YP_002152826.1; Aeromonas hydrophila ABK39477.1; Aeromonas salmonicida YP_001142146.1; Sodalis glossinidius YP_456010.1; Vibrio cholerae YP_001217562.1; Pseudomonas aeruginosa NP_251807.1; Erwinia carovora atrosepticum YP_052242.1.
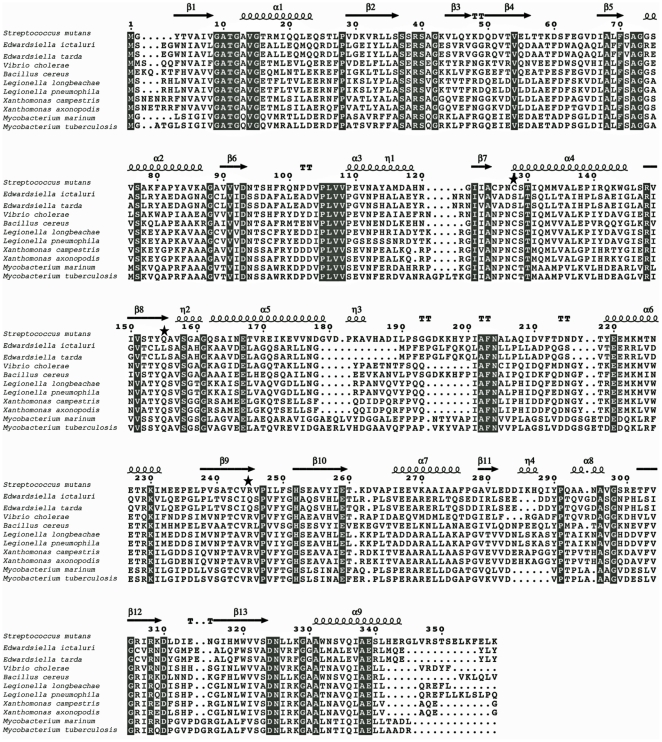
The sequence and structural alignment between representative bacterial AsdB proteins reveals that 52 amino acid residues (∼15%) are strictly conserved out of 336 in E. ictaluri AsdB (Fig. 2). The E. ictaluri AsdB has 30%, 32%, 40%, 75%, and 99% amino acid similarity to the AsdB of Streptococcus mutans, Mycobacterium marinum, Vibrio cholerae, Y. pestis, and E. tarda, respectively. In contrast to AsdA, the overall domain organization of E. ictaluri AsdB is similar to other Gram-positive Asd-family members. However, E. ictaluri AsdB lacks key functional groups in the active sites (Cys-135, Gln-162, and Arg-267) and likely has no catalytic activity.
Figure 2. Sequence alignment among representative members of the AsdB family.
The secondary structure at the top of the alignment corresponds to the S. mutans AsdB enzyme (spirals represent α-helix; arrows represent β-sheet). Conserved amino acids residues are indicated in grey. The stars indicated the key catalytic active site residues not present in AsdB from Edwardsiella. The AsdB sequences were obtained from NCBI's Entrez Protein database for Streptococcus mutans NP_721384.1; Edwardsiella ictaluri YP_002934124; Edwardsiella tarda YP_003296462; Vibrio cholerae YP_001217630.1; Bacillus cereus YP_085142.1; Legionella longbeachae CBJ10915; Legionella pneumophila YP_096311.1; Xanthomonas axonopodis NP_643032.1; Xanthomonas campestris NP_637897.1; Mycobacterium tuberculosis NP_218225.1; Mycobacterium marinum YP_001853481.1.
The guanine plus cytosine (G+C) content found in the E. ictaluri asdA gene was 62%, significantly higher than the 54% of G+C found in the Escherichia coli asdA gene. Overall DNA comparison of the asdA gene showed that the E. ictaluri asdA gene shared 72% identity with the Escherichia coli asdA gene.
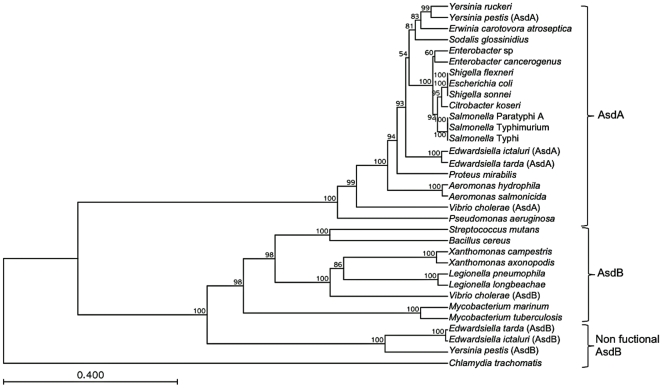
In terms of phylogeny, the bacterial Asd family is subdivided into two structural branches consisting of the enzymes from Gram-negative and Gram-positive bacteria [28] (Fig. 3). The E. ictaluri AsdA enzyme belongs to the Gram-negative branch, in contrast to AsdB that belongs to the Gram-positive branch (Fig. 3). Edwardsiella species comprise a linage that diverged from the ancestral trunk before the divergence of some other enteric bacteria, such as Salmonella and Escherichia [29], [30]. The phylogenetic position of the E. ictaluri AsdA enzyme corresponds with the E. ictaluri genome phylogenetic position (Fig. 3). Inside of the AsdB branch, a non-functional AsdB branch composed of Edwardsiella and Yersinia AsdB sequences was identified (Fig. 3), indicating that these non-functional AsdB proteins may have a common origin.
Figure 3. Phylogenetic tree constructed by the unweighted pair group method with arithmetic mean.
Bootstrap values indicate the number of times that a given node was detected out of 100. The Asd sequences were obtained from NCBI's Entrez Protein database for Edwardsiella ictaluri YP_002935083.1; Edwardsiella tarda YP_003297386.1; Escherichia coli AP_004358.1; Salmonella Typhi NP_807591.1; Salmonella Paratyphi A YP_152515.1; Salmonella Typhimurium AAB69392.1; Shigella flexnieri YP_690789.1; Shigella sonnei YP_312455.1; Citrobacter koseri YP_001456333.1; Enterobacter cancerogenus ZP_05969786.1; Enterobacter sp. YP_001178547.1; Yersinia pestis NP_671174.1; Yersinia ruckeri ZP_04615435.1; Proteus mirabilis YP_002152826.1; Aeromonas hydrophila ABK39477.1; Aeromonas salmonicida YP_001142146.1; Sodalis glossinidius YP_456010.1; Vibrio cholerae YP_002810714.1; Pseudomonas aeruginosa NP_251807.1; Erwinia carovora atrosepticum YP_052242.1; Streptococcus mutans NP_721384.1; Edwardsiella ictaluri YP_002934124; Edwardsiella tarda YP_003296462; Vibrio cholerae YP_001217630.1; Bacillus cereus YP_085142.1; Legionella longbeachae CBJ10915; Legionella pneumophila YP_096311.1; Xanthomonas axonopodis NP_643032.1; Xanthomonas campestris NP_637897.1; Mycobacterium tuberculosis NP_218225.1; Mycobacterium marinum YP_001853481.1; Chlamydia trachomatis YP_002887982.1.
Construction and characterization of asdA mutants
The construction of E. ictaluri ΔasdA mutants was performed first by using pEZ101, a pR112 (Cm) base suicide vector (Tables 1 and 2). pEZ101 was conjugated from Escherichia coli χ7213 to E. ictaluri J100 and E. ictaluri J102 using the methods described for E. ictaluri [31] and Escherichia coli [32]. The selection of transconjugants was carried out in BHI agar supplemented with Col, DAP, and Cm. We did not recover transconjugants by using pEZ101. Therefore, we constructed and used pEZ102, a pMEG-375 (Cm, Amp) base suicide vector (Table 2). The selection of transconjugants was carried out in BHI agar supplemented with Col, DAP, Amp or Cm. Transconjugants were recovered in the presence of Amp, but not in the presence of Cm. Transconjugants Ampr, harboring pEZ102 (Amp, Cm), were sensitive to Cm. We determined that E. ictaluri is highly sensitive to Cm. Small colonies (>0.5 mm) harboring pEZ102 were recovered in a Cm concentration below 1 µg/ml. Using BHI agar supplemented with Col, DAP, and Cm (1 µg/ml), transconjugants were not recovered using pEZ101 (Cm) or pEZ102 (Amp, Cm). Certainly, these results indicate that Cm selection and Cm-base suicide vectors are not useful to genetically manipulate E. ictaluri.
