Summary
In the course of combating infectious agents, type I Interferon (IFN) needs a timely down-regulation mechanism to avoid detrimental overreaction. Here we showed a mechanism for restraining type I IFN responses, which relied on a HECT domain ubiquitin (Ub) E3 ligase, RAUL. RAUL limited type I IFN production by directly catalyzing lysine 48-linked polyubiquitination of both interferon regulatory factor 7 (IRF7) and IRF3 followed by proteasome-dependent degradation. Suppression of RAUL by dominant negative RAUL or siRNA augmented both basal and virus-induced production of type I IFN, which resulted in reduced viral replication. The Kaposi’s sarcoma-associated herpes virus immediate-early lytic cycle trigger protein RTA recruited this mechanism to augment its countermeasures against the host antiviral response. These results unveil a previously unrecognized “brake mechanism” for type I IFN that maintains proper low amounts of type I IFN under physiological conditions and restrains its magnitude when the antiviral response intensifies.
Introduction
Interferon regulatory factor 7 (IRF7) and IRF3 are master transcriptional factors that regulate type I interferon (IFN) gene (IFN-α and IFN-β) induction and innate immune defenses after virus infection (Honda et al., 2005). IRF7, the master regulator for type I IFN, is induced by toll-like receptor 7 (TLR7) and TLR9, while IRF3 is activated through TLR3 and TLR4 (Doyle et al., 2002). These factors need to be destroyed in a timely manner after their activation, and this task is fulfilled by the ubiquitin (Ub)-proteasome system (Kodadek et al., 2006). Recent studies indicate that both IRF7 and IRF3 proteins undergo ubiquitination-mediated proteolysis: prolyl isomerase Pin1, E3 ligase Ro52, Ub ligase (E3) RBCC protein interacting with PKC1 (RBCK1) and human immunodeficiency virus (HIV)1 accessory proteins Vpr and Vif have been implicated indirectly for IRF3 (Barrington et al., 1998; Hemmi et al., 2004; Higgs et al., 2008; Kawai et al., 2004; Nakagawa and Yokosawa, 2000; Okumura et al., 2008; Saitoh et al., 2006; Yu et al., 2005; Zhang et al., 2008), whereas the Kaposi’s sarcoma-associated virus (KSHV) immediate-early lytic cycle trigger protein, RNA Transcriptional Activator (RTA, also known as ORF50) and the rotavirus nonstructural protein NSP1 have been shown to target IRF7 for proteolysis (Barro and Patton, 2007; Yu et al., 2005). Yet the endogenous Ub E3 ligase for IRF7 is still unknown (Kaisho and Tanaka, 2008). Although Pin1 and RBCK1 have been implicated in the ubiquitination of activated IRF3, an endogenous E3 ligase that directly ubiquitinates IRF3 has also not been reported. The fact that the usually low expression of short half-lived IRF7 in normal cells can be boosted by treatment with proteasome inhibitors (Yu et al, 2005) clearly implies that there exists a cellular ubiquitination mechanism for controlling the abundance of the IRF7 protein, and this remains a key missing component in understanding regulation of the type I IFN pathway.
Ubiquitination requires the sequential actions of three enzymes: Ub-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub ligase (E3), and the E3 dictates which target protein gets ubiquitinated. Different Ub chain structures tend to direct distinctive functional consequences, which is best manifested by lysine 48 versus lysine 63-linked Ub chains, resulting in proteolysis or functional activation, respectively (Chen, 2005; Hershko and Ciechanover, 1998). There are two types of E3 ligase, HECT (Homologous to E6-Associated Protein C-Terminus) domain and The Really Interesting New Gene (RING) finger domain E3 Ub ligases. Similar to phosphorylation, ubiquitination is a reversible process, and this is accomplished by deubiquitinases (DUBs; also known as deubiquitinating enzymes), which remove Ub moieties from Ub-protein conjugates, leading to reduced ubiquitination signaling (Komander et al., 2009).
The ubiquitous nature of pathogens and stress proteins are such that if left unchecked the host will be overwhelmed by immune activation. Therefore, the immune system needs to constantly create (or maintain) a balance between activation and inhibition to avoid detrimental overreaction. Although immunologists have long been focusing on activation machinery, a picture of how immune balance is achieved is beginning to emerge (Liew et al., 2005). The present work set out to investigate the other side of the coin, the mechanisms that restrain type I IFN responses. RAUL (RTA-Associated Ubiquitin Ligase) (also known as KIAA10 or UBE3C) is an established HECT domain E3 Ub ligase that is able to catalyze a variety of unanchored polyUb chain types in vitro and targets the putative transcriptional factor TIP120B for proteolysis, yet its physiological functions are largely unknown (Garcia-Gonzalo and Rosa, 2005; Wang et al., 2006; You and Pickart, 2001; You et al., 2003).
The first implication that this particular ligase might play a role in the IFN pathway came when RAUL, together with IRF7, were identified in our yeast two-hybrid screening as binding partners for the Kaposi’s sarcoma-associated virus (KSHV) immediate-early lytic cycle trigger protein, RTA. We also observed that RTA, as a viral E3 Ub ligase, promotes direct Ub-proteasome-dependent proteolysis of IRF7 the master regulator of type I IFN (Yu et al., 2005). In addition, we observed that the viral E3 RTA can cooperate with cellular E3 RAUL to enhance proteolysis of both IRF7 and IRF3. These observations, together with the fact that IRF7 and IRF3 share structural and functional similarities as well as phosphorylation pathways (Sharma et al., 2003), prompted us to hypothesize that RAUL itself might regulate both the IRF7 and IRF3 proteins, thus negatively regulating type I IFN. We further hypothesized that KSHV RTA not only copies but also recruits this cellular negative regulatory mechanism of type I IFN to mute antiviral innate responses.
Here we showed RAUL regulates type I IFN by targeting both IRF7 and IRF3 for lysine 48-linked ubiquitination and proteolysis, which represents an important pathway in maintaining proper low amounts of type I IFN under physiological conditions, and for restraining its magnitude when the antiviral innate response intensifies. We also showed that RAUL was positively regulated by a deubiquitinating enzyme Ubiquitin Specific Processing Protease 7 (Usp7 also known as HAUSP) and that KSHV RTA utilized and enhanced this mechanism to augment its countermeasures against the host antiviral response, which may represent a common strategy adopted by viruses to counteract innate immunity.
Results
RAUL associates with IRF7 and IRF3
A necessary step for a HECT domain E3 ligase to initiate ubiquitination is physical binding to its target protein. To determine whether and how RAUL binds to either the IRF7 or IRF3 proteins under more physiological conditions in mammalian cells, we first used immunoprecipitation to confirm that endogenous binding between RAUL and IRF7 or IRF3 occurred in the mouse bone marrow dendritic cell line DC 2.4 (Fig 1A). Next reciprocal immunoprecipitation and immunoblot experiments indicated that RAUL indeed interacted with IRF7 and IRF3 (but not IRF1) in human cells after exogenous coexpression (Fig S1A and 1B available online). Third, we verified the associations in vitro by glutathione-S-transferase (GST) pull down assays and showed that both IRF7 and IRF3 interacted with RAUL through their N-terminal regions (Fig S1C and S1D). Fourth, further deletion analysis showed that the amino-terminal regions of IRF7 (amino acids 103-151) (Fig S1E) and IRF3 (amino acids 1-128) (Fig S1F) were required for mediating the interaction with full length RAUL protein, and that RAUL associated with IRF7 and IRF3 via its N-terminal region (1-655) (Fig S1G). Fifth, we tested whether the binding is dependent upon IRF7 phosphorylation status and found that all three IRF7 phosphorylation-deficient mutants (IRF7S483A, IRF7Δ477-479, and IRF7D490A) retained their ability to bind RAUL (Fig S1H, lanes 1, 2 and 4). A similar pattern was observed with IRF3 phosphorylation mutants (data not shown), indicating that the binding between IRF7 or IRF3 and RAUL are independent of their phosphorylation status. Finally, we examined whether RAUL catalytic activity affects the binding and found that its catalytically inactive mutant (C1051A, designated as Myc-RAUL-M) was unchanged in its binding ability for IRF7 (Fig S1H, lane 3). Collectively, these results show that RAUL binds specifically to both IRF7 and IRF3 both inside cultured cells and in cell-free extracts.
