Abstract
OBJECTIVE
Insulin receptor (IR) translocates to the nucleus, but its recruitment to gene loci has not been demonstrated. Here, we tested the hypothesis that IR and its downstream mitogenic transducers are corecruited to two prototypic insulin-inducible genes: early growth response 1 (egr-1), involved in mitogenic response, and glucokinase (Gck), encoding a key metabolic enzyme.
RESEARCH DESIGN AND METHODS
We used RNA and chromatin from insulin-treated rat hepatic tumor cell line expressing human insulin receptor (HTC-IR) and livers from lean and insulin-resistant ob/ob glucose-fed mice in quantitative RT-PCR and chromatin immunoprecipitation studies to determine gene expression levels and associated recruitment of RNA polymerase II (Pol II), insulin receptor, and cognate signaling proteins to gene loci, respectively.
RESULTS
Insulin-induced egr-1 mRNA in HTC-IR cells was associated with corecruitment of IR signaling cascade (IR, SOS, Grb2, B-Raf, MEK, and ERK) to this gene. Recruitment profiles of phosphorylated IR, B-Raf, MEK, and Erk along egr-1 transcribed region were similar to those of elongating Pol II. Glucose-feeding increased Gck mRNA expression in livers of lean but not ob/ob mice. In lean mice, there was glucose feeding-induced recruitment of IR and its transducers to Gck gene synchronized with elongating Pol II. In sharp contrast, in glucose-fed ob/ob mice, the Gck recruitment patterns of active MEK/Erk, IR, and Pol II were asynchronous.
CONCLUSIONS
IR and its signal transducers recruited to genes coupled to elongating Pol II may play a role in maintaining productive mRNA synthesis of target genes. These studies suggest a possibility that impaired Pol II processivity along genes bearing aberrant levels of IR/signal transducers is a previously unrecognized facet of insulin resistance.
The insulin receptor (IR) is a member of a family of receptor tyrosine kinases (RTKs) that include the epidermal growth factor receptor, fibroblast growth factor receptor, and several others (1). Studies have shown that RTKs are translocated to the nucleus upon stimulation, including fibroblast growth factor receptor (2,3) and epidermal growth factor receptor (4). Importantly, there is evidence for recruitment of RTKs to chromatin and gene loci, and for several RTKs the recruitment increases transcription (2,5,6,7). RTKs are not the only kinases found along genes. For instance, active ERK1/2, MEK, p38, and AMPK are also recruited to genes (8,9,10,11).
IR also translocates to the nucleus after binding its ligand (12,13,14). For instance, mice fed a glucose meal showed an increase in nuclear IR, which correlated with the glucose-stimulated rise of serum insulin (13,15). The fact that RTKs and the terminal mitogen-activated protein kinases (MAPKs) can be inducibly recruited to chromatin presents the possibility that canonical insulin signaling pathways are recapitulated along gene loci. Here, we provide the first evidence that not only the IR but also most of the ERK cascade components, as well as ERK dual-specificity phosphatase (16), are corecruited to inducible genes.
RESEARCH DESIGN AND METHODS
Reagents.
BSA (cat. no. A2153), salmon sperm DNA (cat. no. D1626), and protein A (cat. no. P7837) were from Sigma, proteinase K was from Invitrogen (cat. no. 25530-015), and Humulin N was from Eli Lilly. Matrix chromatin immunoprecipitation (ChIP) 96-well polystyrene plates were from Corning (Costar cat. no. 9018), and polypropylene plates were from Bioexpress (T-3060).
Tissue culture and insulin treatment.
HTC and HTC-IR cells were maintained and insulin treated as in ref (17) with the exception that 1 × 10−8 M insulin was used.
Mice and glucose feeding experiments.
Male C57BL/6 and ob/ob mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and were maintained, glucose fed, and killed as described previously (13). Blood was collected just prior to sacrifice. Livers were removed after whole-animal perfusion with sterile, cold PBS and flash frozen in liquid nitrogen. Blood glucose was measured using One Touch Ultra system (LifeScan Inc.). Plasma insulin levels were measured using the Linco insulin ELISA (Millipore). All procedures were done in accordance with current National Institutes of Health guidelines and approved by the Animal Care and Use Committee of the University of Washington.
RNA extraction and cDNA synthesis.
RNA was extracted from cell pellets or tissue fragments using Trizol reagent as per the manufacturer's protocol. To synthesize cDNA, 400 ng of Trizol extracted total RNA was used in reverse transcription reactions with 200 units of MMLV reverse transcriptase (Invitrogen) and random hexamers. RT reactions were diluted 100-fold prior to running quantitative PCR (18).
Matrix ChIP.
Chromatin shearing was done using either Misonix 3000 microprobe (1 ml IP buffer [8], six rounds of sonication power 5, 15 s, on ice) or Diagenode Bioruptor (100 μl IP buffer, 30 rounds 30 s ON/30 s OFF, high power, 4°C). The suspension was cleared by centrifugation at 12,000g (10 min at 4°C), and the supernatant, representing sheared chromatin, was aliquoted and stored at −80°C. ChIP assays were done using the Matrix ChIP platform in 96-well microplates as described before (8,19). ChIP DNA samples were assayed by quantitative PCR. PCR calibration curves were generated for each primer pair from a dilution series of total rat or mouse sheared genomic DNA. The PCR primer efficiency curve was fit to cycle threshold versus log (genomic DNA dilutions) using an r2 best fit. DNA concentration values for each ChIP and input DNA samples were calculated from their respective average cycle threshold values. Final results are expressed as a fraction of input DNA (8). Matrix ChIP PCR primers are shown in supplementary Table 1 and the list of antibodies are shown in supplementary Table 2, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-1806/DC1.