Table 1. Bacterial strains and plasmids.
| Strain | Relevant characteristics | Source or reference |
| Escherichia coli | ||
| χ6212 | F− Δ(argF-lacZYA)-U169 glnV44 l− deoR f80dlacZΔM15 gyrA96 recA1 relA1 endA1 ΔasdA4 Δ(zhf-2::Tn10) thi-1 hsdR17; Tetr | [60] |
| χ7213 | thr-1 leuB6 fhuA21 lacY1 glnV44 recA1 ΔasdA4 D(zhf-2::Tn10) thi-1 RP4-2-Tc::Mu [λpir]; Kmr | [53] |
| χ7232 | endA1 hsdR17 (rK-, mk+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 λpir deoR (f80dlacΔ(lacZ)M15) | Lab collection |
| Edwardsiella ictaluri | ||
| J100 | Wild-type; pEI1+; pEI2+ API20E 40040057; smooth LPS; Colr DAP+ | [59] |
| J102 | Wild-type; pEI1+; pEI2+ API20E 40040057; smooth LPS; Colr DAP+ | ATCC 33202 |
| J111 | J102 derivative; ΔasdA01; pEI1+; pEI2+ API20E 40040057; smooth LPS; Colr DAP− | This study |
| J112 | J100 derivative; ΔasdA01; pEI1+; pEI2+ API20E 40040057; smooth LPS; Colr DAP− | This study |
| Salmonella enterica | ||
| χ3761 | S. Typhimurium UK-1; wild-type | [19] |
| χ8958 | S. Typhimurium UK-1 ΔasdA33 | Lab collection |
| χ9112 | S. Typhi ISP1820 ΔasdA33 | Lab collection |
| χ9124 | S. Typhi Ty2 ΔasdA33 | Lab collection |
| Yersinia pestis | ||
| χ10006 | ΔasdA12 | Lab collection |
Table 2. Plasmid used in this study.
| Plasmids | Relevant characteristics | Source or reference |
| pYA248 | 3,000 bp, contains 1,071 bp of S. mutans asdA gene; p15A ori | [18] |
| pYA575 | 5,730 bp, contains ∼1,330 bp of S. mutans DNA inserted between the EcoRI and HindIII sites of pBR322 plasmid, Amp, Tet, pBR ori | [36] |
| pYA3341 | 2595 bp, plasmid Asd+; pUC ori | [19] |
| pYA3493 | 3113 bp, plasmid Asd+; pBR ori β-lactamase signal sequence-based periplasmic N- terminal secretion plasmid | [19] |
| pYA3620 | 3169 bp, plasmid Asd+; pBR ori β-lactamase signal sequence-based periplasmic N- and C- terminal secretion plasmid | [19] |
| pYA3994 | pBR ori, Asd+, GFP+ 3113 bp, | Lab collection |
| pYA3840 | 323 bp DNA encoding the LcrV in pYA3493 | [38] |
| pYA4088 | 852 bp DNA encoding the α-helical region of PspA aa 3-285 in pYA3493 | [39] |
| pRE112 | 5,173 bp, Cm, sacB, oriV, oriT | [27] |
| pMEG-375 | 8,142 bp, Cm, Amp, lacZ, R6K ori, mob incP, sacR sacB | [52] |
| pACYC184 | 4,245 bp, Tet, Cm, p15A ori | [54] |
| pEZ101 | ΔasdA01, pR112 | This study |
| pEZ102 | ΔasdA01, pMEG-375 | This study |
| pEZ140 | SD-asdA, Cm, pACYC184 | This study |
| pEZ142 | PasdA-asdA, Cm, pACYC184 | This study |
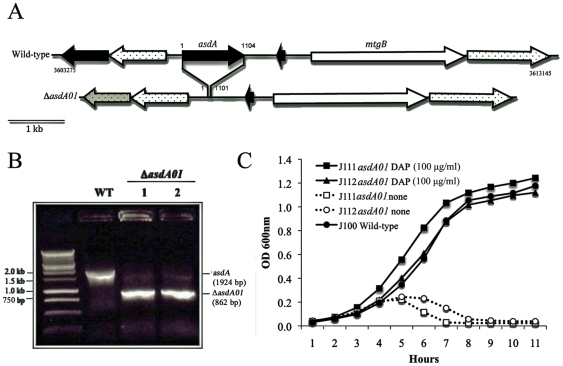
Single colonies of E. ictaluri transconjugants harboring pEZ102 (Colr, Ampr), were grown in BHI, TSB or LB supplemented with DAP and Col at 28°C for 6 h with aeration (180 r.p.m.). The selection was performed in BHI, TSA and LB agar plates supplemented with DAP, Col, and 5% sucrose at 28°C for 4–5 days. BHI sucrose selection agar did not provide selection, due to E. ictaluri overgrowth. TSA and LB sucrose selection agar presented a satisfactory selection. Positive mutants were screened for Colr, Amps, and DAP–. Several E. ictaluri ΔasdA mutants were recovered from TSA and LB sucrose-selection agar plates. The genotype was verified by PCR, and the phenotype by growth in presence of DAP and no growth in absence of DAP (Fig. 4). The biochemical profile, evaluated by API20E, did not present any difference between the wild type and ΔasdA01 mutant strains. E. ictaluri strains were identified as Edwardsiella sp (code 4004000). These results confirmed that the AsdB present in E. ictaluri is non-functional, since deletion of asdA is enough to preclude cell growth in the absence of DAP. Thus, asdB can be considered a pseudo gene in E. ictaluri.
Figure 4. Deletion of asdA gene in E. ictaluri.
A. Deletion map of ΔasdA01; B. Genotype verification of J112 ΔasdA01 by PCR; C. Phenotype of E. ictaluri J111 ΔasdA01 and J112 ΔasdA01 mutants. The strains were grown in BHI at 28°C with agitation (180 r.p.m.).
We evaluated reutilization of DAP by the E. ictaluri ΔasdA01 mutants released from lysed ΔasdA cells grown in absence of DAP. Washed cells of E. ictaluri J112 ΔasdA01 were diluted from 101 to 1010 CFU/ml in BHI Col. The estimated minimum number of E. ictaluri ΔasdA01 cells needed to support growth in absence of DAP was 1.3×108–2.7×108 CFU/ml. This is because of DAP-less death and reuse of DAP to permit growth on media without DAP.
The amount of DAP in the cell wall of Escherichia coli has been estimated at ∼3.5×106 molecules [33]. Based on the results obtained for the minimum number of E. ictaluri ΔasdA01 cells needed to support growth in the absence of DAP, and the calculated amount of DAP molecules per cell of Escherichia coli, we estimated that the minimum number of DAP molecules to support growth is ∼4.5×1014–9.5×1014 molecules of DAP/ml in the growth media. We evaluated the growth of E. ictaluri ΔasdA01 in 1010 to 1020 molecules of DAP/ml in BHI Col. E. ictaluri ΔasdA01 did not grow in concentrations below 1014 molecules of DAP/ml. Our previous estimation about the minimum number of DAP molecules required to support growth was confirmed, indicating that the amount of DAP in the cell wall of E. ictaluri is similar to Escherichia coli.