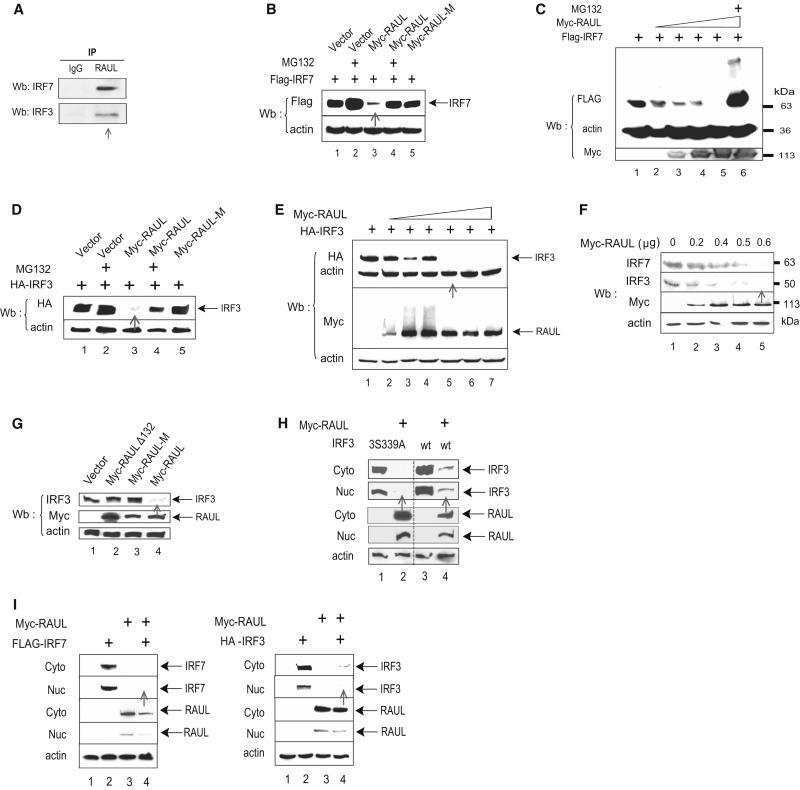
Figure 1. RAUL directs IRF7 and IRF3 for proteolysis.
(A): Interaction between endogenous RAUL and IRF7 or IRF3. Lysates of DC2.4 mouse bone marrow dendritic cells treated with the proteasome inhibitor MG132 were immunoprecipitated with anti-RAUL and immunoblotted with anti-IRF7 or anti-IRF3. (B, C, D and E): RAUL, but not its calalytically inactive mutant, causes loss of IRF7 and IRF3 proteins. Flag-IRF7 or HA-IRF3 was cotransfected with Myc-RAUL or Myc-RAUL-M (point mutant C1051A) into 293T cells in the presence of MG132 (0.5 μM for 12 hr). The amounts of IRF7 and IRF3 proteins were evaluated by immunoblot. (C and E): Dose response with increasing amounts of Myc-RAUL. (F): Enforced RAUL expression causes loss of endogenous IRF7 and IRF3 proteins. The indicated increasing amounts of Myc-RAUL plasmid were transfected into 293T cells, and the endogenous IRF7 or IRF3 protein were assessed by IRF7 or IRF3 immunoblotting. (G): RAUL-mediated IRF3 proteolysis is dependent on its substrate binding and catalytic domains. WT RAUL or its substrate binding domain deletion mutant (Myc-RAULΔ132, also designated as Myc-RAUL-DN) or its catalytically inactive mutant (Myc-RAUL-M) were transfected into 293T cells and endogenous IRF3 protein was evaluated by IRF3 immunoblotting. (H): RAUL-mediated loss of IRF3 occurs independently of IRF3 phosphorylation status. Myc-RAUL and WT IRF3 or its phosphorylation-deficient mutant IRF3S339A were cotransfected into 293T cells and cytoplasmic and nuclear portions were isolated and amounts of IRF7, IRF3 and RAUL were determined by immunoblotting. (I): Subcellular distribution pattern. 293T cells were transfected with Myc-RAUL together with Flag-IRF7 or HA-IRF3 plasmids. Cytoplasmic and nuclear fractions were isolated and IRF7, IRF3 and RAUL proteins were detected by immunoblot with Flag, HA or RAUL antibodies, respectively.
RAUL directs IRF7 and IRF3 for proteolysis
We next sought to determine whether RAUL, like KSHV E3 RTA, might promote IRF7 or IRF3 proteasome-dependent proteolysis. Indeed, coexpression of wild-type (WT) RAUL but not the catalytic domain point mutant of RAUL, with IRF7 or IRF3 led to a loss of both proteins in a dose-dependent manner, which was prevented by treatment with the proteasome inhibitor MG132 (Fig. 1B, C, D and E). We also observed that the N-terminal deletion mutant (RAULΔ132, designated as RAUL-DN in this article) as well as its catalytically inactive mutant (Myc-RAUL-M) functionally acted in a dominant negative fashion to inhibit RAUL function (data not shown). These results indicate that RAUL mediates IRF7 and IRF3 proteolysis in a ubiquitination and proteasome-dependent manner. To determine whether RAUL also regulates endogenous IRF7 and IRF3, we transfected increasing amounts of RAUL into 293T cells, which caused a dose-dependent loss of the endogenous IRF7 and IRF3 proteins (Fig 1F). To determine which domains of RAUL are involved in this proteolysis, we cotransfected RAUL mutants, which either lack the substrate binding domain (RAULΔ132) or the catalytic cysteine (RAUL-M), together with IRF3 plasmids into 293T cells. Both the substrate-binding and intact catalytic C-terminal domains of RAUL proved to be required (Fig 1G) suggesting that the degradation involves both specific-target recognition and the enzymatic activity of RAUL. We also tested whether RAUL-mediated proteolysis depends on IRF3’s phosphorylation status. However, Fig 1H shows that RAUL promoted degradation of both WT IRF3 and its phosphorylation deficient mutant (IRF3S339A) in a cotransfection system, indicating that the proteolysis is independent of IRF3 phosphorylation status. A similar pattern was observed for IRF7 phosphorylation mutants (data not shown).
We also examined whether the proteolysis occurs in the nucleus or cytoplasm. A sub-cellular localization experiment showed that when expressed alone IRF7 and IRF3 were largely localized in the cytoplasm, but that when coexpressed with RAUL a large proportion of both forms of IRF7 and IRF3 could no longer be detected (Fig 1I, lane 4). RAUL exhibited a similar sub-cellular expression pattern as IRF3 and IRF7 (Fig 1I, lane 3). Therefore, RAUL is capable of regulating both the resting (cytoplasmic) and activated (nuclear) forms of both IRF7 and IRF3, the key transcription factors mediating innate type I IFN responses to cell stress, and RAUL directs them for proteasome-dependent proteolysis in a manner that is dependent on both the substrate-binding and catalytic domains of RAUL.