RESULTS
Insulin stimulates egr-1 transcription in hepatocyte culture.
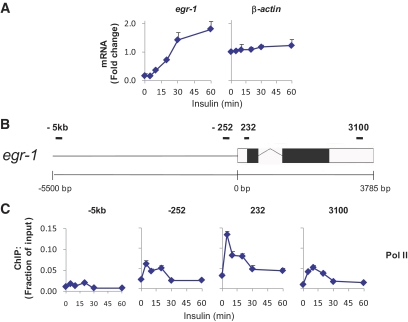
We examined expression of the mitogen-sensitive immediate early gene, early growth response 1 (egr-1), as a model of insulin-induced transcription in rat HTC-IR cells overexpressing human IR (20). Figure 1A (left) shows that the egr-1 transcript increased by >10-fold after 60 min of insulin treatment. β-actin transcript levels did not change (Fig. 1A, right).
FIG. 1.
Insulin-induced early growth response 1, egr-1, expression, and Pol II recruitment in HTC-IR cells overexpressing IR. A: Serum-deprived hepatocyte HTC-IR cells were treated with insulin (10−8 M) for 5, 10, 20, 30, and 60 min or left untreated. Total cellular RNA from these cells was used in RT reactions with random hexamers. cDNAs were used in real-time PCR with primers to the last exon of egr-1 or the first exon of gapdh. Data represent the mean ± SEM of three independent experiments. B: Schematic of the egr-1 locus. Boxes represent exons (black boxes are translated regions and white boxes are untranslated regions), the zigzag line represents an intron, and the straight line represents upstream sequence. Regions amplified by egr-1 primers are represented by black bars. C: Cells were treated as in (A), cross-linked, and then used in ChIP with antibodies to Pol II. ChIPed DNA was used in PCR with primers to −5.3 kb, −252 bp, +232 bp, and +3.1 kb with respect to the TSS of egr-1. Data represent the mean ± SEM for three independent experiments. (A high-quality color representation of this figure is available in the online issue.)
To determine whether the increase in egr-1 transcript was due to increased transcription, Matrix ChIP (8) for polymerase II (Pol II) was used with primers to or near the egr-1 locus. Pol II density increased within the body of the gene beginning as early as 5 min after treatment (Fig. 1C). Peak Pol II binding at the end of egr-1 coincided with the beginning of the increase in its transcript, 10 min after insulin treatment (Fig. 1A, left panel), suggesting that transcription upregulation was at least partly responsible for the induction of egr-1 transcript.
Insulin induces recruitment of the IR transducers to the egr-1 locus in hepatocyte culture.
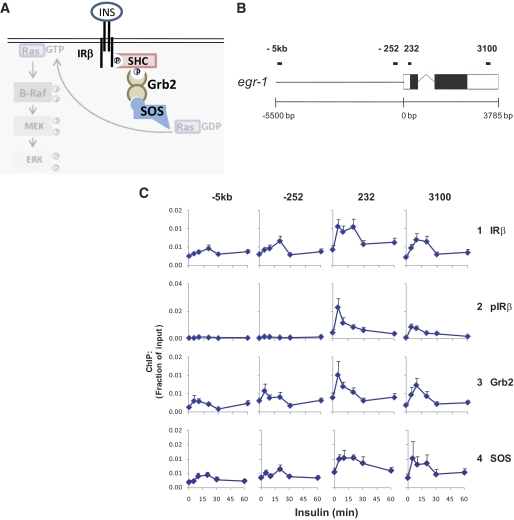
We used antibodies to the β-subunit of IR with Matrix ChIP to determine whether IR was recruited to egr-1 in insulin-treated HTC-IR cells. IR was recruited to egr-1 within the transcribed region, beginning 5 min after insulin treatment and remaining at the same level for 15 min (Fig. 2C, row 1).
FIG. 2.
Insulin receptor signaling complex at the egr-1 locus in HTC-IR cells. A: Schematic of one arm of the IR signaling pathway including some components of the IR signaling complex (highlighted) and MAPK pathway components. B: Schematic of the egr-1 locus. C: Cells were treated as in Fig. 1A, cross-linked, and used in ChIP with antibodies to the β subunit of IR (IR-β), IR-β phosphorylated at Y1146 (pIR), Grb2, and SOS. ChIPed DNA was used in PCR as in Fig. 1C. Data represent mean ± SEM for three independent experiments.
To determine whether the recruited IR was activated, we used Matrix ChIP with an antibody to IR phosphorylated at tyrosine 1146, a site autophosphorylated upon binding of the receptor to insulin (21). Interestingly, phospho-IR had a large transient increase at both sites within the coding region of egr-1, with kinetics similar to those of Pol II (Fig. 2C, row 2). The insulin-induced total and phospho-IR ChIP signals were much less intense in the rat parental HTC cell line without human IR (20), while the histone H3 lysine 27 trimethylation, H3K27m3, ChIP signals were the same (supplementary Fig. 1, available in an online appendix). These results confirm that the antibodies detect total and phospho-IR. The more robust insulin-induced activation of IR at egr-1 in HTC-IR cells paralleled higher levels of recruitment of Pol II and phospho-Erk (supplementary Fig. 1), suggesting that these insulin-induced changes were linked.