It has been reported that lysine, threonine, methionine, and isoleucine are essential amino acids in the diet of teleostei fish [12], [13], [14], [15], [16], [17], suggesting the absence of the DAP/lysine synthesis pathway in fish cells. We tested the growth of E. ictaluri J112 ΔasdA01 in different catfish broths (1% of catfish liver, spleen, kidney and meat in BHI) in presence and absence of DAP. E. ictaluri J112 ΔasdA01 was not able to grow in fish broth not supplemented with DAP. E. ictaluri J100 wild-type, used as control, grew in all fish broth conditions (data not shown). This result supports the idea that as mammalian cells, fish cells neither synthesize nor use DAP as substrate in any metabolic pathway.
Complementation of E. ictaluri asdA gene and E. ictaluri ΔasdA01 mutant
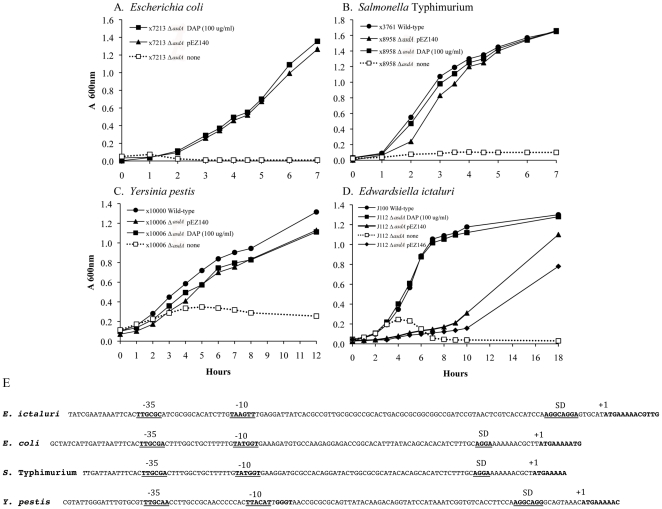
The structural analysis of E. ictaluri AsdA indicated that the overall domain organization is similar to other AsdA-family members and has the same set of key active-site functional groups and therefore the same catalytic mechanism as other Asd enzymes (Fig. 1). To evaluate the likely broad functionality of E. ictaluri AsdA enzyme, asdA mutants of Escherichia coli, Salmonella enterica (serovars Typhimurium, and Typhi), Y. pestis, and E. ictaluri, were complemented with the E. ictaluri asdA gene. Because overproduction of AsdA enzyme increases generation times [19], [34] and synthesis of Asd enzyme is proportional to the copy number of the complementing plasmid, asdA mutants were complemented with E. ictaluri asdA gene with (PasdA- asdA) and without its promoter (SD-asdA), this last to decrease Asd synthesis, cloned into p15A ori plasmid (pACYC184; Table 2).
Escherichia coli, S. enterica and Y. pestis ΔasdA mutants complemented with E. ictaluri SD-asdA presented similar growth rates compared to wild type (Fig. 5), indicating full complementation. E. ictaluri ΔasdA01 mutants complemented with SD-asdA presented a significantly lower growth rate than the wild type (Fig. 5). This could be due to overproduction or underproduction of Asd. It has been reported that SD-asd constructions do not enable ΔasdA strains to survive in absence of DAP if the origin of plasmid replication (ori) is from pSC101 or p15A. In other words, with these lower-copy-number replicons, the amount of Asd enzyme synthesized is insufficient to enable growth in absence of lysis [19]. To evaluate if the decrease in the generation time of the SD-asdA complemented E. ictaluri ΔasdA01 strain was due to overproduction or underproduction of AsdA, complementation with PasdA-asdA (pEZ142) was performed. Complementation of E. ictaluri ΔasdA01 mutants with pEZ142 decreased the growth rate even more than complementation with SD-asdA. These results suggest that the decreased growth rate in the E. ictaluri ΔasdA01 complemented with its own asdA gene is due to overproduction of AsdA. There are differences in the SD regions that could justify part of the difference in the growth rate of E. ictaluri ΔasdA01 complemented with its own SD-asd gene. The SD region of E. ictaluri asdA gene has an optimal spacing (6 nt) between the SD region and the ATG initiation codon of the mRNA [35] in contrast to the other bacterial species complemented with E. ictaluri SD-asdA (Fig. 5).
Figure 5. Complementation of representative ΔasdA mutant strains with E. ictaluri asdA gene.
(A–D) Growth of representative ΔasdA mutant strains complemented with asdA from E. ictaluri. pEZ140 (SD-asdA); pEZ142 (PasdA-asdA); The strains were grown in BHI at 28°C with agitation (180 r.p.m.); (E) Promoter region of asdA gene from E. ictaluri and representative strains.
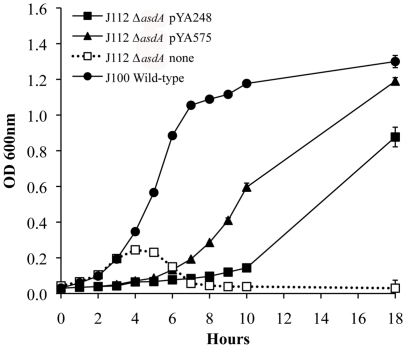
Complementation of E. ictaluri ΔasdA01 mutants by Gram-positive AsdB enzyme was also evaluated. Streptococcus mutans asdB region (including the full promoter), cloned into pYA575 [36] and S. mutans SD-asdB, cloned into pYA248 [18] complemented E. ictaluri ΔasdA01 mutants. However these strains presented lower growth rates than the wild type (Fig. 6). E. ictaluri ΔasdA01 mutants complemented with SD-asdB (pYA248), presented the lowest growth rate, suggesting that S. mutans AsdB is probably required in higher levels to fully complement E. ictaluri or S. mutans AsdB do not interact efficiently with E. ictaluri aspartokinase enzymes to transfer the β-aspartyl phosphate to Asd.
Figure 6. Growth of E. ictaluri ΔasdA01 complemented with asdB from Streptococcus mutans.
The strains were gown in BHI at 28°C with agitation (180 r.p.m.).
Complementation by Asd+ vectors to develop a balanced-lethal system in E. ictaluri
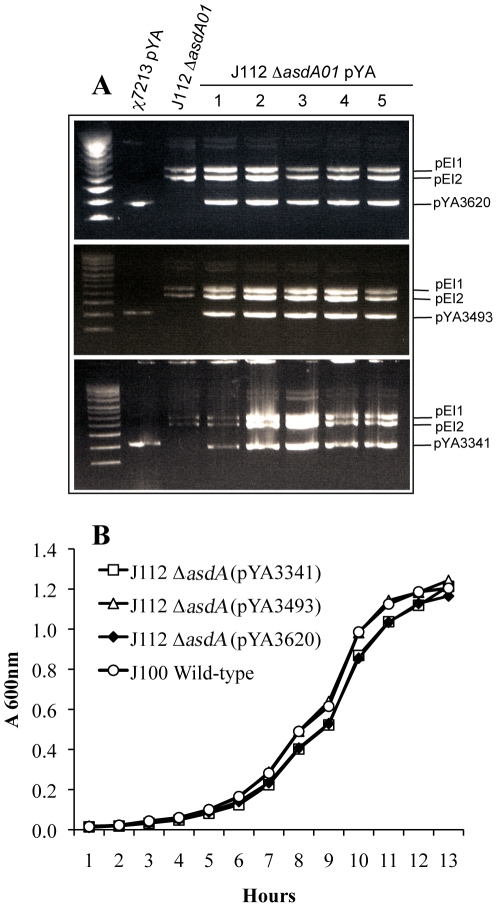
The asdA gene from E. ictaluri complemented S. enterica ΔasdA mutants, in addition the Asd enzymes from E. ictaluri and S. enterica share 81% similarity. Therefore, we used the Asd+ vectors utilized in live recombinant attenuated Salmonella vaccines [19] to develop a balanced-lethal system in E. ictaluri. The Asd+ vectors utilized in this study possess only the SD-asdA gene from S. Typhimurium with a modified start codon from ATG to GTG. E. ictaluri ΔasdA01 mutants were complemented with the asdA gene from S. Typhimurium (Fig. 7). The growth rate of E. ictaluri ΔasdA01 complemented with different copy number of Asd+ vectors was similar to the wild type in all cases (Fig. 7). The Asd+ vectors were compatible with the native plasmids of E. ictaluri (Fig. 7) and stable for at least 80 generations. These results show the first balanced-lethal system in E. ictaluri.