RAUL ubiquitinates IRF7 and IRF3 in vivo and in vitro
We then investigated whether RAUL-mediated IRF7 and IRF3 proteolysis occurs through ubiquitination. Initially, we asked whether RAUL can induce the addition of ubiquitin chains on endogenous IRF7 and IRF3 proteins and observed that ectopic expression of WT RAUL, but not of its catalytically inactive mutant, produced polyubiquitinated forms of both proteins in the human DG7 lymphoma cell line (Fig 2A and 2B). These results indicate that RAUL overexpression induced endogenous IRF7 and IRF3 ubiquitination. To determine whether endogenous IRF7 undergoes ubiquitination in resting conditions, dendritic cells DC2.4 were electroporated with hemagglutinin (HA)-Ub and treated with MG132. IRF7 was readily polyubiquitinated (Fig 2C), but this was substantially reduced in the absence of either HA-Ub or MG132. Notably, depletion of RAUL by specific small interfering RNA (siRNA) (see below) in the same cells abolished IRF7 ubiquitination, suggesting that IRF7 undergoes ubiquitination and proteasome-mediated degradation events, even in the absence of pathogen induction stresses, in which RAUL appears to be essential. We proceeded to determine whether RAUL is required for IRF7 and IRF3 polyubiquitination under stress conditions. Primary mouse bone marrow dendritic cells were isolated (Tseng et al., 2001) and electroporated with a dominant negative form of RAUL (RAUL-DN) to suppress RAUL functions and the cells were treated with lipopolysaccharide (LPS) to mimic TLR4 activation. As shown in Fig 2D, LPS readily stimulated IRF7 ubiquitination, whereas suppression of RAUL by this approach abolished the induced IRF7 ubiquitination. Likewise, when a stable 293T cell line ectopically expressing toll-like receptor 3 (293T-TLR3), which constitutively express TLR3, was stimulated with poly I:C to mimic TLR3 activation, polyubiquitinated forms of IRF3 were induced, but silencing RAUL abrogated the effect (Fig 2E). Therefore, these results suggest that RAUL is also required for polyubiquitination of the much higher stress-induced amounts of IRF7 and IRF3.
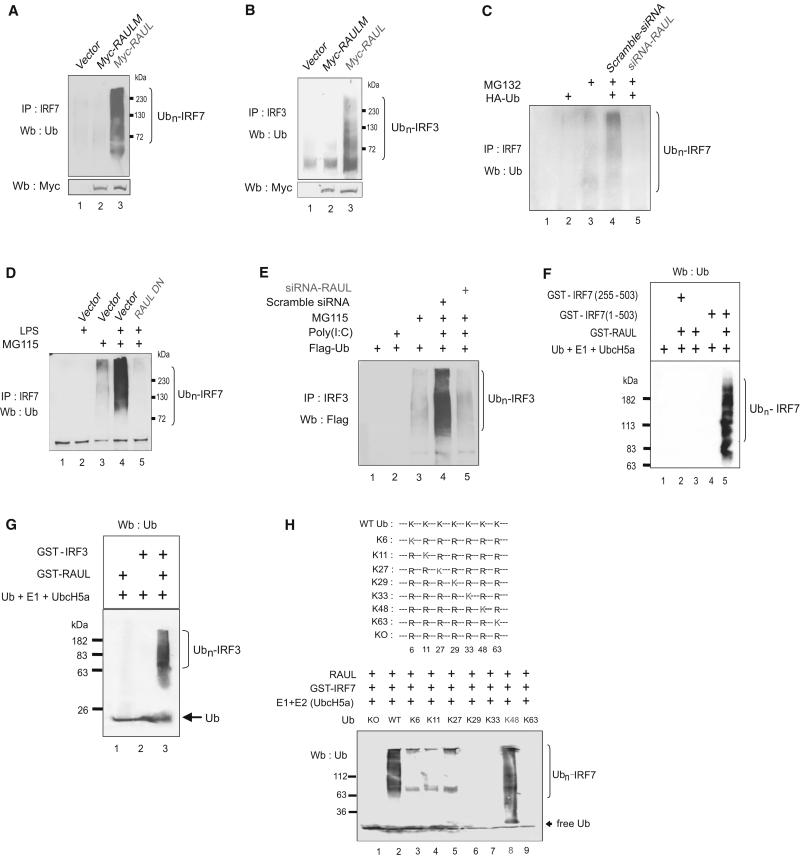
Figure 2. RAUL ubiquitinates IRF7 and IRF3 in vivo and in vitro.
(A and B): RAUL mediates ubiquitination of endogenous IRF7 and IRF3. DG75 B-lymphocyte cells were electroporated withWT Myc-RAUL or its catalytically inactive mutant (Myc-RAUL-M) plasmids in the presence of proteasome inhibitor MG115 and lysates were immunoprecipitated with anti-IRF7 and IRF3 and ubiquitinated forms of IRF7 and IRF3 were determined by Ub immunoblotting. (C): RAUL is required for endogenous IRF7 ubiquitination. Mouse bone marrow dendritic cells DC2.4 were electroporated with siRNA-RAUL or scramble siRNA together with HA-Ub plasmid and the indicated cells were treated with MG132. The cell lysates were immunoprecipitated with anti-IRF7 followed by Ub immunoblotting. (D): RAUL is required for ubiquitination of endogenous IRF7 following TLR4 activation. Mouse primary bone marrow dendritic cells were isolated (Tseng et al., 2001) and electroporated with dominant negative (Myc-RAUL-DN) plasmid or control vector and the indicated cells were treated with LPS (100 ng/ml for 9 hr) and proteasome inhibitor MG115. Conjugated IRF7 was determined by immunoprecipitation with anti-IRF7 followed by Ub immunoblotting. (E): RAUL is required for ubiquitination of IRF3 following TLR3 activation. 293-TLR3 cells were transfected with Flag-Ub plasmids and indicated cells were treated with poly I:C (25μg/ml for 9 hr) or MG115 (0.5μM for 12 hr). Conjugated IRF3 was evaluated by immunoprecipitation with anti-IRF3 followed by immunoblot with anti-Flag. (F and G): RAUL catalyzes the conjugation of polyUb chains on IRF7 and IRF3 in vitro. Purified GST-RAUL protein was incubated with various combinations of WT or truncated GST-IRF7 or GST-IRF3 plus E1, UbcH5a (E2) and Ub at 37° C for 1 hr. Following the ubiquitination reaction, the samples were subjected to immunoblot with Ub antibody for detection of polyubiquitinated forms of IRF7 and IRF3. Only a low catalytic concentration of GST-RAUL (20 nM, as an E3 source) was used compared to 400nM of target proteins to minimize interference from RAUL autoubiquitination (lane 3). (H): Determining IRF7 and IRF3 Ub chain structure by single lysine mutant assay in vitro. Illustration of Ub lysine mutants (upper panel). PurifiedWT Ub or its single lysine mutants, plus E1, E2 (Ubc5a), GST-IRF7 and RAUL were employed in in vitro cell-free Ub assays and ubiquitinated forms of the proteins were evaluated by Ub immunoblotting (lower panel).
To determine whether RAUL is able to directly catalyze the conjugation of ubiquitination on both proteins, we reconstituted the ubiquitination of IRF7 and IRF3 in a cell-free system using purified glutathione-S-transferase (GST)-IRF7 or GST-IRF3, E1, UbcH5a (E2) and Ub proteins. RAUL efficiently catalyzed the conjugation of polyUb chains on both IRF7 and IRF3 in cooperation with the E2 enzyme UbcH5a. Omission of any one of these ingredients or by using an IRF7 N-terminal deletion mutant as substrate abolished ubiquitination (Fig 2F, lane 5; Fig 2G, lane 3). Collectively, these results provide strong evidence that RAUL catalyzes polyubiquitination on IRF7 and IRF3 both in vitro and in cells under both physiologically normal and induced stress settings. Therefore, we conclude that RAUL represents a bona fide endogenous Ub E3 ligase that directly catalyses ubiquitination of both IRF3 and IRF7.
RAUL catalyzes lysine 48-linked polyUb chains on IRF7 and IRF3
To determine what type of Ub chain structure was formed on the IRF7 and IRF3 proteins, first we employed a set of mutants in which six out of the seven lysine residues (lysine 6, −11, −27, −29, −33, −48 and −63) in the Ub molecule were mutated to arginine, leaving just one lysine for polymerization. These recombinant Ub point mutants plus the Ub mutant with all seven lysines mutated to arginine (KO) were tested in a cell-free Ub assay to determine which lysine is utilized (Fig 2H, upper panel). As shown in Fig 2H (lower panel) and Fig S2A, RAUL catalyzed the conjugation of polyUb chains on both proteins only with the Ub mutant retaining a lysine at position 48 (K48), indicating that RAUL catalyzes lysine 48-linked Ub chains on both proteins in vitro. We then tested these Ub single lysine mutants plus a lysine 48 point mutation (K48R) in transfected cells. In accord with the in vitro cell-free data, IRF7 and IRF3 polyubiquitination was observed only with Ub mutants containing lysine at position 48 (K48), but not with any mutants where lysine 48 was eliminated (K48R) (Fig S2B, S2C and S2D). Therefore, these findings provide evidence that the polyUb chains on IRF7 and IRF3 by RAUL are linked primarily via lysine 48 of Ub.