Upon activation and autophosphorylation of IR, the insulin receptor substrates (IRSs) and Src homology 2 domain containing protein (Shc) are recruited to phospho-tyrosines on the receptor and then become phosphorylated at tyrosines themselves (21,22). These IRSs recruit several proteins including the transducing adapter, Grb2 (22). Grb2 in turn recruits the guanine-nucleotide exchange factor, SOS (21,23), which then activates Ras (22,24). Several of these transducers including IRS-1 (25), Shc (26), and Grb2 (27) have been found in the nucleus. Thus, antibodies to IRS-1, Shc, Grb2, SOS, and Ras were used next in Matrix ChIP to determine which if any of these components of the insulin receptor complex were recruited to egr-1. We observed recruitment of Grb2 and SOS but not IRS-1, Shc, or Ras to the egr-1 gene (Fig. 2C, rows 3 and 4). These data are the first example of corecruitment of the IR and downstream signaling components to a gene locus and, importantly, in a manner associated with activation of the gene by insulin. The failure to detect IRS-1, Shc, and Ras may reflect either their absence at the egr-1 locus or that the antibodies used do not work in ChIP assays.
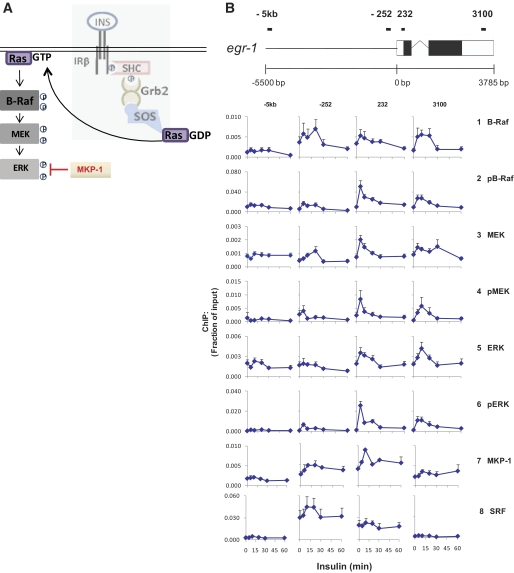
Insulin induces the recruitment and activation of the Raf–MEK–ERK MAPK pathway at the egr-1 locus in hepatocyte culture.
The fact that we were able to demonstrate recruitment of SOS to egr-1 and that SOS activates the ERK MAPK pathway through the small GTP-binding protein, Ras, led us to probe recruitment of the Raf–MEK–ERK pathway to egr-1. We used antibodies to B-Raf phosphorylated at T598 and S601 (pB-Raf) (28), MEK1/2 phosphorylated at S217/221 (pMEK), and ERK1/2 phosphorylated at T202 and Y204 (pERK) in Matrix ChIP. All three phosphorylated kinases were transiently but robustly recruited to both sites within the transcribed region of egr-1 and with kinetics that were very similar to those of the phosphorylated IR (Fig. 3B, rows 2, 4, and 6). ChIP with antibodies to the total kinases, however, showed less pronounced and less transient patterns (Fig. 3B, rows 1, 3, and 5).
FIG. 3.
MAPK Erk pathway at the egr-1 locus in HTC-IR cells. A: Schematic of one arm of the IR signaling pathway with MAPK pathway highlighted. B: Cells were treated as in Fig. 1A, cross-linked, and used in ChIP with antibodies to Raf-B, Raf-B phosphorylated at T598 and S601 (pRaf-B), MEK1/2, MEK1/2 phosphorylated at S217/S221 (pMEK), ERK 1, and ERK1/2 phosphorylated at T202/Y204 (pERK). ChIPed DNA was used in PCR as in Fig. 1C. Data represent mean ± SEM for three independent experiments.
The recruitment of insulin-induced signaling proteins to an insulin-responsive gene suggests their involvement in transcription. To examine the relationship between transcribing Pol II and locus-bound kinases, we compared the recruitment kinetics of the total and activated signaling proteins with those of Pol II within the coding regions of egr-1. To normalize the differing efficiencies of pulldown for each antibody, we plotted the ratio of each protein's signal at each time point versus the signal at the last time point (supplementary Fig. 2, available in an online appendix). The nearly identical binding profile of active IRβ, kinases, and Pol II suggests that, after the recruitment of IR and MAPK signaling components, the activation/deactivation of the kinases is tightly coupled to IR, a chain of events that are linked to RNA Pol II elongation. Compared with the insulin-induced levels of binding of total kinases, the phospho-kinase signals exhibited greater inducibility and were short-lived (supplementary Fig. 2). The transient nature of the phospho-kinase signals may reflect rapid dephosphorylation by phosphatases that are recruited to egr-1 in response to insulin. MKP-1 is a member of the dual-specificity family of phosphatases that inactivate Erk1/2 through dephosphorylation of its pThr202 and pTyr204 residues (29) (Fig. 3A). MKP-1 is insulin-responsive (30,31) and is found in the nucleus (32,33). ChIP assays for MKP-1 revealed insulin-induced recruitment to egr-1 (Fig. 3B, row 7). This, together with the corresponding decrease in phospho-ERK, but not total ERK (5–10 min, 232-bp site), suggests the possibility that Erk1/2 found at the egr-1 locus is inactivated in situ and that at least a fraction of the dephosphorylated Erk1/2 remains bound to the gene.