Figure 7. Complementation of asdA gene with Asd+ vectors.
(A) Plasmid profile of E. ictaluri ΔasdA01 complemented with AsdA+ vectors of different copy number. pEI1 (5.7 kb), pEI2 (4.9 kb), pYA3620 (3169 bp), pYA3493 (3113 bp), pYA3341 (2595 bp); Supercoiling ladder, from the top to the bottom: 16210 bp, 14174 bp, 12138 bp, 10102 bp, 8066 bp, 7045 bp, 6030 bp, 5012 bp, 3990 bp, 2972, 2067 bp; (B) Growth of E. ictaluri ΔasdA01 complemented with different AsdA+ vectors; The strains were grown in BHI at 28°C with agitation (180 r.p.m.).
Expression of genes encoding GFP protein in the AsdA+ vector
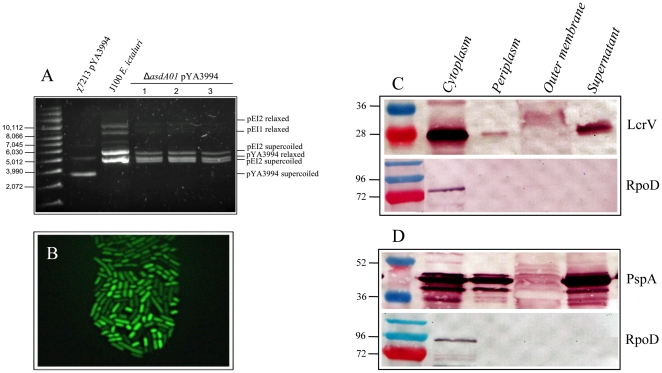
The synthesis of heterologous proteins, for instance GFP, cloned into Asd+ vectors was evaluated in E. ictaluri ΔasdA01 to potentially develop live E. ictaluri recombinant vaccines. First, the synthesis of heterologous proteins was evaluated by using the GFP+ Asd+ vector pYA3994 (Table 2). E. ictaluri ΔasdA01 mutant strains harboring the GFP+ Asd+ vector grew in absence of DAP and synthesized GFP+ as expected (Fig. 8). The GFP+ Asd+ vector was compatible with the native plasmids of E. ictaluri in the relaxed conformation (Fig. 8). The GFP+ Asd+ vector was stable in E. ictaluri ΔasdA01 strains for at least 80 generations. The expression of LcrV and PspA heterologous proteins using AsdA+ vectors was also evaluated (see below).
Figure 8. Synthesis of heterologous antigens in E. ictaluri J112 ΔasdA01 by using AsdA+ expression vectors.
A. Plasmid profile of J112 (pYA3994); B. Expression of GFP J112 (pYA3994); C. Expression and secretion of Y. pestis LcrV antigen by J112 (pYA3840); D. Expression and secretion of S. pneumoniae PspA-Rx1 antigen by J112 (pYA4088).
Secretion of heterologous proteins
Secretion of the heterologous antigens by live attenuated recombinant bacterial vaccines has been shown to enhance immunogenicity against the heterologous antigen [37]. The synthesis and secretion of heterologous proteins was evaluated by using the proteins derived from Gram-positive and Gram-negative bacterial strains. PspA-Rx1 from Streptococcus pneumoniae was utilized as a Gram-positive representative and LcrV from Yersinia pestis was utilized as a Gram-negative representative. The heterologous antigens, PspA-Rx1 and LcrV fused to β-lactamase signal sequence, were expressed from the Asd+ vectors pYA4088 and pYA3841, respectively (Table 2). Both heterologous proteins were secreted through the type II secretion system. No difference in the growth rate was observed between the recombinant E. ictaluri and the wild-type strain J100.
Virulence of E. ictaluri ΔasdA01 strain complemented with the AsdA+ plasmid vector in catfish host and zebrafish host models
The idea to develop a balanced-lethal system in a pathogenic bacterial strain is to synthesize heterologous antigens, without the use of antibiotic-resistant genes, in either the plasmid or in the bacterial chromosome. This is the first step towards developing live recombinant bacterial vaccines. The ideal balanced-lethal system should present nearly the same level of virulence as the wild-type strain with regard to invasion and colonization of lymphoid tissues. We evaluated the virulence of E. ictaluri ΔasdA01 mutants with and without the balanced-lethal system in the catfish and zebrafish hosts (Tables 3 and 4). We used pYA3493 AsdA+ since this vector has been used successfully in live recombinant Salmonella vaccines [19], [38], [39], [40]. E. ictaluri ΔasdA01 was attenuated at the high dose of 108 CFU, but still produced some mortality in catfish (Table 4). E. ictaluri ΔasdA01 at a high dose (108 CFU) was not attenuated in zebrafish and all the fish died (Table 3). However, at lower doses (107-104) E. ictaluri ΔasdA01 was totally attenuated in zebrafish (Table 3). E. ictaluri ΔasdA01 harboring the Asd+ vector pYA3493 increased the LD50 one log-fold, from 104 CFU to 105 CFU in orally infected catfish, and two log-fold, from 103 CFU to 105 CFU, in zebrafish (Tables 3 and 4). Catfish i.p. infected with E. ictaluri ΔasdA01 harboring the Asd+ vector pYA3493 presented the same level of virulence as E. ictaluri wild type (Table 4). From moribund orally infected catfish, E. ictaluri ΔasdA (pYA3493) AsdA+ was recovered from the head-kidney, spleen and liver, indicating that E. ictaluri ΔasdA (pYA3493) AsdA+ colonized these lymphoid tissues.
Table 3. Survival of zebrafish (D. rerio) infected with wild-type and E. ictaluri ΔasdA01 with and without Asd+ vectors.
| Experiment #1i.m. | Experiment #2i.m. | |||
| E. ictaluri strains | Dose (CFU/ml) | Survivors/Total | Dose (CFU/ml) | Survivors/Total |
| J100 wild-type | 1.5×108 | 0/25 | 1.2×107 | 0/25 |
| 1.5×106 | 0/25 | 1.2×103 | 13/25 | |
| 1.5×104 | 2/25 | 1.2×102 | 22/25 | |
| J112 ΔasdA01 | 3.0×108 | 0/10 | 1.7×107 | 1/5 |
| 1.7×106 | 10/10 | |||
| 1.7×104 | 10/10 | |||
| J112 ΔasdA01 (pYA3493) | 2.1×108 | 0/10 | 1.8×108 | 0/5 |
| 2.1×106 | 0/10 | 1.8×06 | 0/5 | |
| 2.1×104 | 10/10 | 1.8×104 | 5/5 | |
| BSG (Control) | None | 10/10 | None | 5/5 |
The zebrafish were infected i.m. with 10 µl of the respective E. ictaluri strain.
Table 4. Survival of catfish (I. punctatus) infected with E. ictaluri wild type and E. ictaluri ΔasdA01 with and without Asd+ vectors.
| Experiment #1i.p. | Experiment #2Oral | |||
| E. ictaluri strains | Dose (CFU/ml) | Survivors/Total | Dose (CFU/ml) | Survivors/Total |
| J100 wild-type | 1.5×108 | 0/6 | 1.2×108 | 1/7 |
| 1.5×106 | 0/6 | 1.2×106 | 2/7 | |
| 1.5×104 | 1/6 | 1.2×104 | 4/7 | |
| J112 ΔasdA01 | 3.0×108 | 3/7* | 1.7×108 | 7/8* |
| J112 ΔasdA01 (pYA3493) | 2.1×108 | 0/6 | 1.8×108 | 3/7 |
| 2.1×106 | 2/6 | 1.8×06 | 4/7 | |
| 2.1×104 | 2/6 | 1.8×104 | 5/7 | |
| BSG (Control) | None | 6/6 | None | 6/6 |
The catfish were infected i.p. with 100 µl and orally with 20 µl of the respective E. ictaluri strain.