RAUL is important of for expression of IRF7 and IRF3 as well as restraining type I IFN responses
To determine whether RAUL is a physiologically relevant regulator for IRF7 and IRF3, we generated a plasmid-based RAUL siRNA. Fig 3A shows that the RAUL-specific siRNA caused a very effective reduction of RAUL protein compared to a randomly scrambled negative control siRNA. Furthermore, silencing RAUL caused a dramatic accumulation of endogenous IRF7 and IRF3 proteins in 293T, Hela and primary mouse spleen cells (Rag2−/− c.c7 TCR transgenic T lymphocyte cells) (Fig 3B and 3C). To eliminate the possibility that the accumulation of IRF7 and IRF3 here was caused by nonspecific double-stranded RNA-mediated activation of IFN responses, IRF1 and IRF2 were included as controls and their amounts were not affected (Fig 3B), confirming the specificity of the siRNA. RAUL silencing also led to accumulation of endogenous IRF3 and IRF7 protein by over 10-fold in a dose-dependent manner (Fig 3D) and extended their half-lives by several-fold in the presence of cycloheximide (CHX) (Fig 3E), indicating a crucial role for RAUL in negative regulation of the resting forms of both proteins.
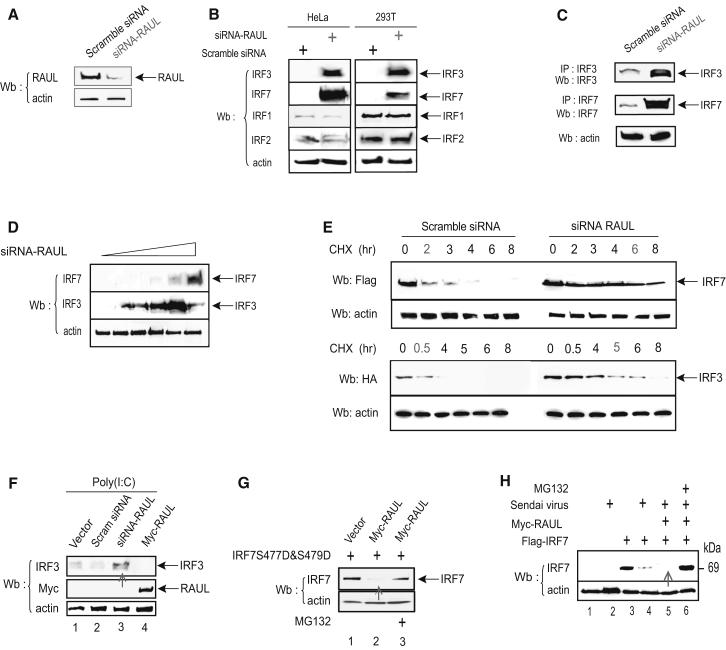
Figure 3. Silencing RAUL increases IRF7 and IRF3 expression.
(A): Immunoblot of lysates with RAUL specific antibody from 293T cells transiently expressing siRNA-RAUL or scramble siRNA. (B and C): Reducing RAUL specifically induces high amounts of endogenous IRF7 and IRF3 proteins in both cell lines and primary cells. (B): Immunoblot with anti-IRF7 or anti-IRF3 of lysates from cells transiently expressing siRNA-RAUL or scramble siRNA in Hela or 293T cells. (C): Isolated primary T-lymphocytes from 5 week-old Rag2−/− 5c.c7 TCR transgenic knockout mice were activated with pigeon cytochrome C (PCC) and expanded for 4 days. After activation, more than 90% of the cell population was TCR-specific transgenic T-lymphocyte cells, which were then transfected with siRNA RAUL or scrambled siRNA. Endogenous IRF3 and IRF7 proteins were determined by IRF3 or IRF7 immunoprecipitation followed by immunoblot. (D): siRNA-mediated reduction of RAUL increases accumulation of both the IRF7 and IRF3 proteins in a dose-dependent manner. 293T cells were transfected with two-fold increasing amounts of siRNA-RAUL, and endogenous IRF7 or IRF3 protein was determined by IRF7 or IRF3 immunoblot. (E): Knocking down RAUL extends the half-lives of IRF7 and IRF3 proteins. Flag-IRF7 or HA-IRF3 were cotransfected with siRNA-RAUL or scrambled siRNA into 293T cells, and the protein synthesis inhibitor cyclohexamide (CHX) was added at a concentration of 30 μg/ml at 36 hr for the indicated times. The IRF7 and IRF3 proteins were determined by Flag or HA immunoblot. (F): Silencing RAUL in 293-TLR3 cells increases the amount of IRF3 protein. Immunoblot analysis of IRF3 protein from 293-TLR3 cells transiently expressing siRNA-RAUL or Myc-RAUL. Poly I:C was used to stimulate TLR3 activation. (G): RAUL also regulates a constitutively active form of IRF7. Immunoblot analysis of lysates with IRF7 antibody from 293T cells transiently coexpressing Myc-RAUL and IRF7S477D and S479D in the presence and absence of MG132. H): Enforced RAUL expression augments the Sendai virus-mediated loss of IRF7 protein. Myc-RAUL and Flag-IRF7 plasmids as indicated were transfected into 293T cells Sendai virus infection (16 hr) with or without treatment with MG132. IRF7 protein was evaluated by immunoblot with a Flag antibody.
To address whether RAUL also regulates the active forms of both proteins, we examined how silencing of RAUL affected the amounts of IRF7 and IRF3 under stress conditions. First, in 293T-TLR3 cells treated with poly I:C to activate IRF3, the reduction of RAUL by siRNA resulted in an accumulation of the endogenous active form of IRF3 protein over the control (Fig 3F). Similarly, coexpression of RAUL with a constitutively active form of IRF7 (S477D and S479D) (Lin et al., 2000) in 293T cells led to the loss of these proteins, which was rescued by treatment with the proteasome inhibitor MG132 (Fig 3G). In Sendai virus-infected cells, coexpression of RAUL with IRF7 caused a further loss of the expected to be fully active form of IRF7, which could be prevented by adding MG132 (Fig 3H). These results demonstrate a role for RAUL in the negative regulation of both TLR3-activated IRF3 and Sendai virus-activated forms of IRF7 proteins. Collectively, these data indicate that RAUL down-regulates both resting and active forms of both proteins.
We next investigated whether suppression of RAUL by either siRNA or dominant negative approaches would translate into an increase of type I IFN production under resting and stress conditions. Indeed, depletion of RAUL by siRNA strongly activated IFN-α1 and IFN-β reporter activities, and Sendai virus infection further increased their activities (Fig 4A). An identical pattern was also observed in poly I:C-stimulated 293T-TLR3 cells (Fig 4B). Likewise, silencing RAUL in DC 2.4 cells augmented both basal and LPS-stimulated IFN-α1 mRNA (Fig 4C). These siRNA and dominant negative studies provide confirmation of the importance of RAUL in regulating the expression of type I IFN.