The serum responsive factor activates egr-1 by binding to several promoter elements (34). Matrix ChIP with an antibody to serum responsive factor showed a high constitutive and a small insulin-inducible component at the −252 bp and +232 bp sites and no binding at the end of the gene (Fig. 3, row 8). Thus, the trend toward higher levels at the transcribed region of egr-1 was specific to the IR signaling and MAPK proteins (Fig. 3B). Together, the above evidence is the first to suggest the recruitment of an entire activated MAPK pathway as well as cognate phosphatase at a gene in mammalian cells across the transcribed region.
Increased recruitment of IR and the MAPK signaling module to the Gck gene in livers of glucose-fed mice.
Glucose feeding of fasted mice was previously shown to cause a rapid increase in serum glucose, serum insulin, and nuclear translocation of IR (13). In the following experiments, fasted C57BL/6 mice were fed a glucose meal and killed at different times later. Figure 4B (top) shows that serum glucose increased rapidly after glucose feeding, peaking at 10 min and then returning to a level twofold above fasted levels 50 min later. Serum insulin also increased quickly, but more transiently (Fig. 4B, middle). Insulin-stimulated expression of six insulin responsive loci was then examined (Fig. 4B and data not shown) including glucokinase (Gck) (Fig. 4A), an insulin-responsive gene whose product is involved in glycolysis (35,36). Gck mRNA was most robustly induced, with transcript levels increasing ninefold 60 min after insulin treatment (Fig. 4B, bottom). egr-1 had a large increase as well; however, this was mostly due to high levels of transcript in the livers of two mice, one at 20 and one at 30 min of glucose feeding (data not shown), while the transcript in most mice remained unchanged. Although the traditional view of IR signaling describes two divergent pathways, the mitogenic one transduced by B-Raf/MEK/Erk and the metabolic one transduced by PI3K/Akt (22), it has become increasingly apparent that MAPK pathways can also regulate metabolic state (16). Gck was chosen to examine recruitment of Pol II and signaling proteins because it had the highest, most consistent increase in transcript levels.
FIG. 4.
Feeding induces an increase in glucose, insulin, Gck expression, and synchronized Pol II and IR pathway recruitment in mice. A: Schematic of the Gck locus. B: Blood and serum from glucose-fed, fasted mice were used to determine glucose and insulin concentrations, respectively (top and middle). Total RNA from livers of glucose-fed, fasted mice was used in RT reactions with random hexamers. cDNA was used in PCR with primers to glucokinase (Gck). Data represent mean ± SEM of five mice for each time point. C: Chromatin from livers of glucose-fed, fasted mice was used in ChIP assays with antibodies to the indicated proteins. ChIPed DNA was used in PCR with primers to the upstream (first TSS) and downstream (second TSS) transcription start sites and the extreme 3′ end of Gck. Data represent mean fraction of input ± SEM of five mice for each time point. (A high-quality color representation of this figure is available in the online issue.)
To show that the increase in Gck transcript was due to increased transcription, we used Matrix ChIP to determine Pol II levels along the locus. Gck has two transcription start sites (TSSs), 34,957 bp apart, with the downstream site being the one used by cells in the liver (the length from the start of the second TSS to the 3′ end is 14,301 bp) (35) (Fig. 4A). As expected, Pol II levels increased at the second TSS and the 3′ end of the gene (Fig. 4C, row 1). The 30-min peak at the 3′ end of the gene coincided with the first increase in Gck mRNA, suggesting that increased transcription was at least partially responsible for Gck induction (compare Fig. 4C, row 1, frame 3 with Fig. 4B, bottom).
Next we looked at the recruitment of IR complex components to Gck using Matrix ChIP. IRβ, Grb2, and SOS were recruited to the second TSS and 3′ end of Gck (Fig. 4C, rows 2, 4, and 5). Importantly, the phosphorylated, active form of IRβ was more robustly induced than the total receptor (Fig. 4C, row 3). These results are consistent with those seen in cell culture Figures 2 and 3.
Next we used Matrix ChIP to probe the recruitment of B-Raf, MEK, and ERK and their activated forms at the Gck locus in liver chromatin. Similar to the results in cells, the phosphorylated forms of all three kinases showed robust increases at all of the transcribed regions of the gene with kinetics similar to that of the phosphorylated insulin receptor (Fig. 4C, rows 7, 9, and 11). Also, as with the results in cells, ChIP with antibodies to the total kinases showed smaller responses compared with phosphorylated kinases (Fig. 4C, rows 6, 8, and 10). We also found a transient increase in the levels of MKP-1 at the second TSS and 3′ end of Gck. These data confirm that the insulin-induced recruitment and activation of the MAPK pathway seen in hepatocyte culture also occurs during insulin-induced expression of Gck in mammalian tissue.
Next, we compared Pol II and the phosphorylated kinases at first TSS and second TSS of Gck to the profiles at the TSS of gapdh, which, while highly expressed, was not stimulated in response to feeding within the 60-min time course (supplementary Fig. 3 [available in an online appendix], first row, compare frame 3 to frames 1 and 2). Interestingly, the constitutive Grb2, SOS, pB-Raf, pMEK, and pERK but not the pIRβ levels were much higher at gapdh than at the Gck. Also, unlike the feeding-induced increases in Grb2, SOS, and kinases seen at Gck, no such responses were seen at gapdh (supplementary Fig. 3). This result suggests that the glucose feeding-induced recruitment/activation of these MAPK signaling proteins at Gck but not gapdh is tightly coupled to the activated insulin receptor. Moreover, the fact that a similar type of inducible recruitment occurs in two different systems, one in vitro along a gene mediating mitogenic responses (Figs. 1–3) and another one in vivo along a gene involved in metabolism (Fig. 4), suggests that recruitment of IR/MAPK pathway in a pattern resembling Pol II elongation could be a general phenomenon of insulin-inducible genes.
