Abstract
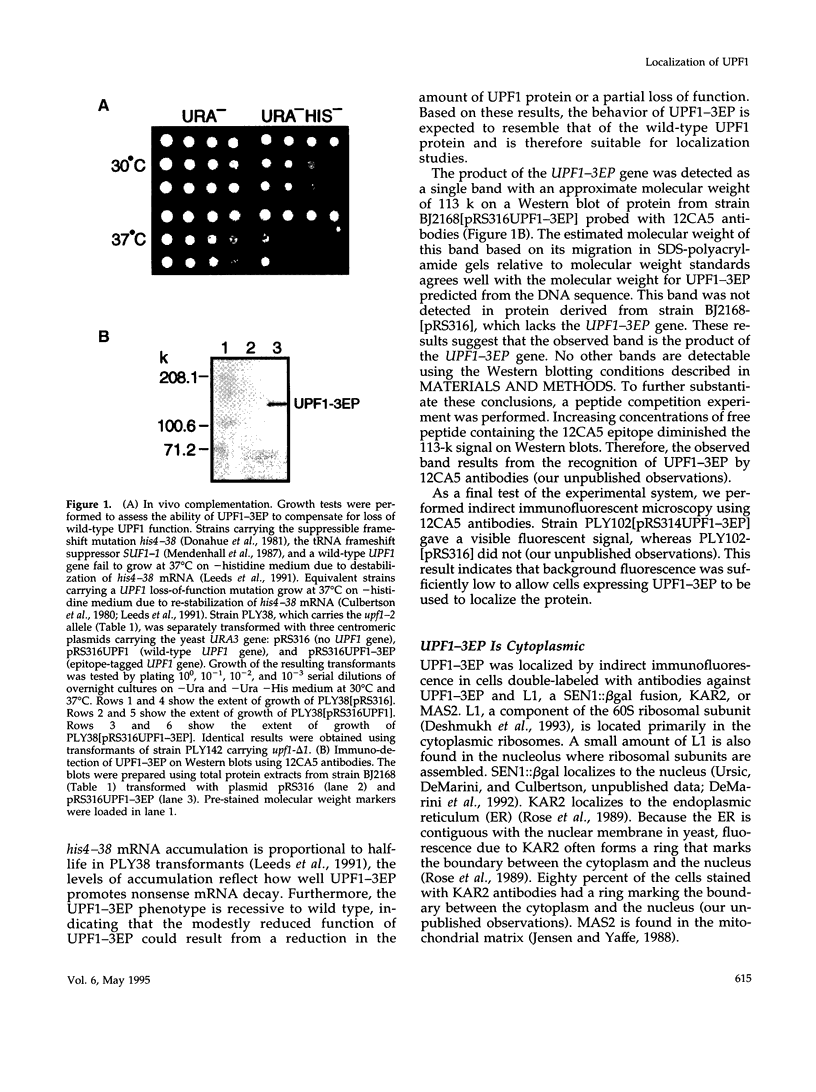
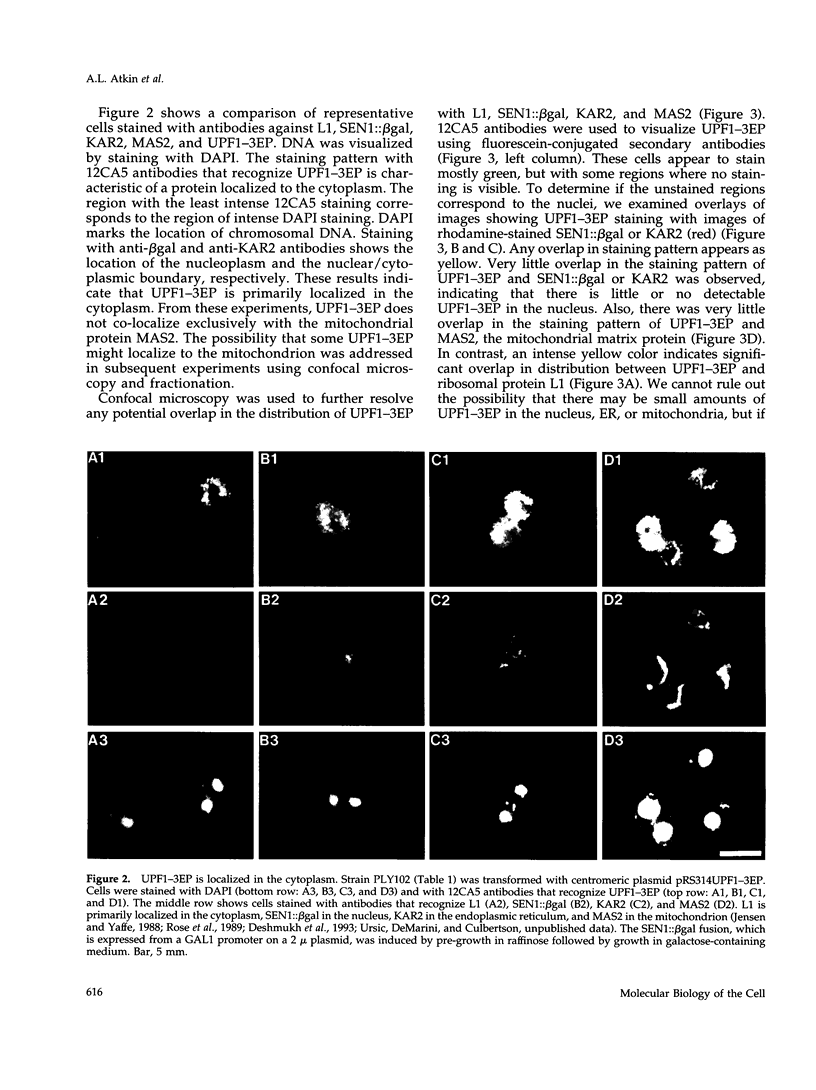
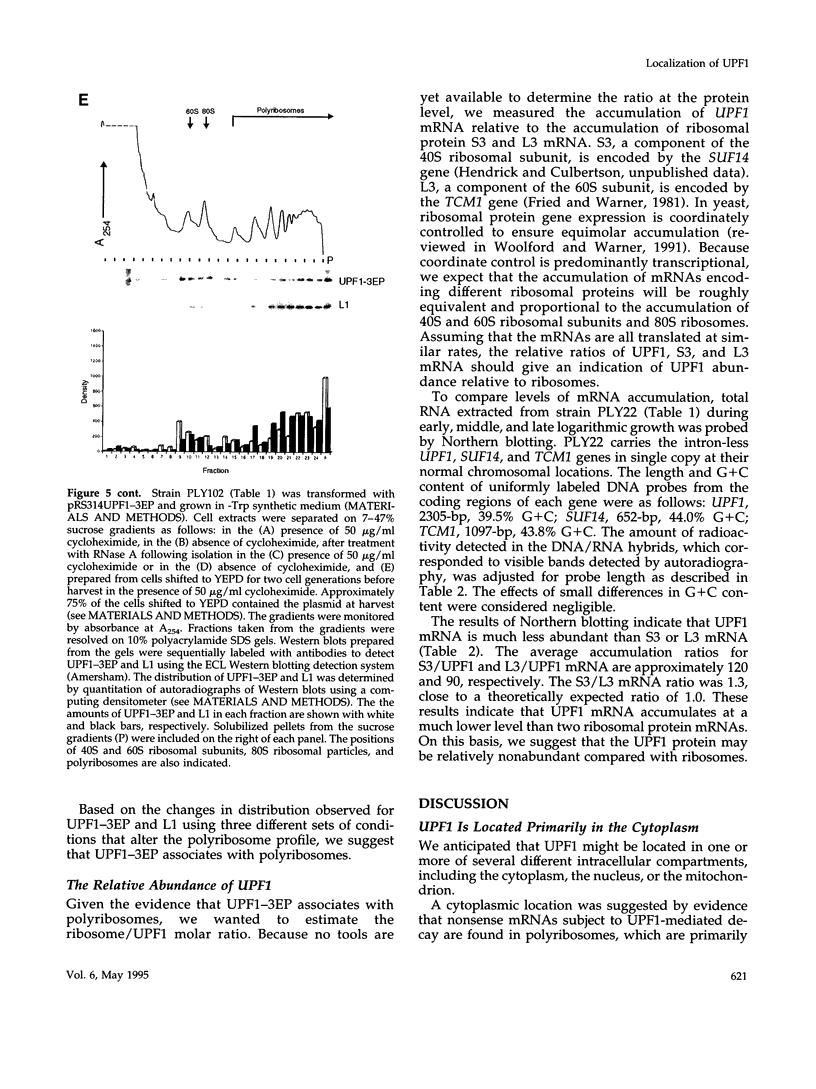
In Saccharomyces cerevisiae the UPF1 protein is required for nonsense-mediated mRNA decay, the accelerated turnover of mRNAs containing a nonsense mutation. Several lines of evidence suggest that translation plays an important role in the mechanism of nonsense mRNA decay, including a previous report that nonsense mRNAs assemble in polyribosomes. In this study we show that UPF1 and ribosomal protein L1 co-localize in the cytoplasm and that UPF1 co-sediments with polyribosomes. To detect UPF1, three copies of the influenza hemagglutinin epitope were placed at the C-terminus. The tagged protein, UPF1-3EP, retains 86% (+/- 5%) of function. Using immunological detection, we found that UPF1-3EP is primarily cytoplasmic and was not detected either in the nucleus or in the mitochondrion. UPF1-3EP and L1 co-distributed with polyribosomes fractionated in a 7-47% sucrose gradient. The sucrose sedimentation profiles for UPF1-3EP and L1 exhibited similar changes using three different sets of conditions that altered the polyribosome profile. When polyribosomes were disaggregated, UPF1-3EP and L1 accumulated in fractions coincident with 80S ribosomal particles. These results suggest that UPF1-3EP associates with polyribosomes. L3 and S3 mRNAs, which code for ribosomal