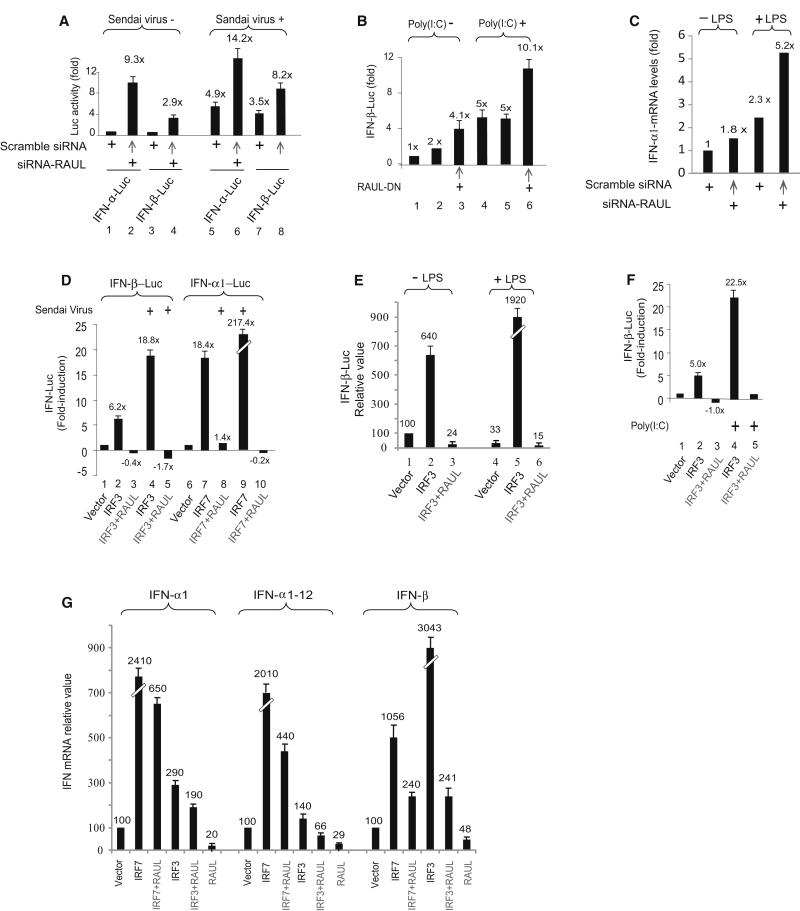
Figure 4. RAUL negatively regulates IRF7 and IRF3-mediated type 1 IFN production.
(A, B and C): Suppression of RAUL by siRNA or dominant negative augments type 1 IFN activity. (A): Luciferase assay of 293T cells transiently expressing siRNA-RAUL together with IFN-α1 or IFN-β luciferase reporter plasmids. The cells were infected with Sendai virus for 16 hr before harvest. The basal IFN-α and IFN-β activities were arbitrarily set to 1.0. (B): Luciferase assay of IFN-β promoter activity from 293-TLR3 cells transiently expressing dominant negative RAUL poly I:C (25 μg/ml) for 9 hr to activate IRF3. (C): Silencing of RAUL increases IFN-α1 mRNA in dendritic cells. DC 2.4 cells were electroporated with siRNA-RAUL or scrambled siRNA in the presence or absence of LPS. Total RNA was isolated and subjected to SYBR green Real time RT-PCR with primers for IFN-α1. (D, E and F): Enforced RAUL expression abolishes IRF7 and IRF3-mediated type I IFN induction under Sendai virus or LPS or poly I:C stimulation. The indicated cells were cotransfected with IFN-α1 or IFN-β luciferase reporters, IRF7 or IRF3 and RAUL and the indicated cells were infected with Sendai virus (16 hr) or stimulated by LPS (100 ng/ml, for 9 hr) or poly I:C. IFN promoter activation was determined by luciferase assay. G: RAUL inhibits IRF7 or IRF3-mediated type I IFN mRNA production. 293T cells were cotransfected with IRF7, IRF3 and RAUL plasmids as indicated. Total RNA was isolated and subjected to SYBR green Real time RT-PCR with primers for IFN-α1, IFNα-1-12 and IFN-β, respectively, to assess type 1 IFN mRNA output.
We also examined whether ectopic expression of RAUL would affect IRF7 and IRF3- dependent type I IFN activation. Coexpression of RAUL together with IRF7 or IRF3 in 293T cells completely abolished the IRF7 and IRF3-mediated type I IFN reporter gene activities in both the absence and presence of Sendai virus or of poly I:C or LPS (Fig 4D, 4E and 4F). These results imply that RAUL exerts a powerful restrictive function on IRF7 and IRF3-dependent type I IFN responses via TLR signaling in a variety of pathogenic settings. Evidently, with the IRF7 and IRF3 proteins degraded, the cells’ ability to launch a type I IFN response was eliminated. As indicated in Fig 4G, coexpression of RAUL with IRF7 or IRF3 completely inhibited their induction of endogenous IFN-α and IFN-β mRNA measured by real-time reverse transcription-polymerase chain reaction (RT-PCR). Notably, ectopic expression of RAUL alone decreased IFN-α and IFN-β to 20-50% of the basal amounts in mock treated cells as well. The results for IFNα and IFN-β mRNA measurements were essentially the same as for IFN-promoter driven luciferase (IFN-LUC) reporter assays for all of the above experiments with poly I:C and LPS induction (not shown, except for Fig 4C).
Collectively, these data demonstrate that RAUL regulates the stability and amount of both the inactive and activated forms of IRF7 and IRF3 proteins and points to a critical role for RAUL both in maintaining proper low amounts of type I IFN in normal conditions, as well as for restraining the magnitude of induced antiviral responses. A previous study demonstrated that Pin1 also indirectly targets IRF3 for destabilization through polyubiquitination and suggested that additional post-translational mechanisms may be involved (Saitoh et al., 2006). RAUL could represent one of them, although our further investigation has revealed that the E3 activity of RAUL toward IRF7 and IRF3 is independent of Pin1 activity and vice versa (data not shown).
The role of RAUL in antiviral innate immunity
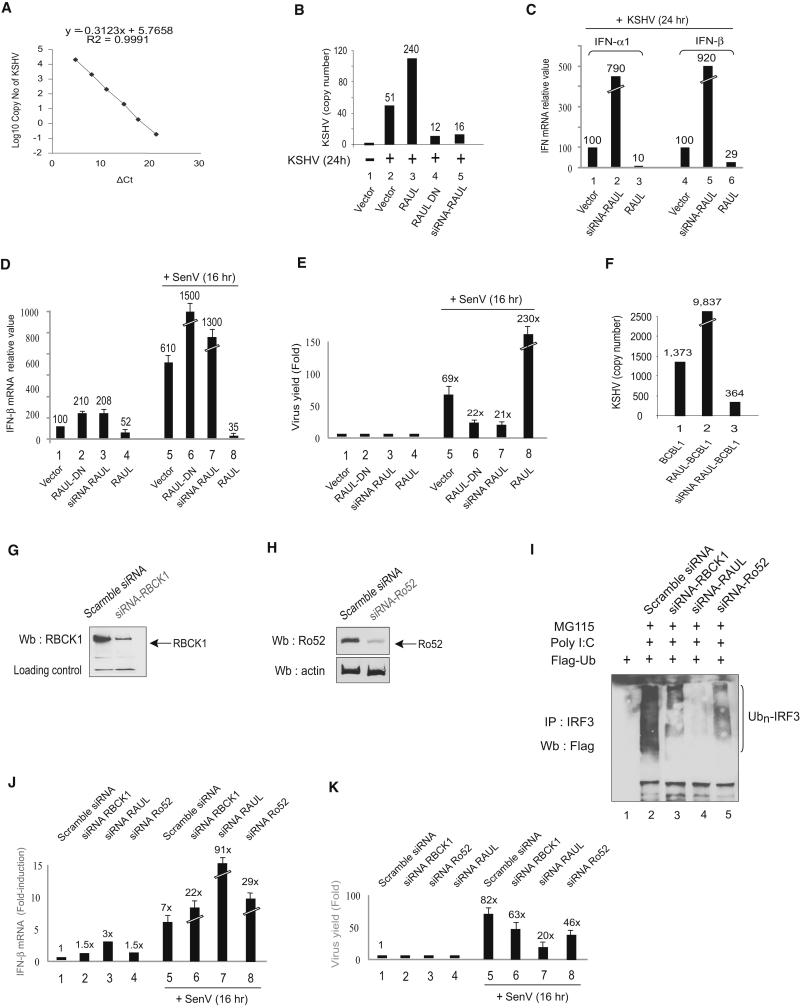
To address the role of RAUL in the innate antiviral response, we infected mouse embryonic fibroblasts (MEFs) with KSHV and monitored alterations in overall viral load in the infected cells using a Real-time DNA PCR-based method, which is able to quantify the copy number of KSHV genomes (Fig 5A). The amount of KSHV DNA generated in these the infected cells were measured as 51 copies per 100 ng of cell DNA. Ectopic expression of RAUL, which should suppress IRF3 and IRF7-mediated type I IFN production, boosted the production of KSHV by nearly 5-fold (240 versus 51 copies). In contrast, in the cells expressing dominant negative or siRNA RAUL, the viral copy number was instead decreased by 68-77% (12 and 16 versus 51 copies) (Fig 5B). A parallel type I IFN assay by Real-time PCR confirmed a reduction of type I IFN mRNA production subsequent to ectopic expression of RAUL and an increase following expression of siRNA RAUL (Fig 5C).