*death within 48 h.
Discussion
To develop a balanced-lethal system in E. ictaluri, we first characterized the asdA and asdB genes present in the genome of E. ictaluri (Fig. 1). Deletion of the asdA gene precluded the growth of E. ictaluri in absence of DAP (Fig. 4), indicating that asdB does not encode for a functional protein related to DAP synthesis. This is consistent with the bioinformatic analysis (Fig. 2), which showed that the AsdB enzyme lacked several key amino acid residues at the catalytic active site.
The phylogeny of Asd has two branches, AsdA related with Gram-negatives and AsdB related with Gram-positives [28]. We found a particular group of non-functional AsdB genes in Edwardsiella and Yersinia. The common origin of AsdB in these bacteria suggests that the genes might have lost their activity through evolution, and that asdB could be considered a pseudogene in Edwardsiella and Y. pestis.
Suicide vector technology has been successfully used in several enteric bacteria to develop antibiotic-sensitive mutants [27]. Using this technology it was possible to construct defined deletion mutations in the absence of antibiotic-resistance markers for the first time in E. ictaluri (Fig. 3). During this process, we determined that E. ictaluri is extremely sensitive to Cm, even in the presence of the cat gene. The cat gene confers high-level resistance to Cm in most bacterial species. It codes for an enzyme called chloramphenicol acetyltransferase which inactivates Cm by covalently linking one or two acetyl groups, derived from acetyl-S-coenzyme A, to the hydroxyl groups on the chloramphenicol molecule [41]. This might indicate that chloramphenicol acetyltransferase is not functional or inefficient in E. ictaluri. Further studies are required to answer this.
The current live attenuated E. ictaluri vaccine is a rifampicin-resistant strain [22]. Antibiotic resistance in live attenuated bacterial vaccines present a threat to both the animal and to human health, due to the horizontal transmission of genes, in this case by transduction. Recently lytic bacteriophages have been isolated from catfish ponds against E. ictaluri [42]. This suggests that temperate phages for E. ictaluri that can establish lysogeny might be present in these environments and could spread rifampicin resistance to native environmental bacterial flora. Here we have described a methodology to genetically engineer E. ictaluri without the use of antibiotic-resistance genes in the final strain. This advancement opens up the field of E. ictaluri live attenuated vaccine development and will provide opportunities for further research into the pathogenesis of this important organism.
Although, E. ictaluri ΔasdA01 is complemented with its own asdA gene, the complemented strain did not grow at the same rate as the parental wild-type strain, presenting a higher growth rate. To achieve the right amount of native AsdA in E. ictaluri using Asd+ vectors requires further studies. However, E. ictaluri ΔasdA01 was fully complemented by the Salmonella SD-asdA gene, allowing the development of a balanced-lethal system.
One of the major difficulties in the construction of a balanced-lethal system in E. ictaluri is the incompatibility of the Asd+ vectors with cryptic plasmids present in the bacterial strain. E. ictaluri possesses two native autonomous small plasmids, pEI1 and pEI2 [43], that have been implicated in virulence [44]. The Asd+ expression vectors were compatible with pEI1 and pEI2 native plasmids of E. ictaluri, indicating that the origin of replication of these plasmids, ColE1 ori and ColE2 ori-like, respectively [43], are compatible with p15A ori, pBR ori and pUC ori.
E. ictaluri was described by Hawke in 1979 [45], and recently sequenced (NCBI's Entrez Genome database NC_012779). Most of its genes encode for putative functions. E. ictaluri possesses the machinery for the type II secretion system in its genome. Therefore we evaluated the secretion of proteins by using a β-lactamase signal sequence at the N-terminal end of a recombinant protein [19], a signal required for a protein to be secreted through the system mentioned above. Recombinant proteins, cloned in the AsdA+ vector and using the β-lactamase signal sequence, were secreted in a similar fashion (Fig. 8) as for a Salmonella recombinant vaccine [37], suggesting that the type II secretion system in E. ictaluri is fully functional.
Salmonella ΔasdA mutants are totally attenuated in mice orally infected with 108 CFU per dose [19]. E. ictaluri ΔasdA01 mutants were not fully attenuated in catfish i.p. or orally infected (Table 4). Zebrafish i.m. infected with doses of 108 CFU succumbed to E. ictaluri ΔasdA01 mutant infection (Table 3). Lower doses of E. ictaluri ΔasdA01 mutants (106-104 CFU) were totally attenuated (Table 3). It has been reported that E. ictaluri contain toxins, like hemolysin [46], [47]. We believe that the mortality caused by E. ictaluri ΔasdA01 mutants, either in catfish or zebrafish, is due to a toxic shock-like effect caused by the toxins realized after this DAP dependent mutant lyse in vivo. These toxins probably are not LPS related, since fish and amphibians are resistant to the toxic effects of LPS [48], [49]. E. ictaluri ΔasdA01 (pAsdA+) was attenuated by one log-fold in catfish animal host model (orally infected), and two log-fold in zebrafish. The next step in the construction and design of a live recombinant E. ictaluri vaccine is the attenuation of the bacterial strain without altering colonization of lymphoid tissues and immunogenicity. From moribund orally infected catfish, E. ictaluri ΔasdA (pAsdA+) were recovered from the head kidney, spleen and liver, indicating that E. ictaluri asdA (pAsdA+) colonize lymphoid tissues. The increase in attenuation in catfish orally infected with E. ictaluri ΔasdA (pAsdA+) could be used together with other genetic modifications to attenuate E. ictaluri in regard to constructing a live RAEV.
In summary, we have described methods to genetically engineer E. ictaluri without the use of antibiotic-resistant genes in the final strain. This opens up the field of RAEV development and will provide opportunities for further research into E. ictaluri pathogenesis. We have developed an antibiotic-sensitive recombinant E. ictaluri strain, using suicide vector technology [27] and Asd+ expression vectors [19]. This first balanced-lethal vector-host system in E. ictaluri is key in constructing antibiotic-sensitive live RAEV for the catfish industry.
Materials and Methods
Ethics statement
All research involving fish was conducted as per Protocol #09-1042R, approved by the Arizona State University Institutional Animal Care and Use Committee.
Bacterial strains, plasmids, media, and reagents
The bacterial strains and plasmids are listed in Table 1 and 2. Bacteriological media and components are from Difco (Franklin Lakes, NJ). Antibiotics and reagents are from Sigma (St. Louis, MO). LB broth (tryptone, 10 g; yeast extract 5 g; NaCl 10 g; dextrose 1 g, ddH2O 1 L) [50], Bacto-Brain Heart Infusion broth (BHI), and Trypticase Soy Broth (TSB), were used routinely. When required, the media were supplemented with 1.5% agar, 5% sucrose, colistin sulphate (Col; 12.5 µg/ml), ampicillin (Amp; 100 µg/ml), chloramphenicol (Cm; 25 µg/ml), or kanamycin (Km; 50 µg/ml). Fish broths were prepared with fresh homogenized catfish tissues (liver, spleen, kidney, and meat; catfish specific pathogen free were from University of Arkansas at Pine Bluff) to 1% in BHI and filter sterilized (0.22 µm). Bacterial growth was monitored spectrophotometrically and/or by plating. Oligonucleotides were from IDT (Coralville, IA). Restriction endonucleases were from New England Biolabs. Taq DNA polymerase (New England Biolabs) was used in all PCR tests. Qiagen products (Hilden, Germany) were used to isolate plasmid DNA, gel-purify fragments or purify PCR products. T4 ligase, T4 DNA polymerase and shrimp alkaline phosphatase (SAP) were from Promega.