proteins of the 60S and 40S ribosomal subunits, respectively, were on average about 100-fold more abundant than UPF1 mRNA. Assuming that translation rates for L3, S3, and UPF1 mRNA are similar, this result suggests that there are far fewer UPF1 molecules than ribosomes per cell. Constraints imposed by the low UPF1 abundance on the functional relationships between UPF1, polyribosomes, and nonsense mRNA turnover are discussed.
Full text
PDF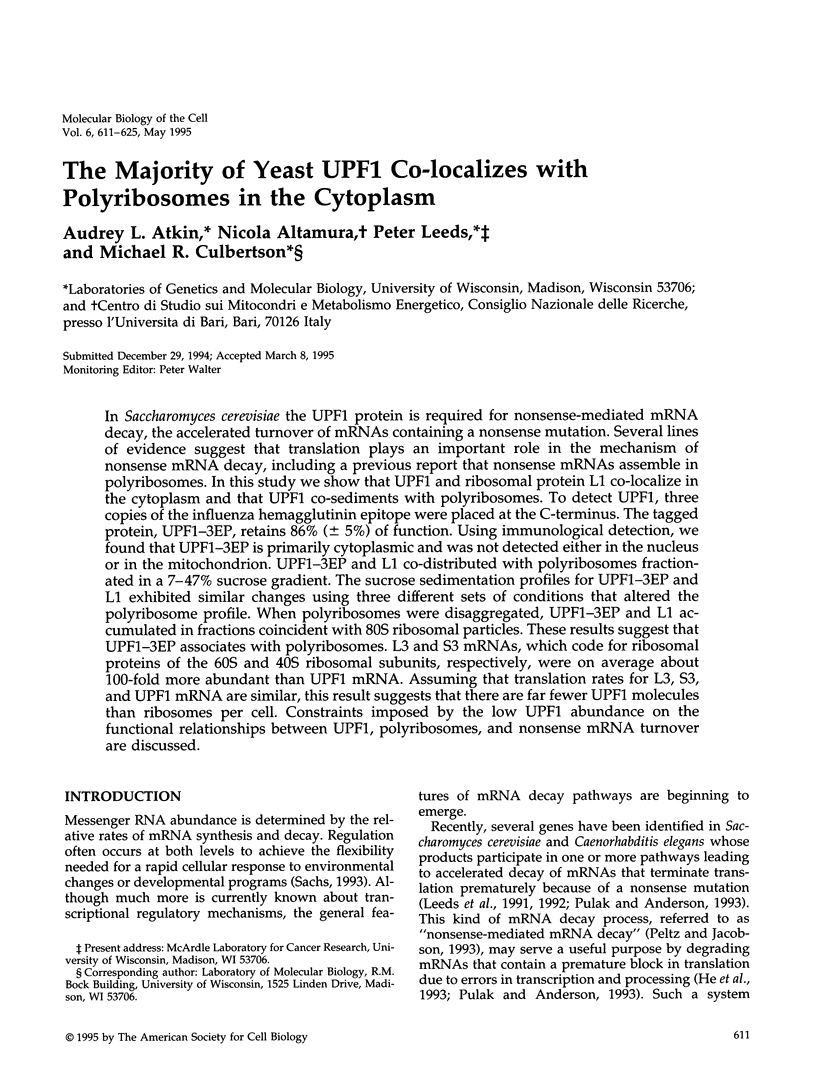










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altamura N., Dujardin G., Groudinsky O., Slonimski P. P. Two adjacent nuclear genes, ISF1 and NAM7/UPF1, cooperatively participate in mitochondrial functions in Saccharomyces cerevisiae. Mol Gen Genet. 1994 Jan;242(1):49–56. doi: 10.1007/BF00277347. [DOI] [PubMed] [Google Scholar]
- Altamura N., Groudinsky O., Dujardin G., Slonimski P. P. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992 Apr 5;224(3):575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- Asher E. B., Groudinsky O., Dujardin G., Altamura N., Kermorgant M., Slonimski P. P. Novel class of nuclear genes involved in both mRNA splicing and protein synthesis in Saccharomyces cerevisiae mitochondria. Mol Gen Genet. 1989 Feb;215(3):517–528. doi: 10.1007/BF00427051. [DOI] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L. E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Zhou X., Stephenson L. S., Maquat L. E. