Abstract
The dynamic protein interactions required for transcription are functionally important yet poorly understood; in this issue, Zobeck et al. (2010) resolve the sequential recruitment and selective recycling of transcription factors at an actively transcribing locus in Drosophila.
Transcription of protein-coding genes by RNA polymerase II (Pol II) is a highly dynamic process considered to have several distinct steps in which Pol II function is regulated by transient protein interactions. These steps include preinitiation complex assembly, transcription initiation, promoter escape, processive transcription elongation, and transcription termination (Selth et al., 2010). Much of our knowledge about the assembly and movement of the Pol II transcription machinery comes from relatively static biochemical and molecular studies, in which transcription reactions have been initiated, allowed to proceed for a certain amount of time, and then stopped for offline analysis (Voss and Hager, 2008). While these studies have collectively provided many mechanistic insights into transcription, they are fundamentally static snapshots of a highly dynamic process. However, recent technological advances in microscopy now allow for live cell imaging of Pol II during transcription with greatly enhanced temporal and spatial resolution (Yao et al., 2007). In this issue of Molecular Cell, Lis and colleagues use this technology for real-time kinetic analyses of transcription factor (TF) recruitment to an actively transcribing locus.
The structural dynamics that accompany the transcription process are readily visualized in polytene chromosomes from Drosophila salivary glands. When these large, multistrand chromosomes form, multiple copies of the same gene remain closely associated, spatially amplifying sites of active transcription (Lis, 2007). One of the best-characterized models to study transcription in Drosophila are the Hsp70 loci, which form chromosomal puffs upon heat shock (HS) that are indicative of rapid and robust activation of transcription (Lis, 2007). While many regulators of Hsp70 expression have been identified (Fuda et al., 2009), a complete understanding of regulation itself requires an understanding of how these factors are recruited and assembled onto the activated genes.
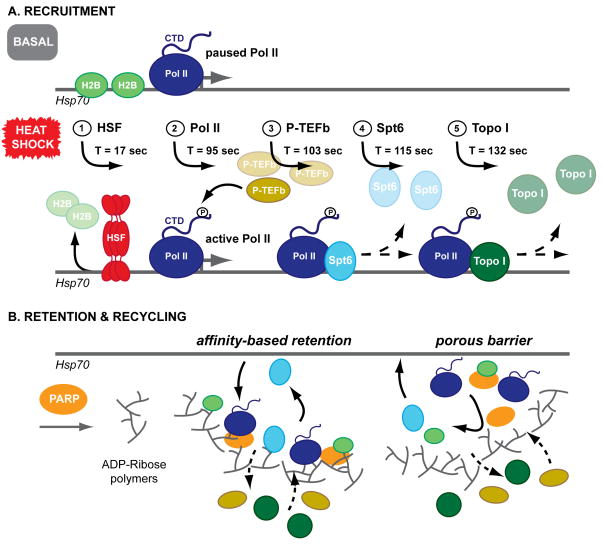
To study the dynamic recruitment of TFs to the Hsp70 loci in real-time, Zobeck et al. (2010) utilized transgenic fly lines that each express an eGFP- or mRFP-tagged TF involved in HS gene activation and monitored TF recruitment to HS-induced chromosomal puffs using various microscopy methods. Three proteins were studied: P-TEFb, a kinase that targets Pol II; Spt6, a nucleosome chaperone; and Topo-I, which unwinds supercoiled DNA. Each of these transgenic lines coexpress a subunit of Pol II (Rpb3), tagged to be complementary to the TF (either eGFP or mRFP). Initial laser scanning confocal microscopy (LSCM) experiments confirmed that all three TFs are recruited together with Pol II to the Hsp70 loci in living Drosophila salivary gland cells upon HS treatment (hence studying them “on the fly, in the fly”). The authors then used spinning disk confocal microscopy (SDCM) to acquire images at threefold higher frame rates than LSCM, improving the temporal resolution up to 6 frames/min to refine the precise timing of recruitment for each TF. Data were collected from several independent salivary gland nuclei over a period of 20 min after HS. Remarkably, there was little cell-to-cell variation in the calculated times and rates of recruitments for each factor. By fitting the buildup of fluorescence intensities to obtain rates of TF recruitment, the authors found that the process initiated with the arrival of the heat shock factor (HSF, Fig. 1A) seconds after heat shock starts (17±1 s). P-TEFb and Pol II were recruited at similar times after HS induction (95±3 s and 103±2 s, respectively). Spt6 and Topo-I followed afterwards, at 115±2 s post-HS for Spt6 and 132±4 s for Topo-I. Together, these data demonstrate that TFs follow a sequential order of recruitment to the activated Hsp70 loci and that this process occurs synchronously within a population of cells.
Figure 1. Sequential recruitment and selective retention of transcription factors at the Hsp70 locus after heat shock.
(A) Heat shock induces sequential recruitment of TFs to Hsp70, beginning with HSF. Additional TFs are recruited in synchronous waves across independent nuclei and to stoichiometric excess over chromatin binding sites. (B) FRAP data demonstrate that TFs are selectively retained at the Hsp70 loci throughout HS. PARP activity is required for TF retention in this “transcription compartment,” possibly through the formation of a matrix of polymerized ADP-ribose. This matrix could participate in TF retention and recycling by affinity-based retention of TFs for the ADP-ribose polymers, formation of a porous barrier, or some combination of both models.