Sequence analysis
Nucleotide Basic Local Alignment Search Tool (BLAST) was performed based on the sequences of the putative asd genes present in the genome sequence of E. ictaluri 93-146 accessed from NCBI's Entrez Genome database (NC_012779).
Asd sequences used were obtained from NCBI's Entrez Protein database. Amino acid sequence alignments were performed using the CLC Free Workbench software tool (v. 6.1 CLC bio A/S, Aarhus, Denmark). Protein structural-based alignments were performed by using the web-based interface for ESPript v.2.2 located at http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi [51]. Phylogenetic position of E. ictaluri AsdA protein was performed with CLC Free Workbench version using the unweighted pair group method with arithmetic mean (UPGMA). Bootstrap analysis was performed with 100 resamplings.
Construction and characterization of asdA mutants
The recombinant suicide vector pEZ102 (Table 2) carrying the linked flanking regions (5′ 361 bp and 3′ 422 bp) to generate an in-frame deletion of the asdA gene was constructed as described in [52]. The ΔasdA01 defined deletion mutation encompasses a 1,104 base pair deletion including the ATG start codon but not including the TAG stop codon. Primers (primer 1) 5′- ACATGCATGCAATGCCGTCAACGCCGCAGAAT-‘3 and (primer 2) 5′- CCGCTCGAGATGCACTCCTGCCTTGGATGGTGA -‘3 were designed to amplify the upstream asdA flanking region (361 bp). A SphI site was included in the primer 1 (underlined) and a XhoI site was included in primer 2 (underlined). The downstream asdA flanking region (422 bp) was amplified by primers (primer 3) 5′- CCGCTCGAGTGAGGCTACTGCTCTAGCCCGTGC -‘3 and (primer 4) 5′- TCGTCTAGAGCCAGATAGATTTGATGTTGTCTCTGCTGC -‘3. A XhoI site was included in primer 3 (underlined) and XbaI site was included in primer 4. The flanking regions were amplified from E. ictaluri J100, ligated, cloned into pRE112 and pMEG-375, and then digested with SphI and XbaI. The resulting plasmids were designated pEZ101 and pEZ102, respectively. To construct the E. ictaluri ΔasdA01 mutant, the suicide plasmid was conjugationally transferred from Escherichia coli χ7213 [53] to E. ictaluri wild-type strains J100 and J102. Strains containing single-crossover plasmid insertions (E. ictaluri asdA::pEZ102) were isolated on BHI agar plates containing Col, Amp, and DAP. Loss of the suicide vector after the second recombination between homologous regions (i.e., allelic exchange) was selected by using the sacB-based sucrose sensitivity counter-selection system [27]. The colonies were screened for Amps, Colr and for growth only in presence of DAP. DAP– colonies were screened by PCR using primer 1 and 4. Biochemical profiles of E. ictaluri strains were determined using the API 20E system (bioMériux, Marcy I'Etoile, France).
Complementation of asdA gene
The asdA gene of E. ictaluri, with and without its promoter, was cloned into a pAYCY184 vector [54] by inactivating the Tet cassette at the BamHI and XbaI restriction sites. The primers used to amplify asdA with its promoter (PasdA-asdA) were 5′ – TCGTCTAGATCTTGTAAGTTTGAGGATTA – 3′ (upstream) and 5′ – CGGGATCCTCAGCATGCGGCGCAACGGCTC – 3′ (downstream). An XbaI and BamHI site were included in these primers, respectively (underlined). To amplify the E. ictaluri Shine-Dalgarno (SD)-asdA promoter-less the upstream primer 5′ – TCGTCTAGAAGGCAGGAGTGCATATGAAAAA – 3′ was used with the downstream primer previously described. An XbaI site was included in this primer (underlined). The E. ictaluri promoter-less asdA includes the SD AGGA region, 6 bp upstream from the ATG start codon (SD-asdA). The resulting plasmids, pEZ140 (SD-asdA) and pEZ146 (Pasd-asdA) were used to complement different ΔasdA mutant strains. Also asd from Streptococcus mutans, cloned into pYA575 [36] and pYA248 [18], was used to evaluate complementation of E. ictaluri ΔasdA01 mutants.
To create a balanced-lethal system in E. ictaluri, several Asd+ expression vectors harboring the SD-asdA gene sequence from Salmonella Typhimurium UK-1 with different origins of replication, (Table 2) [19] were transformed into E. ictaluri ΔasdA01 to evaluate their complementation and stability. The growth rate of the complementing strains was evaluated in the absence of DAP. Plasmid stability was evaluated for fifty generations as described by Konjufca et al. [55].
Expression of heterologous antigens by E. ictaluri ΔasdA01
Asd+ expression vectors encoding different heterologous proteins (Table 2) were transformed into E. ictaluri ΔasdA01 to evaluate the expression and secretion of foreign proteins. First, the green fluorescent protein (GFP) was used to evaluate protein synthesis in the E. ictaluri ΔasdA01 strain. The vector pYA3994 AsdA+ GFP+ without a peptide secretion signal sequence was transformed into E. ictaluri ΔasdA01 (Table 2). The synthesis of GFP was evaluated by fluorescent microscopy. The synthesis of LcrV and PspA was evaluated by western blot and the secretion was evaluated by subcellular fractionation [37].
Western blot analysis
To evaluate the synthesis of heterologus proteins by E. ictaluri, the strains were grown in 3 ml of BHI at 28°C with aeration (180 r.p.m.). The samples were collected when the culture reached the absorbance of 1.0 (O.D600 1.0∼1×108 cfu/ml). One ml of culture was collected and prepared for Western blot analysis [56]. The total proteins were normalized by using a nanodrop spectrophotometer (ND-1000, NanoDrop) at 25 mg/µl and separated by 10% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes [56]. Fat-free milk powder solution (5%, wt/vol) in PBS supplemented with 0.05% of Tween 20 (PBS-T) was used for blocking. The membrane was incubated individually with a primary mouse anti-RpoD monoclonal antibody (1∶1,000) (Neoclone), rabbit anti-LcrV polyclonal antibody (1∶1,000) [57], or rabbit anti-PspA polyclonal antibody (1∶10,000) [40], for 1 h at room temperature, washed three times with PBS-T, and then incubated with a 1∶10,000 dilution of alkaline phosphatase-conjugated anti-mouse immunoglobulin G (IgG) (Sigma) or anti-rabbit immunoglobulin G (IgG) (Sigma). Color was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Amaresco).
Edwardsiella subcellular fractionation
Cultures were grown in BHI at 28°C static to an OD600 of 0.6 and centrifuged at 5,865 g for 10 min. Periplasmic fractions were prepared by a modification of the lysozyme-osmotic shock method [58] as previously described [37]. The supernatant fluid was saved for analysis of secreted proteins. Equal volumes of periplasmic, cytoplasmic, and supernatant fractions and total lysate samples were separated by SDS-PAGE for western blot analysis.
Determination of LD50 in zebrafish animal host
Zebrafish infections were performed by the methodology described by Petri-Hanson et al. [59] with modifications. The temperature of the water was 26±1°C and the fish were acclimated during 2 weeks prior to the start of the experimentation. Adult zebrafish (average weight, 0.5 g) were sedated in 100 mg/L tricaine methanesulfonate (MS-222, Sigma) and then injected intramuscularly (i.m.). Groups of zebrafish (typically 15 fish per group) were injected i.m. with 10 µl of the bacterial suspension (103–109 CFU) into each fish. A 3/10-cc U-100 ultrafine insulin syringe with a 0.5-in.-long (ca. 1-cm-long) 29-gauge needle (catalog no. BD-309301; VWR) was used to inject the fish. Two sets of controls were used: fish that were injected with 10 µl of sterile phosphate-buffered saline containing 0.01% gelatin (BSG) [60] and fish that were not injected. Moribund fish demonstrating clinical signs were euthanized, necropsied, and bacteria isolated as previously described [59]. The fish were fed twice daily with TetraMin Tropical Fish Flake Feed. During the experiments, the fish were observed daily, and every other day water quality was monitored for pH and NO2 with standard kits. The LD50 was calculated by the method of Reed-Muench [61]. Fish care and use was performed in accordance with the requirements of the Arizona State University, Institutional Animal Care and Use Committee.