Figure 5. RAUL regulates the IFN-mediated antiviral response.
(A): A SYBR Real-time-PCR based quantitative approach for determining KSHV viral load. The standard curve was constructed using KSHV RAP DNA as target by the comparative Ct method. The viral load value, normalized to reference (β-actin) is given by 2 −ΔΔCt in the Applied Biosystems 7500 Real Time PCR system expressed as copy number per 100 ng of infected cell genomic DNA. (B, C, D and E): Role of RAUL in KSHV and Sendai virus-infected mouse embryonic fibroblast cells (MEFs). MEF cells were electroporated with WT RAUL,dominant RAUL-DN or siRNA-RAUL and the cells were infected with KSHV or Sendai virus for the indicated times. The cell lysates were assayed for viral DNA copy number (KSHV) per 100 ng of genomic DNA or relative viral RNA load (Sendai virus) (B and E) as well as for type 1 IFN mRNA production by Real time RT-PCR (C and D). (F):Role of RAUL in KSHV-infected BCBL1 cells. Lysates from BCBL1 derived cell lines stably expressing either RAUL or siRNA-RAUL were assayed for KSHV viral DNA copy number per 100 ng genomic DNA. (G and H): Comparison of the contribution of RBCK1, RAUL and Ro52 to endogenous IRF3 ubiquitination. Immunoblot of lysates with RBCK1 (G) or Ro52 (H) specific antibody from 293T cells transiently expressing siRNA-RBCK1 or siRNA-Ro52 or scramble siRNA. (I): Comparison of siRNA silencing to reduce polyubiquitination of IRF3 in activated cells. 293T-TLR3 cells were transfected with siRNA against RBCK1 or RAUL or Ro52 together with Flag-Ub and the cells were treated with poly I:C to induce polyUb-conjugation of IRF3 in the presence of the proteasome inhibitor MG115. The conjugation of endogenous IRF3 was detected by immunoprecipitation with IRF3 antibody and immunoblot with Flag antibody. (J and K): Relative roles of RBCK1, RAUL and Ro52 in viral replication. 293T cells were transfected with siRNA against RBCK1 or RAUL or Ro52, and then infected with Sendai virus for the indicated times. The cell lysates were assayed for IFN-β mRNA production (J) and for viral RNA load of Sendai virus by real-time RT-PCR (K).
To test the generality of this effect, we examined whether the expression of RAUL also affects the replication of Sendai virus in infected MEFs. A similar pattern was obtained: Sendai virus infection readily induced IFN-β production, while concurrent ectopic expression of RAUL abolished it (Fig 5D lanes 8 vs. 5). As a result, the virus proliferated more efficiently (Fig 5E, lanes 8 vs. 5). In contrast, suppression of RAUL by dominant negative or siRNA boosted IFN-β production (Fig 5D, lanes 6 and 7 vs. 5) and caused a reduction of viral proliferation (Fig 5E, lanes 6 and 7 vs. 5). We also examined whether RAUL affected KSHV DNA concentration in latently infected primary effusion lymphoma (PEL) BCBL1 cell lines selected to stably express either RAUL or siRNA-RAUL. Ectopic expression of RAUL increased the KSHV viral load over the control by 716% (9,837 vs 1,373 copies), and depletion of RAUL by siRNA reduced the viral DNA yield by 73% (364 vs 1,373 copies) (Fig 5F).
Previous studies have suggested that two other Ub E3 ligases RBCK1 and Ro52 also regulate IRF3 ubiquitination and type I IFN, although it is unknown whether the IRF3 ubiquitination mediated by these E3s is direct and whether it occurs with the endogenous forms of these proteins. Therefore, a question arose about their contributions relative to RAUL to IRF3 ubiquitination, to IFN-β production and more importantly to viral replication. To compare their E3 activity toward catalyzing IRF3 conjugation, we first silenced all three E3s individually using RNA interference in 293T-TLR3 cells, and examined the conjugation of endogenous IRF3 in response to poly I:C. Figs 5G and Fig 5H depict the efficacy of siRNA-RBCK1 and siRNA-Ro52 in silencing RBCK1 and Ro52 genes in 293 cells. As shown in Fig 5I, this resulted in different degrees of reduction in IRF3 conjugation with the largest reduction being observed after silencing RAUL. Next, we evaluated the effects of these siRNAs on IFN-β production. Silencing RBCK1 or RAUL or Ro52 in Sendai virus infected 293T cells caused an increase of IFN-β mRNA production by 3-, 12- and 4-fold, respectively, with lower effects on mock-infected cells (Fig 5J). Again, silencing RAUL produced the greatest increase both with and without Sendai virus infection. Last, we compared the impact of silencing these E3s on Sendai virus replication. Compared to the control, depletion of RBCK1 or RAUL or Ro52 led to a reduction in Sendai virus yield by 24%, 76% and 56%, respectively (Fig 5K). Collectively, these data suggest that all three E3s play some role in the negative regulation of IFN-β production through directly or indirectly catalyzing proteolytic IRF3 ubiquitination. Nevertheless, RAUL evidently has by far the more predominant role, possibly because of its unique capability to directly ubiquitinate both IRF3 and IRF7. Overall, our findings offer biological evidence that RAUL, through negatively controlling IRF7 and IRF3 protein stability, strongly limits the magnitude of the innate antiviral response.
KSHV recruits and enhances the function of RAUL
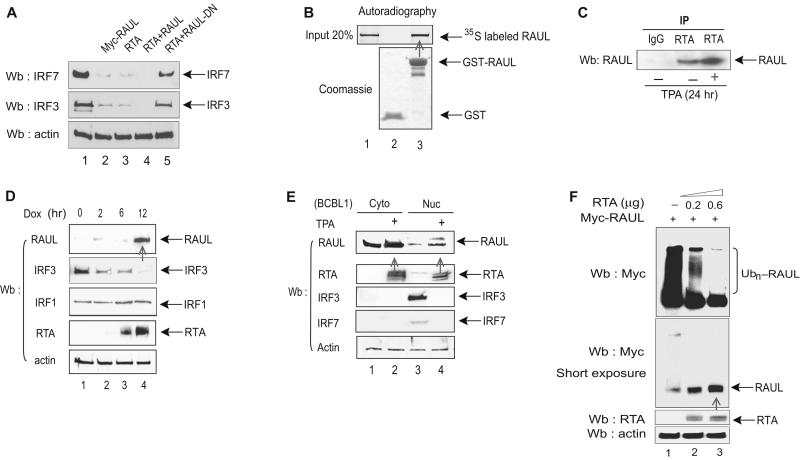
Both IRF7 and RAUL are binding partners for KSHV RTA, and we also observed that RTA can enhance the ability of RAUL to degrade both IRF7 and IRF3 in a cotransfection system (Fig 6A). These observations prompted us to investigate the possibility that RTA utilizes or modifies the RAUL regulatory mechanism for type I IFN to augment its own direct effects on blocking the antiviral response. We first confirmed the interaction between RTA and RAUL in vitro by a cell-free GST pull-down assay (Fig 6B). By using specific antibodies to both proteins, we also detected this binding with the endogenous protein in KSHV-infected BCBL1 cells, especially after TPA-treatment, which induced the expression of RTA (Fig 6C). To address the functional consequences of this binding, we examined whether RTA affected the stabilization of RAUL in both TRExRTA BCBL1 and standard BCBL1 PEL cell lines. Induction of RTA expression by either doxycycline (Dox) or TPA, respectively, substantially increased the steady-state amount of endogenous RAUL (Fig 6D and 6E), which in turn decreased the abundance of IRF3 and IRF7 (Fig 6E), but it had no effect on the amounts of IRF1 (Fig 6D), whose stability is also regulated independently by a ubiquitination-mediated pathway (Nakagawa and Yokosawa, 2000). To address whether this change in the abundance of RAUL might involve post-translational modification, we coexpressed RTA with RAUL in 293T cells. RTA dramatically reduced the degree of RAUL self-ubiquitination, with a concomitant increase in unconjugated RAUL (Fig 6F), implying that RTA-mediated deubiquitination of RAUL rescues it from self-induced protein degradation.