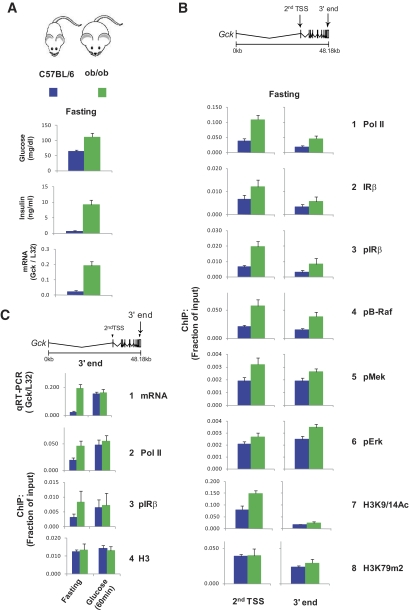
Absence of feeding-induced Gck mRNA response in ob/ob mice is associated with unsynchronized induction of IR-MAPK components at the Gck locus.
Obese (ob/ob) C57BL/6J mice are insulin resistant and have higher nuclear insulin receptor levels in the liver (13). Moreover, unlike the lean C57BL/6J mice, the ob/ob mice express constitutively high hepatic Gck activity that is unresponsive to either fasting or feeding (37,38). Thus, comparison of the lean and obese strains provides an opportunity to explore the relationship between direct recruitment of IR and MAPK components and Gck gene expression. We carried out side-by-side fasting followed by glucose feeding and performed the same measurements as in Fig. 4 in both lean and obese mice (Figs. 5–6). Compared with fasting lean mice, fasting ob/ob mice had twice the serum glucose levels and >10-fold higher insulin levels (Fig. 5A). The fasting Gck mRNA and Pol II levels (Fig. 5A bottom and 5B, row 1) were also higher in the livers of the ob/ob mice, suggesting higher rates of transcription in the obese compared with the lean mice (Figs. 5A and B). The constitutive differences in Pol II levels at Gck gene (Fig. 5B row 1) in lean and obese mice matched the levels of IR (rows 2 and 3) and MAPK components (rows 4–6), suggesting that the baseline transcription and IR-MAPK activation is coupled in both strains. Histone H3 lysine 9 acetylation (H3K9/14Ac), a mark of open chromatin, is typically high at the TSS and low at the 3′ end of actively transcribed genes (39). Interestingly, at the Gck TSS, H3K9/14Ac but not H3 lysine 79 dimethylation (H3K79m2) (Fig. 5B rows 7 and 8) (40), a mark associated with transcription elongation, was higher in the ob/ob mice. Taken together, these results suggest that, at baseline, in the ob/ob mice, the hepatic chromatin structure at Gck is different from the lean mice and consistent with the higher transcription rates.
FIG. 5.
Fasted obese mice have higher glucose, insulin, Gck expression, and Pol II and IR pathway recruitment than lean mice. A: Blood and serum from fasted lean and ob/ob mice were used to determine glucose and insulin concentrations, respectively (top and middle). Total RNA from livers of fasted lean and ob/ob mice was used in RT reactions with random hexamers. cDNA was used in PCR with primers to Gck and ribosomal protein L32 gene as a control for total RNA. B: Chromatin from the livers of fasted lean and ob/ob mice was used in ChIP assays with antibodies to the indicated proteins and histone H3 covalent modification. ChIPed DNA was used in PCR with primers to the indicated regions. C: Comparison of RT-PCR and ChIP results for Gck of fasted and 60-min glucose-fed lean and obese mice. Data (A–C) represent mean ± SEM of four mice for each time point.
FIG. 6.
Glucose feeding fails to induce Gck expression and synchronized Pol II and IR pathway recruitment in obese mice. A: Blood glucose, serum insulin, and liver Gck expression were measured in fasted, glucose-fed lean and obese mice as in Fig. 5A. B: Chromatin from the livers of fasted, glucose-fed lean and ob/ob mice was used in ChIP assays as in Fig. 5B.
After glucose feeding, the transient increase in serum glucose was greater in the ob/ob mice but glycemic differences were not nearly as dramatic as the increases in insulin levels, which were >25-fold higher in the obese compared with the lean mice (Figs. 4 and 6A, middle). Unlike in the lean mice, the ob/ob mRNA levels did not respond to glucose feeding (Fig. 6A, bottom). Interestingly, after glucose feeding the Gck mRNA levels in the lean mice reached the unresponsive, constitutively high levels in the obese mice. The absent mRNA response is in agreement with the lack of induction in the 3′ end Pol II levels (Fig. 5C row 2), where unlike the lean mice, little or no activation of IR and MAPK components was detected in the ob/ob strain (Fig. 5C, row 3, and 6B, rows 2–6).