Determination of LD50 in catfish animal host
Specific-pathogen-free channel catfish (Ictalurus punctatus) fingerlings were used with a mean weight of 18.5±1.3 g. The animals were randomly assigned to treatment groups of 6 to 8 fish each in 100 liter tanks. Each tank was equipped with a self-contained, recirculating, biofiltered, mechanical filtered, and U.V. water treated system with 12 h of illumination daily. The water temperature was 28±1°C during the 2 weeks of acclimatization and during the experiments. The fish were fed daily with commercial Aquamax grower 400 (Purina Mills Inc., St. Louis, MO). During the experiments, the fish were observed daily, and every other day water quality was monitored for pH and NO2 with standards kits. Catfish were infected with 103 to 109 CFU of E. ictaluri strains (fish were not fed until 1 h after infection) orally and intra peritoneal (i.p.). The fish were anesthetized with tricaine methanesulfonate (MS-222, Sigma; 100 mg/L of water) prior to handling. The LD50 was calculated by the method of Reed-Muench [61]. Moribund animals were necropsied to evaluate presence of E. ictaluri in kidney, spleen and liver. Fish care and use was performed in accordance with the requirements of the Arizona State University, Institutional Animal Care and Use Committee.
Bacteria preparation
Bacterial strains were grown overnight in standing cultures that were diluted 1∶20 in prewarmed BHI broth and grown with mild aeration (180 r.p.m.) at 28°C to an OD600 of 0.8 to 0.9 (∼108 CFU/ml). Bacteria were sedimented 10 min by centrifugation (5,865 g) at room temperature and resuspended in BSG [60] to densities appropriate for the inoculation.
Acknowledgments
We thank Greg Golden for his assistance with the manuscript editing. We also thank Dr. Wei Sun for his assistance with the manipulation of Y. pestis strains, and Rebin Kader, Dr. Maria Dolores Juarez-Rodriguez and Dr. Ascensión Torres-Escobar for their suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was supported by United States Deparment of Agriculture (USDA) grant CRIS-ARZR-2009-01801 and Comisión Nacional de Investigación Científica y Tecnológica (CONICYT), Gestión Propia Fellowship, Chile.
References
- 1.Paidhungat M, Setlow B, Driks A, Setlow P. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol. 2000;182:5505–5512. doi: 10.1128/jb.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavelka MS, Jr, Jacobs WR., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. . J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viola RE. The central enzymes of the aspartate family of amino acid biosynthesis. Acc Chem Res. 2001;34:339–349. doi: 10.1021/ar000057q. [DOI] [PubMed] [Google Scholar]
- 4.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harb OS, Abu Kwaik Y. Identification of the aspartate-beta-semialdehyde dehydrogenase gene of Legionella pneumophila and characterization of a null mutant. Infect Immun. 1998;66:1898–1903. doi: 10.1128/iai.66.5.1898-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galan JE, Nakayama K, Curtiss R., 3rd Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene. 1990;94:29–35. doi: 10.1016/0378-1119(90)90464-3. [DOI] [PubMed] [Google Scholar]
- 7.Cardineau GA, Curtiss R., 3rd Nucleotide sequence of the asd gene of Streptococcus mutans. Identification of the promoter region and evidence for attenuator-like sequences preceding the structural gene. J Biol Chem. 1987;262:3344–3353. [PubMed] [Google Scholar]
- 8.Vogel HJ. On biochemical evolution: lysine formation in higher plants. Proc Natl Acad Sci U S A. 1959;45:1717–1721. doi: 10.1073/pnas.45.12.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson AO, Bless C, Macedo P, Chatterjee SP, Singh BK, et al. Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways. Biochim Biophys Acta. 2005;1721:27–36. doi: 10.1016/j.bbagen.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Burns-Keliher LL, Portteus A, Curtiss R., 3rd Specific detection of Salmonella typhimurium proteins synthesized intracellularly. J Bacteriol. 1997;179:3604–3612. doi: 10.1128/jb.179.11.3604-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cersini A, Salvia AM, Bernardini ML. Intracellular multiplication and virulence of Shigella flexneri auxotrophic mutants. Infect Immun. 1998;66:549–557. doi: 10.1128/iai.66.2.549-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halver JE, Delong DC, Mertz ET. Nutrition of salmonoid fishes.V. Classification of essential amino acids for chinook salmon. J Nut. 1957;63:95–105. doi: 10.1093/jn/63.1.95. [DOI] [PubMed] [Google Scholar]
- 13.Halver JE, Shanks WE. Nutrition of salmonoid fishes. VIII. Indispensable amino acids for sockeye salmon. J Nut. 1960;72:340–346. doi: 10.1093/jn/72.3.340. [DOI] [PubMed] [Google Scholar]
- 14.Dupree HK, Halver JE. Amino acids essential for growth of channel catfish, Ictalurus punctatus. Trans Am Fish Soc. 1970;99:90–100. [Google Scholar]
- 15.Nose T, Arai S, Lee DL, Hashimot Y. Note on amino-acids essential for growth of young carp. Bull Jap Soc Sci Fish. 1974;40:903–908. [Google Scholar]
- 16.Mazid MA, Tanaka Y, Katayama T, Simpson KL, Chichester CO. Metabolism of amino-acids in aquatic animals.III. Indispensable amino-acids for tilapia-zillii. Bull Jap Soc Sci Fish. 1978;44:739–742. [Google Scholar]
- 17.Li MH, Robinson EH. Effects of supplemental lysine and methionine in low protein diets on weight gain and body composition of young channel catfish Ictalurus punctatus. Aquaculture. 1998;163:297–307. [Google Scholar]
- 18.Nakayama K, Kelly SM, Curtiss R., 3rd Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotechnol. 1988;6:693–697. [Google Scholar]
- 19.Curtiss RI, Zhang X, Wanda SY, Kang HY, Konjufca V, et al. Induction of host immune responses using Salmonella-vectored vaccines; In: Brogden KA, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemuehler MJ, editors. Washington, D.C: ASM Press; 2007. [Google Scholar]
- 20.Russo R, Shoemaker CA, Panangala VS, Klesius PH. In vitro and in vivo interaction of macrophages from vaccinated and non-vaccinated channel catfish (Ictalurus punctatus) to Edwardsiella ictaluri. Fish Shellfish Immunol. 2009;26:543–552. doi: 10.1016/j.fsi.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Arias CR, Shoemaker CA, Evans JJ, Klesius PH. A comparative study of Edwardsiella ictaluri parent (EILO) and E. ictaluri rifampicin-mutant (RE-33) isolates using lipopolysaccharides, outer membrane proteins, fatty acids, Biolog, API 20E and genomic analyses. J Fish Dis. 2003;26:415–421. doi: 10.1046/j.1365-2761.2003.00475.x. [DOI] [PubMed] [Google Scholar]
- 22.Klesius PH, Shoemaker CA. Development and use of modified live Edwardsiella ictaluri vaccine against enteric septicemia of catfish. Adv Vet Med. 1999;41:523–537. doi: 10.1016/s0065-3519(99)80039-1. [DOI] [PubMed] [Google Scholar]
- 23.Shoemaker CA, Klesius PH, Evans JJ. Immunization of eyed channel catfish, Ictalurus punctatus, eggs with monovalent Flavobacterium columnare vaccine and bivalent F. columnare and Edwardsiella ictaluri vaccine. Vaccine. 2007;25:1126–1131. doi: 10.1016/j.vaccine.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence M, Banes M, Williams M. Phenotype and virulence of a transposon- derived lipopolysaccharide O side-chain mutant of Edwardsiella ictaluri. J Aquat Anim Health. 2001;13:291–299. [Google Scholar]