Figure 6. RTA recruits and enhances RAUL.
(A): RTA enhances the ability of RAUL to degrade IRF7 and IRF3. 293T cells were cotransfected with RTA, Myc-RAUL or Myc-RAUL-DN, IRF7 and IRF3 as indicated and the amounts of IRF7 and IRF3 proteins were detected by IRF7 or IRF3 immunoblot. (B and C):RTA binds to RAUL in vitro and vivo. (B): A GST-pull down assay was carried out with 35S-labeled in vitro translated RAUL and GST-RTA proteins. (C): Cell lysates from BCBL1 PEL cells were immunoprecipitated with anti-RTA followed by RAUL immunoblot to detect endogenous RTA-RAUL interaction with TPA. (D and E): Induction of RTA stabilizes RAUL protein. (D): TREx BCBL1 cells treated with doxycycline (Dox) for the indicated times to induce RTA expression, and RAUL, IRF3, IRF1 and RTA proteins were monitored by immunoblot using the indicated antibodies. (E): BCBL1 PEL cells were treated with TPA to induce RTA expression and cytoplasmic and nuclear fractions were isolated. The RAUL, IRF7, IRF3 and RTA proteins were evaluated by immunoblot using the indicated antibodies. (F):RTA mediates RAUL deubiquitination. RTA and Myc-RAUL plasmids were cotransfected into 293T cells and ubiquitinated forms of RAUL were determined by immunoblot with anti-Myc. To show both ubiquitinated and nonubiquitinated forms of RAUL, a short-exposure film was included (lower panel).
KSHV RTA mediates RAUL deubiquitination via Usp7
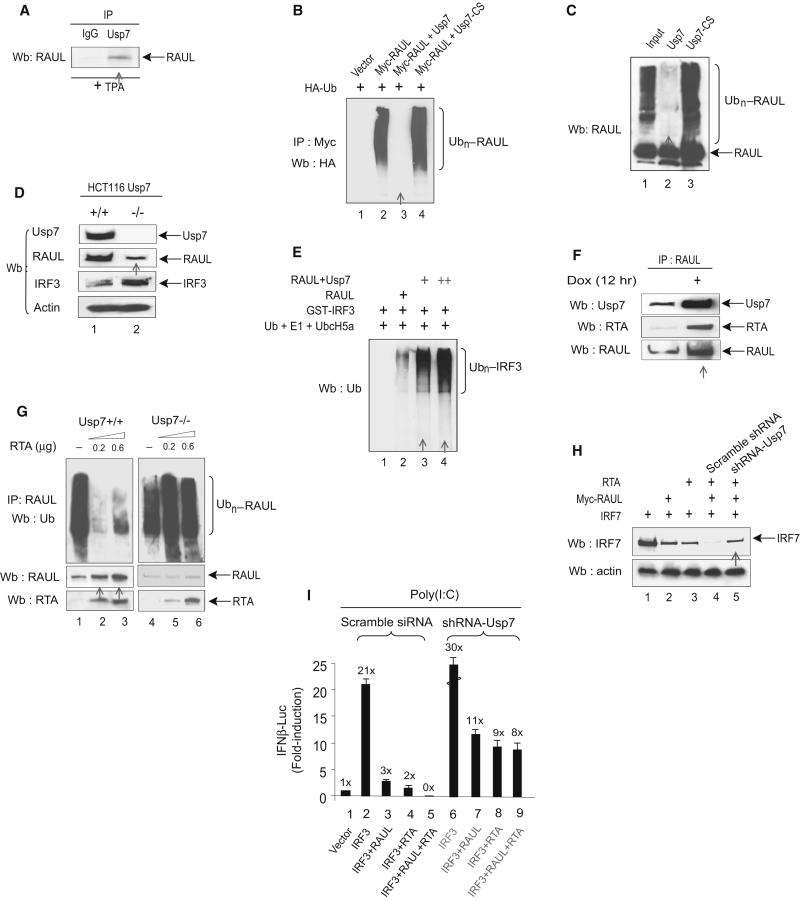
RAUL, as an E3 ligase, mediates the ubiquitination of not only its substrates but also itself (self-ubiquitination), leading to proteolysis of RAUL. We set out to investigate the mechanism underlying KSHV RTA-mediated RAUL deubiquitination and found that RTA promotes RAUL deubiquitination through a deubiquitinating enzyme Usp7 (formerly known as HAUSP) (Everett et al., 1997), which is best known for stabilizing p53 through deubiquitination (Li et al., 2002).
First, we investigated whether Usp7 can directly interact with RAUL. Using specific Usp7 and RAUL antibodies, we detected an interaction between endogenous RAUL and endogenous Usp7 proteins in BCBL1 cells (Fig 7A). To test the specific deubiquitination activity of Usp7 on RAUL, we examined whether Usp7 regulated RAUL ubiquitination in transfected cells. As shown in Fig 7B, a large amount of conjugated RAUL was detected in cells transfected with RAUL and HA-Ub; however, RAUL ubiquitination was completely abolished by Usp7 expression, and the Usp7 catalytically inactive mutant Usp7-cs failed to deubiquitinate RAUL. We then examined whether Usp7 can directly deubiquitinate RAUL in a purified protein system. A polyubiquitinated form of RAUL was generated in vitro Ub-conjugation system in the presence of E1, E2 (UbcH5a) and Ub. The resulting purified product was then subjected to in vitro deubiquitination in the presence of the WT or mutant Usp7 proteins purified from insect cells (Everett et al., 1997). RAUL alone generated high molecular weight moieties in the presence of E1, E2 and Ub, but including the Usp7 protein efficiently eliminated the polyUb moieties attached to RAUL, whereas its catalytically inactive point mutant Usp7-cs was functionally defective in deubiquitinating RAUL (Fig 7C, lanes 2 and 3). These data collectively indicate that Usp7 can deubiquitinate RAUL both inside cultured cells and in cell-free systems.
Figure 7. RTA mediates RAUL deubiquitination through Usp7.