Once initiated, TF recruitment continues for 8–10 min post-activation, as revealed by total fluorescence intensity measurements of P-TEFb, Spt6, and Topo-I. Notably, this is longer than needed to fully saturate the binding of these same factors to their chromatin binding sites – chromatin immunoprecipitation (ChIP) data for P-TEF-b and Spt6 show saturation by ~3 min. Similar results were also obtained for Pol II, confirming the initial report of this recruitment phenomenon (Yao et al., 2007), thus supporting a model where both general and gene-specific factors are recruited in stoichiometric excess and retained at sites of active transcription.
To investigate a possible kinetic basis for this retention effect, Zobeck et al. measured the local exchange dynamics of the three TFs at the Hsp70 loci using fluorescence recovery after photobleaching (FRAP) experiments. Their data show that these dynamics vary considerably as a function of the TF and time post-HS. For example, at a point early in the HS treatment, both P-TEFb and Topo-I rapidly recover after photobleaching (~80% recovery within 2 min), while Spt6 recovery is slower (~50% recovery). However, at later times (40–60 min into HS), recruitment of all of these proteins is dramatically slowed (~40% recovery of P-TEFb and Topo-I; no recovery for Spt6). Thus, as transcription progresses, a portion of the TF population remains associated with the Hsp70 loci, suggesting they are actively retained and that diffusion into the area is limited. Similar results obtained for Pol II itself led to the proposal of a so-called “transcription compartment” that could generate high local concentrations of TFs readily available for subsequent rounds of transcription (Yao et al., 2007).
How might such a “compartment” be dynamically formed near actively transcribing genes? One candidate protein that could regulate this process is the enzyme poly(ADP)-ribose polymerase (PARP), which attaches ADP-ribose molecules to target proteins and creates polymers of ADP-ribose (Tulin and Spradling, 2003). Given that PARP is required after HS to induce expression of Hsp70 genes in Drosophila and for the formation of chromosomal puffs at this locus (Tulin and Spradling, 2003), Zobeck et al. directly examined the role of PARP activity in the local retention of TFs. Strikingly, treatment with the PARP inhibitor PJ34 significantly reduced Pol II retention near the Hsp70 locus, effectively eliminating the reduced FRAP observed in late HS without PJ34 treatment. These results suggest that PARP’s generation of ADP-ribose polymers is essential for retaining Pol II at the transcription site. Analogous non-covalent matrices that restrain protein diffusion have been proposed for other biological processes (Buchan and Parker, 2009), although differing in composition and mechanism of assembly.
Collectively, these data favor a model where individual TFs are recruited to activated genes in a series of sequential steps, implying that controlling the recruitment efficiency of any of these TFs could significantly impact transcriptional yield. In contrast, studies done in other experimental systems suggest that activated genes are recruited to foci enriched in pre-assembled complexes containing phospho-Pol II and various TFs (Chakalova et al., 2005). It is not yet known under what circumstances each of these model mechanisms is favored over the other, but one could imagine that it might be gene or cell type specific. Interestingly, just as polytene chromosomes have multiple copies of genes that allow amplified transcription of regulated genes, it has been proposed that colocalization to these “transcription factories” in diploid cells boosts the expression of co-regulated genes (Chakalova et al., 2005).
Determining what TFs are specifically ADP-ribosylated by PARP and to what extent will be an important step to evaluate what factors make up the “transcription compartment” and to establish how ADP-ribose polymers cause retention of factors (Fig. 1B). To this end, additional FRAP experiments combined with PARP inhibitor treatments could assess whether the mobilization of the other TFs are more or less affected than Pol II by PARP inactivation. Because Spt6 was almost completely retained at the Hsp70 loci after HS, one would expect this protein to be the most affected by PARP inactivation. If the ADP-ribose polymer/transcription compartment model is correct, then the efficiency of Hsp70 gene transcription should be dependent on the formation of this compartment; therefore, an inhibitor-mediated decrease in PARP activity should also reduce Hsp70 transcript levels. Moreover, the fact that PARP remains associated at the Hsp70 loci at the same level before and after HS indicates that the rate-limiting step in ADP-ribose polymerization (and by extension, formation of the compartment) is not the recruitment of PARP itself, but rather the activation of PARP molecules already localized to the Hsp70 loci. Insight into what triggers PARP activity during HS, and conversely, what signals repress PARP activity awaits further studies.
The work presented by Zobeck et al., as well as work by others (Voss and Hager, 2008), successfully combines biochemical and biophysical methods to study the mechanistic details of dynamic biological processes. While several features of complexes at the heart of such processes challenge existing methods in their size, dynamic composition, and interactions with surrounding matrices, improved structural and kinetic characterization cannot be far behind.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Buchan JR, Parker R. Mol Cell. 2009;36:932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakalova L, Debrand E, Mitchell JA, Isborne CS, Fraser P. Nat Rev Genet. 2005;6:669–77. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Biochim Biophys Acta. 2008;1783:2044–51. doi: 10.1016/j.bbamcr.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ. Annu Rev Biochem. 2010;79:271–93. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Science. 2003;299:560–2. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Mol Cell. 2007;28:978–90. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]