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994 Dec;14(12):8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic M., Maquat L. E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990 Oct;10(10):5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Maquat L. E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993 Mar;13(3):1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Underbrink K. M., Fink G. R. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980 Aug;95(4):833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D. J., Winey M., Ursic D., Webb F., Culbertson M. R. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992 May;12(5):2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M., Tsay Y. F., Paulovich A. G., Woolford J. L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993 May;13(5):2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci U S A. 1981 Jan;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the Group II his4-206 and leu 2-3 frameshift mutations. Genetics. 1982 Jul-Aug;101(3-4):345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalbo D., Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990 Jan;17(1):77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- Grant P., Sánchez L., Jiménez A. Cryptopleurine resistance: genetic locus for a 40S ribosomal component in Saccharomyces cerevisiae. J Bacteriol. 1974 Dec;120(3):1308–1314. doi: 10.1128/jb.120.3.1308-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Peltz S. W., Donahue J. L., Rosbash M., Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988 Feb;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Yaffe M. P. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988 Dec 1;7(12):3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. A new group of putative RNA helicases. Trends Biochem Sci. 1992 Dec;17(12):495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- Leeds P., Peltz S. W., Jacobson A., Culbertson M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P., Wood J. M., Lee B. S., Culbertson M. R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992 May;12(5):2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall M. D., Leeds P., Fen H., Mathison L., Zwick M., Sleiziz C., Culbertson M. R. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Saccharomyces cerevisiae. J Mol Biol. 1987 Mar 5;194(1):41–58. doi: 10.1016/0022-2836(87)90714-5. [DOI] [PubMed] [Google Scholar]
- Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993 Oct;7(10):1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Hinnebusch A. G. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992 Jul;118(1):95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989 Jun 30;57(7):1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]