- 25.Shoemaker CA, Klesius PH, Arias CR, Evans JJ. Uses of modified live vaccines in the aquaculture. J World Aqua Soc. 2008;40:573–585. [Google Scholar]
- 26.Shoemaker CA, Klesius PH, Bricker JM. Efficacy of a modified live Edwardsiella ictaluri vaccine in channel catfish as young as seven days post hatch. Aquaculture. 1999;176:189–193. [Google Scholar]
- 27.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 28.Viola RE, Liu X, Ohren JF, Faehnle CR. The structure of a redundant enzyme: a second isoform of aspartate beta-semialdehyde dehydrogenase in Vibrio cholerae. Acta Crystallogr D Biol Crystallogr. 2008;64:321–330. doi: 10.1107/S0907444907068552. [DOI] [PubMed] [Google Scholar]
- 29.Brenner DJ, Fanning GR, Knutson JKL, Steigerwalt AG, Krichevsky MI. Attempts to classify herbicola group-enterobacter-agglomerans strains by deoxyribonucleic-acid hybridization and phenotypic tests. Int J Syst Bacteriol. 1984;34:45–55. [Google Scholar]
- 30.Wang QY, Yang MJ, Xiao JF, Wu HZ, Wang X, et al. Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. Plos One. 2009;4:e7646. doi: 10.1371/journal.pone.0007646. doi: 7610.1371/journal.pone.0007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer KJ, Lawrence ML, Fernandez DH, Thune RL. Evaluation and Optimization of a DNA Transfer System for Edwardsiella ictaluri. J Aquat Anim Health. 2001;13:163–167. [Google Scholar]
- 32.Miller JH. New York: Cold Spring Harbor; 1972. Experiments in Molecular Genetics. [Google Scholar]
- 33.Wientjes FB, Pas E, Taschner PE, Woldringh CL. Kinetics of uptake and incorporation of meso-diaminopimelic acid in different Escherichia coli strains. J Bacteriol. 1985;164:331–337. doi: 10.1128/jb.164.1.331-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HY, Curtiss R., 3rd Immune responses dependent on antigen location in recombinant attenuated Salmonella typhimurium vaccines following oral immunization. FEMS Immunol Med Microbiol. 2003;37:99–104. doi: 10.1016/S0928-8244(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagusztyn-Krynicka EK, Smorawinska M, Curtiss R., 3rd Expression of Streptococcus mutans aspartate-semialdehyde dehydrogenase gene cloned into plasmid pBR322. J Gen Microbiol. 1982;128:1135–1145. doi: 10.1099/00221287-128-5-1135. [DOI] [PubMed] [Google Scholar]
- 37.Kang HY, Srinivasan J, Curtiss R., 3rd Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun. 2002;70:1739–1749. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branger CG, Fetherston JD, Perry RD, Curtiss R., 3rd Oral vaccination with different antigens from Yersinia pestis KIM delivered by live attenuated Salmonella typhimurium elicits a protective immune response against plague. Adv Exp Med Biol. 2007;603:387–399. doi: 10.1007/978-0-387-72124-8_36. [DOI] [PubMed] [Google Scholar]
- 39.Xin W, Wanda SY, Li Y, Wang S, Mo H, et al. Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect Immun. 2008;76:3241–3254. doi: 10.1128/IAI.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi H, Santander J, Brenneman KE, Wanda SY, Wang S, et al. Live recombinant Salmonella Typhi vaccines constructed to investigate the role of rpoS in eliciting immunity to a heterologous antigen. Plos One. 2010;5:e11142. doi: 10.1371/journal.pone.0011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leslie AG, Moody PC, Shaw WV. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A. 1988;85:4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walakira JK, Carrias AA, Hossain MJ, Jones E, Terhune JS, et al. Identification and characterization of bacteriophages specific to the catfish pathogen, Edwardsiella ictaluri. J Appl Microbiol. 2008;105:2133–2142. doi: 10.1111/j.1365-2672.2008.03933.x. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez DH, Pittman-Cooley L, Thune RL. Sequencing and analysis of the Edwardsiella ictaluri plasmids. Plasmid. 2001;45:52–56. doi: 10.1006/plas.2000.1499. [DOI] [PubMed] [Google Scholar]
- 44.Thune RL, Fernandez DH, Benoit JL, Kelly-Smith M, Rogge ML, et al. Signature-tagged mutagenesis of Edwardsiella ictaluri identifies virulence-related genes, including a Salmonella pathogenicity island 2 class of type III secretion systems. Appl Environ Microbiol. 2007;73:7934–7946. doi: 10.1128/AEM.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawke JP, McWhorter AC, Steigerwalt AG, Brenner DJ. Edwardsiella ictaluri sp. nov., the causative agent of enteric septicemia of catfish. Int J Syst Bacteriol. 1981;31:396–340. [Google Scholar]
- 46.Williams ML, Azadi P, Lawrence ML. Comparison of cellular and extracellular products of virulent and attenuated strains of Edwardsiella ictaluri. J Aquat Anim Health. 2003;15:264–273. [Google Scholar]
- 47.Williams ML, Lawrence ML. Identification and characterization of a two-component hemolysin from Edwardsiella ictaluri. Vet Microbiol. 2005;108:281–289. doi: 10.1016/j.vetmic.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 48.Berczi I, Bertok L, Bereznai T. Comparative studies on the toxicity of Escherichia coli lipopolysaccharide endotoxin in various animal species. Can J Microbiol. 1966;12:1070–1071. doi: 10.1139/m66-143. [DOI] [PubMed] [Google Scholar]
- 49.Iliev DB, Roach JC, Mackenzie S, Planas JV, Goetz FW. Endotoxin recognition: in fish or not in fish? FEBS Lett. 2005;579:6519–6528. doi: 10.1016/j.febslet.2005.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 52.Santander J, Wanda SY, Nickerson CA, Curtiss R., 3rd Role of RpoS in fine-tuning the synthesis of Vi capsular polysaccharide in Salmonella enterica serotype Typhi. Infect Immun. 2007;75:1382–1392. doi: 10.1128/IAI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roland K, Curtiss R, 3rd, Sizemore D. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43:429–441. [PubMed] [Google Scholar]
- 54.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konjufca V, Wanda SY, Jenkins MC, Curtiss R., 3rd A recombinant attenuated Salmonella enterica serovar Typhimurium vaccine encoding Eimeria acervulina antigen offers protection against E. acervulina challenge. Infect Immun. 2006;74:6785–6796. doi: 10.1128/IAI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Russell W. Cold Spring Harbor Press; 2001. Molecular Cloning; A Laboratory Manual. Editor: [Google Scholar]
- 57.Branger CG, Sun W, Torres-Escobar A, Perry R, Roland KL, et al. Evaluation of Psn, HmuR and a modified LcrV protein delivered to mice by live attenuated Salmonella as a vaccine against bubonic and pneumonic Yersinia pestis challenge. Vaccine. 2010. In Press, Available online 24 October 2010. [DOI] [PMC free article] [PubMed]
- 58.Witholt B, Boekhout M, Brock M, Kingma J, V Heerikhuizen H, et al. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]
- 59.Petrie-Hanson L, Romano CL, Mackey RB, Khosravi P, Hohn CM, et al. Evaluation of zebrafish Danio rerio as a model for enteric septicemia of catfish (ESC). J Aquat Anim Health. 2007;19:151–158. doi: 10.1577/H06-026.1. [DOI] [PubMed] [Google Scholar]
- 60.Curtiss R., III Chromosomal aberrations associated with mutations to bacteriophage resistance in Escherichia coli. J Bacteriol. 1965;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson DP. Fish Immunology. In: Hua SZaD., editor. Fish Immunology. Beijin: China Agriculture Press; 1984. pp. 110–113. [Google Scholar]