Remarkably, glucose feeding in ob/ob mice induced second TSS Pol II, IR, and MAPK changes, but unlike the lean mice, these changes appeared unsynchronized and did not propagate to the 3′ end of the gene (Fig. 6B, rows 1–6). These results show that, unlike the lean mice, in the obese mice activation of Mek/Erk at Gck TSS is uncoupled from its canonical activator, B-Raf.
In contrast to the IR/MAPK changes (Fig. 6B, rows 1–6), the second TSS H3K9/14Ac levels in both lean and obese mice were largely unaffected by glucose feeding (Fig. 6B, row 7), indicating that this mark of open chromatin remains stable even though Pol II, IR, and MAPK activities are changing.
DISCUSSION
Here, using two inducible genes as examples, egr-1 in rat hepatocyte cultures and mouse Gck in liver tissues, we provide evidence that components of the IR signaling complex and downstream ERK MAPK cascade are directly recruited to insulin inducible loci. Our results suggest that effective mRNA synthesis/processing depends on coordinated activation of IR/MAPK cascade along Pol II transcribed regions.
Nuclear translocation of IR.
Nearly 25 years ago, Olefsky and coworkers demonstrated nuclear translocation of IR and suggested that it may interact with specific targets in the genome to mediate insulin's long-term effects on transcription (14). Our ChIP-based observations are consistent with this and other previous IR nuclear translocation studies (14,15). Nuclear translocation of insulin has also been demonstrated. Interestingly, insulin's transit through the nuclear pore may be independent of IR and instead be mediated by other cytoplasmic proteins (41). Using three different types of insulin antibodies, we could not detect a ChIP signal specific to insulin at either egr-1 or Gck (data not shown). Thus, it is conceivable that a large fraction of chromatin-bound IR is insulin-free.
Recruitment of IR and MAPK components to inducible gene loci.
The IR/MAPK kinetics that resemble the Pol II pattern (Figs. 2–4, 6) suggest that these kinases either dictate or follow the Pol II initiation/elongation at the TSS. This raises the following question: what are the functional implications for this synchrony? Because many factors involved in chromatin rearrangements, gene transcription, and RNA processing are regulated by phosphorylation, coordinated action of signaling kinases at discrete points of time and space likely plays a critical role in synchronizing these events. The distribution of insulin-responsive kinases in the nuclear milieu could be too diffuse for the spatiotemporal specificity required to coordinately induce these processes (42,43). Thus, precisely timed recruitment and activation of kinases at induced gene loci could provide the requisite spatiotemporal specificity. For example, activated IR could serve to bring together and activate downstream components at target genes in a spatiotemporal manner that matches Pol II elongation. This tight spatiotemporal coupling would serve to compartmentalize kinase signals to specifically regulate productive Pol II mRNA synthesis from insulin-responsive gene loci. The side-by-side comparison of Gck responses in glucose-fed lean and obese mice lends further support to this suggestion (Fig. 6).
Differences and similarities of IR/MAPK recruitment to the Gck locus in lean and obese mice.
Gck is a critical regulator of glucose metabolism, including hepatic control of glycogen synthesis and glycolysis (44). Thus, it would not be surprising if a chronic hyperglycemic state in the ob/ob mice were to trigger a homeostatic mechanism that maintains elevated constitutive Gck transcription to meet the metabolic burden posed by the high glucose load (Fig. 5A). The higher Gck mRNA levels in ob/ob mice, at least in part, reflect increased transcription as evidenced by higher Pol II density (Fig. 5B, row 1). The higher rates of transcription are further supported by the more open chromatin structure revealed by H3K9/14Ac differences between the two strains (Fig. 5B, row 7). The constitutive IR and MAPK ChIP signals at Gck in the lean and obese mice seem to match Pol II levels (row 1), suggesting that, in both strains, fasting activities of Pol II, IR (rows 1–3), and MAPK components (rows 4–6) are functionally coupled. After glucose feeding, Gck mRNA and 3′ end Pol II, IR, and MAPK densities in ob/ob mice remained unchanged. Interestingly, in lean mice, the mRNA and 3′ end values reached those of obese mice (Fig. 5C). This observation lends further support to the suggestion that 3′ end IR/MAPK cascade activities, coupled to Pol II, tune the mRNA output from the Gck locus to match the glucose load (Fig. 6A).
In sharp contrast to the strong similarity between IR/MAPK activities and Pol II density at the 3′ end during fasting in lean and obese mice (Fig. 5B), the changes in the levels of these proteins at the Gck TSS during feeding were dramatically different between the two strains (Figs. 4C and 6B). Specifically, the synchronous, feeding-induced increase in TSS pIR/B-Raf/MEK/ERK activities and Pol II in the lean mice (Figs. 4C and 6B) was absent in the ob/ob strain (Fig. 6B). Instead, in the obese mice at the TSS, there was a wave of Pol II, IR, and B-Raf activities, a pattern different from the steady MEK and Erk increase. The discrepant TSS kinase patterns in the obese mice suggest that at least a fraction of the total pool of MEK/Erk (rows 5 and 6) is uncoupled from the upstream activator, B-Raf, at this site (row 4). The apparent B-Raf–MEK/Erk uncoupling could reflect heterogeneous hepatocyte populations in the liver where activation of the MEK/Erk axis could be mediated by alternative upstream MEK activators such as C-Raf, A-Raf, or PAK (45,46,47).
Release of proximally paused Pol II, seen as a decrease in ChIP signal at the TSS, is an important mode of transcriptional activation (48). Whereas the initial feeding-induced decline (10 min) in TSS Pol II density in the ob/ob mice is consistent with this model (Fig. 6B, row 1), the increase that follows (20 min) may represent de novo promoter initiation. Remarkably, the TSS Pol II wave does not elongate to the 3′ end; instead it seems that the feeding-induced polymerase either falls off or is stalled downstream. Thus, the unsynchronized activities of IR and MEK/Erk at the Gck TSS in ob/ob mice may be associated with low processivity (i.e., premature termination of RNA synthesis) (49) of the feeding-induced Pol II wave. Taken together, these observations are consistent with the suggestion that coupled activation of IR/B-Raf/MEK/Erk along a gene could play a role in productive mRNA synthesis by Pol II.
In summary, we have shown for the first time that the IR signaling complex and downstream ERK MAPK cascade components are recruited and activated along insulin-induced loci. These observations raise many questions about how IR and the MAPK pathway are targeted to insulin-responsive genes and the role of this recruitment. Thus, it is clear that this facet of insulin- and IR-regulated gene expression needs to be explored. In this regard, the present study represents a new paradigm to study these processes at the chromatin level in health and disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants DKR-3745978 and GMR-O145134 (K.B.), HL-079382 (R.C.L.), and Seattle MMPC (U24-DK-076126) (R.C.L.).
No potential conflicts of interest relevant to this article were reported.
J.D.N. designed and performed experiments, analyzed the data, and wrote the manuscript. R.C.L. designed the experiments and wrote the manuscript. K.B. came up with the hypothesis, designed and performed experiments, analyzed the data, and wrote the manuscript.
We thank Steve Flanagin (University of Washington) for developing GraphGrid software tools. We would like to acknowledge the technical assistance of Phuong-Oanh T. Mai, Johnny Bushey, and Mark Caldwell (University of Washington).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Fantl WJ, Johnson DE, Williams LT: Signalling by receptor tyrosine kinases. Annu Rev Biochem 1993;62:453–481 [DOI] [PubMed] [Google Scholar]
- 2.Reilly JF, Maher PA: Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol 2001;152:1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK: Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell 2009;20:2401–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offterdinger M, Schöfer C, Weipoltshammer K, Grunt TW: c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol 2002;157:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G: Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 2006;127:185–197 [DOI] [PubMed] [Google Scholar]
- 6.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC: Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog 2006;45:10–17 [DOI] [PubMed] [Google Scholar]
- 7.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC: Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 2005;7:575–589 [DOI] [PubMed] [Google Scholar]
- 8.Flanagin S, Nelson JD, Castner DG, Denisenko O, Bomsztyk K: Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events. Nucleic Acids Res 2008;36:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL: p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 2004;36:738–743 [DOI] [PubMed] [Google Scholar]
- 10.Lawrence MC, McGlynn K, Shao C, Duan L, Naziruddin B, Levy MF, Cobb MH: Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc Natl Acad Sci U S A 2008;105:13315–13320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL: Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 2010;329:1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seol KC, Kim SJ: Nuclear matrix association of insulin receptor and IRS-1 by insulin in osteoblast-like UMR-106 cells. Biochem Biophys Res Commun 2003;306:898–904 [DOI] [PubMed] [Google Scholar]
- 13.Gletsu NA, Field CJ, Clandinin MT: Obese mice have higher insulin receptor levels in the hepatocyte cell nucleus following insulin stimulation in vivo with an oral glucose meal. Biochim Biophys Acta 1999;1454:251–260 [DOI] [PubMed] [Google Scholar]
- 14.Podlecki DA, Smith RM, Kao M, Tsai P, Huecksteadt T, Brandenburg D, Lasher RS, Jarett L, Olefsky JM: Nuclear translocation of the insulin receptor. A possible mediator of insulin's long term effects. J Biol Chem 1987;262:3362–3368 [PubMed] [Google Scholar]
- 15.Gletsu N, Dixon W, Clandinin MT: Insulin receptor at the mouse hepatocyte nucleus after a glucose meal induces dephosphorylation of a 30-kDa transcription factor and a concomitant increase in malic enzyme gene expression. J Nutr 1999;129:2154–2161 [DOI] [PubMed] [Google Scholar]
- 16.Wu JJ, Roth RJ, Anderson EJ, Hong EG, Lee MK, Choi CS, Neufer PD, Shulman GI, Kim JK, Bennett AM: Mice lacking MAP kinase phosphatase-1 have enhanced MAP kinase activity and resistance to diet-induced obesity. Cell Metab 2006;4:61–73 [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Higaki Y, Abrass CK, Bomsztyk K: Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proc Natl Acad Sci U S A 2001;98:9044–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson JD, Denisenko O, Sova P, Bomsztyk K: Fast chromatin immunoprecipitation assay. Nucleic Acids Res 2006;34:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naito M, Zager RA, Bomsztyk K: BRG1 increases transcription of proinflammatory genes in renal ischemia. J Am Soc Nephrol 2009;20:1787–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawley DM, Maddux BA, Patel RG, Wong KY, Mamula PW, Firestone GL, Brunetti A, Verspohl E, Goldfine ID: Insulin receptor monoclonal antibodies that mimic insulin action without activating tyrosine kinase. J Biol Chem 1989;264:2438–2444 [PubMed] [Google Scholar]
- 21.White MF: The insulin signalling system and the IRS proteins. Diabetologia 1997;40(Suppl. 2):S2–S17 [DOI] [PubMed] [Google Scholar]
- 22.Biddinger SB, Kahn CR: From mice to men: insights into the insulin resistance syndromes. Annu Rev Physiol 2006;68:123–158 [DOI] [PubMed] [Google Scholar]
- 23.Montagnani M, Ravichandran LV, Chen H, Esposito DL, Quon MJ: Insulin receptor substrate-1 and phosphoinositide-dependent kinase-1 are required for insulin-stimulated production of nitric oxide in endothelial cells. Mol Endocrinol 2002;16:1931–1942 [DOI] [PubMed] [Google Scholar]
- 24.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J: Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 1993;363:85–88 [DOI] [PubMed] [Google Scholar]
- 25.Wu A, Chen J, Baserga R: Nuclear insulin receptor substrate-1 activates promoters of cell cycle progression genes. Oncogene 2008;27:397–403 [DOI] [PubMed] [Google Scholar]
- 26.Song RX, Barnes CJ, Zhang Z, Bao Y, Kumar R, Santen RJ: The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor alpha to the plasma membrane. Proc Natl Acad Sci U S A 2004;101:2076–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero F, Ramos-Morales F, Domínguez A, Rios RM, Schweighoffer F, Tocqué B, Pintor-Toro JA, Fischer S, Tortolero M: Grb2 and its apoptotic isoform Grb3–3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. J Biol Chem 1998;273:7776–7781 [DOI] [PubMed] [Google Scholar]
- 28.Zhang BH, Guan KL: Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J 2000;19:5429–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens DM, Keyse SM: Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007;26:3203–3213 [DOI] [PubMed] [Google Scholar]
- 30.Kusari AB, Byon J, Bandyopadhyay D, Kenner KA, Kusari J: Insulin-induced mitogen-activated protein (MAP) kinase phosphatase-1 (MKP-1) attenuates insulin-stimulated MAP kinase activity: a mechanism for the feedback inhibition of insulin signaling. Mol Endocrinol 1997;11:1532–1543 [DOI] [PubMed] [Google Scholar]
- 31.Byon JC, Dadke SS, Rulli S, Kusari AB, Kusari J: Insulin regulates MAP kinase phosphatase-1 induction in Hirc B cells via activation of both extracellular signal-regulated kinase (ERK) and c-Jun-N-terminal kinase (JNK). Mol Cell Biochem 2001;218:131–138 [DOI] [PubMed] [Google Scholar]
- 32.Wu JJ, Zhang L, Bennett AM: The noncatalytic amino terminus of mitogen-activated protein kinase phosphatase 1 directs nuclear targeting and serum response element transcriptional regulation. Mol Cell Biol 2005;25:4792–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, Charles CH, Lau LF, Tonks NK: MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 1993;75:487–493 [DOI] [PubMed] [Google Scholar]
- 34.Wu SQ, Minami T, Donovan DJ, Aird WC: The proximal serum response element in the Egr-1 promoter mediates response to thrombin in primary human endothelial cells. Blood 2002;100:4454–4461 [DOI] [PubMed] [Google Scholar]
- 35.Magnuson MA, Shelton KD: An alternate promoter in the glucokinase gene is active in the pancreatic beta cell. J Biol Chem 1989;264:15936–15942 [PubMed] [Google Scholar]
- 36.Iynedjian PB, Gjinovci A, Renold AE: Stimulation by insulin of glucokinase gene transcription in liver of diabetic rats. J Biol Chem 1988;263:740–744 [PubMed] [Google Scholar]
- 37.Meglasson MD, Matschinsky FM: New perspectives on pancreatic islet glucokinase. Am J Physiol 1984;246:E1–E13 [DOI] [PubMed] [Google Scholar]
- 38.Yen TT, Stamm NB: Constitutive hepatic glucokinase activity in db/db and ob/ob mice. Biochim Biophys Acta 1981;657:195–202 [DOI] [PubMed] [Google Scholar]
- 39.Berger SL: Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002;12:142–148 [DOI] [PubMed] [Google Scholar]
- 40.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K: High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823–837 [DOI] [PubMed] [Google Scholar]
- 41.Harada S, Smith RM, Jarett L: Mechanisms of nuclear translocation of insulin. Cell Biochem Biophys 1999;31:307–319 [DOI] [PubMed] [Google Scholar]
- 42.Yao Z, Seger R: The ERK signaling cascade–views from different subcellular compartments. Biofactors 2009;35:407–416 [DOI] [PubMed] [Google Scholar]
- 43.McKay MM, Morrison DK: Integrating signals from RTKs to ERK/MAPK. Oncogene 2007;26:3113–3121 [DOI] [PubMed] [Google Scholar]
- 44.Iynedjian PB: Molecular physiology of mammalian glucokinase. Cell Mol Life Sci 2009;66:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roskoski R, Jr: RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399:313–317 [DOI] [PubMed] [Google Scholar]
- 46.Park ER, Eblen ST, Catling AD: MEK1 activation by PAK: a novel mechanism. Cell Signal 2007;19:1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee M: Raf-1 kinase activation is uncoupled from downstream MEK/ERK pathway in cells treated with Src tyrosine kinase inhibitor PP2. Biochem Biophys Res Commun 2006;350:450–456 [DOI] [PubMed] [Google Scholar]
- 48.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA: c-Myc regulates transcriptional pause release. Cell 2010;141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mason PB, Struhl K: Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 2005;17:831–840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.