(A): Endogenous RAUL and Usp7 interact. BCBL1 cells were treated with TPA for 24 hr and cell lysates were immunoprecipitated with anti-Usp7 followed by RAUL immunoblot to detect bound protein. (B and C): Usp7 deubiquitinates RAUL in vivo and in vitro. 293T cells were transfected with Myc-RAUL, WT Usp7, Usp7-cs and HA-Ub. The cells were treated with MG132 for 6 hr before harvest. Polyubiquitinated forms of RAUL were detected by immunoprecipitation with anti-Myc followed by HA immunoblot. (C): Purified RAUL protein (400 nM) plus E1, E2 (UbcH5a) and Ub were incubated at 37°C for 1 hr to catalyze self-ubiquitination. The Ub-RAUL protein was then purified by immunoprecipitation with anti-RAUL antibody and incubated with purified Usp7 protein or its catalytically inactive mutant (Usp7-cs) in deubiquitination buffer at 37°C for 2 hr. The resulting reactions were subjected to immunoblot with RAUL. (D): Usp7 upregulates RAUL protein amounts. Colon cancer cell lines HCT116 Usp7+/+ or Usp7−/− cells were immunblotted with the indicated antibodies to evaluate both RAUL and its target substrate IRF3. (E): Usp7 enhances the E3 activity of RAUL toward IRF3 in vitro. Immunoprecipitated purified RAUL protein from either RAUL expressing 293T cells or RAUL+Usp7 expressing 293T cells were incubated with its substrate GST-IRF3, plus E1, UbcH5a and Ub in vitro at 37°C for 1 hr. Conjugated IRF3 was determined by Ub immunoblot. (F): RTA increases the recruitment of Usp7 to RAUL complexes. TRExBCBL1 cells were treated with Doxycyclin (Dox) for 12 hr to activate RTA expression. Endogenous RTA-Usp7-RAUL interaction was evaluated by immunoblot with the indicated antibodies. (G):RTA promotes RAUL deubiquitination. HCT116 (Usp7+/+ or Usp7−/−) cells were electroporated with the indicated amounts of RTA plasmid and RAUL self-ubiquitition was determined by Ub immunoblotting. Unconjugated RAUL and RTA were measured by RAUL or RTA immunoblotting. (H): RTA enhances the ability of RAUL to degrade IRF7 via Usp7. 293T cells were cotransfected with IRF7, Myc-RAUL, RTA and shRNA-Usp7 as indicated, and IRF7 protein amounts were evaluated by IRF7 immunoblotting. (I): RTA and RAUL cooperate in suppressing IRF3-mediated IFNβ reporter activity. 293-TLR3 cells were transfected with the indicated combinations of IRF3, RAUL, RTA, shRNA- Usp7 and IFNβ-LUC plasmids and the cells were treated with poly I:C for 9 hr to mimic TLR3 activation. IRF3-mediated IFN-β reporter activity in the cells that were pretreated shRNA-RAUL or scrambled IFN-β activation was determined by luciferase assays.
We then investigated the functional consequences of the deubiquitination of RAUL. To determine the effect of Usp7 on RAUL protein, we examined the endogenous RAUL protein in Usp7 WT and silenced cells. Ablation of Usp7 gave rise to a decrease in RAUL protein and concomitantly increase in its substrate IRF3 (Fig 7D). This result indicates that Usp7 expression plays an important role in maintaining the stability of RAUL and presumably also of its cellular functions. To assess the effect of Usp7 toward the E3 activity of RAUL, we purified RAUL protein by immunoprecipitation from cells that either expressed RAUL alone or coexpressed RAUL together with Usp7, and incubated them in vitro with purified GST-IRF3, E1, UbcH5a and Ub to evaluate the activity of RAUL toward IRF3. Inclusion of RAUL from RAUL + Usp7 expressing cells resulted in a large increase of the ubiquitinated forms of IRF3 compared to in the absence of Usp7 (Fig 7E). This result indicates that Usp7 augments the E3 enzymatic activity of RAUL.
We next investigated whether KSHV RTA mediates RAUL deubiquitination through Usp7. To determine whether RTA assists in recruiting Usp7 to RAUL, we used doxycyclin (DOX) to induce RTA expression in TREx-BCBL1 cells. Using specific antibodies, an increase in the association of endogenous Usp7 with RAUL was observed in the presence of RTA (Fig 7F), suggesting that RTA recruits Usp7 to the RAUL protein to form RTA-Usp7-RAUL complexes, and that this inducible Usp7-RAUL association can be enhanced upon RTA activation. To determine whether Usp7 is required for RTA-mediated RAUL deubiquitination, we transfected RTA into both parental colon cancer HCT116 Usp7+/+ cells and into a derived cell line with the Usp7 gene homozygously disrupted (HCT116 Usp7−/−). As shown in Fig 7G, ectopic expression of RTA in WT Usp7 cells resulted in deubiquitination of endogenous RAUL, whereas added RTA failed to deubiquitinate RAUL in Usp7 null cells (Usp7−/−). Therefore, this result provides strong evidence that the deubiquitination and stabilization of RAUL by RTA occur via Usp7.
Finally, we sought to explore the functional consequence of RTA-promoted, Usp7-mediated RAUL deubiquitination by coexpressing RTA, RAUL and IRF7 in 293T cells. As before, expression of RTA cooperated with RAUL to enhance degradation of the IRF7 protein (Fig 7H); however, concurrently silencing Usp7 severely impaired this effect. Likewise, RTA increased the ability of RAUL to suppress IRF3-mediated IFN-β reporter activation in 293T-TLR3 cells, whereas RTA failed to do so in the same cells after Usp7 was silenced by shRNA (Fig 7I). The Usp7 shRNA also led to reduced inhibition of IRF3-mediated IFN-LUC activity by both RAUL and RTA alone. These results demonstrate that RTA augments RAUL’s activity toward its substrates IRF7 and IRF3 through Usp7 and reinforces the notion that Usp7 acts as a positive regulator for RAUL.
Taken together, these findings reveal an important post-translational regulatory mechanism for RAUL, in which the DUB enzyme Usp7 positively regulates RAUL by preventing it from proteolytic self-ubiquitination, and that KSHV RTA exploits this mechanism whilst amplifying RAUL activity to mute antiviral responses.
Discussion
In this report, we have identified an important negative regulatory mechanism acting on the type I IFN pathway that is likely required for both appropriate cellular homeostasis and for resetting the basal state after induction of antiviral innate immunity. This mechanism relies on the E3 ligase activity of RAUL, which exerts its inhibitory action on type I IFN by directly catalyzing lysine 48-linked polyubiquitination on both IRF7 and IRF3, the two most crucial transcriptional factors for mounting type I IFN responses. In all circumstances tested, increased or decreased amounts of RAUL negatively correlated with either strong negative or positive effects respectively on endogenous type I IFN production.
The implications of this mechanism are threefold: (1) In physiological conditions, it helps to keep type I IFN production at proper low basal amounts through targeting IRF7 and IRF3 ubiquitination and degradation; (2) Under stress conditions, in which type I IFN amounts rise dramatically, it may act as a “speed brake” to prevent detrimental overproduction; (3) Given its crucial role in type I IFN regulation, control of RAUL function may also become a battleground in the war between viruses and the host during infection. High interferon doses administered to patients can cause toxic effects, indicating a need for timely and spatially coordinated down-regulation mechanisms for type I IFN and for effective combat against infectious agents. We propose that RAUL, operating upstream of IRF7 and IRF3, is a key factor for maintaining appropriate cellular homeostasis and for antiviral innate immunity through controlling both the basal amount and intensity of induced type I IFN responses. We speculate that some of the other previously reported viral and cellular proteins that influence the ubiquitination status of both proteins might also exert their actions through the Ub E3 ligase RAUL, which is evidently the first such cellular enzyme identified that has been demonstrated to act directly and specifically on either or both IRF3 and IRF7.
IRF7 and IRF3 comply with all four of the criteria that should be met by specific HECT domain E3 ligase substrates (You et al., 2003): (1) They both bind to the N-terminal substrate binding domain of RAUL; (2) Purified RAUL specifically catalyzes the conjugation of ubiquitin on IRF7 and IRF3 in vitro; and (3) Their protein amounts are reduced by coexpression with RAUL, but not by its catalytically inactive mutant, and this destabilization is proteasome-dependent; (4) Depletion of RAUL increases the stability and hence overall amount of both proteins.
Our investigation into the role of KSHV RTA in RAUL signaling led us to uncover a critical role for deubiquitination in the regulation of RAUL function. Ubiquitination is a dynamic process that involves constantly adding and removing Ub moieties from the substrate. Self-ubiquitination plays an important role in the regulation of RING finger domain ligases TRAF2, TRAF3 and TRAF6 and deubiquitination-mediated protein stabilization has emerged recently as an accompanying regulatory step (Chomvarin et al., 2004; Friedman et al., 2008; Kayagaki et al., 2007; Wang et al., 2001; Yang et al., 2006). HECT domain Ub ligases, once thought to be constitutively active and to recognize their substrates independently of their post-translational modification are also subjected to post-translational regulation (Gao et al., 2004; Wiesner et al., 2007). It appears that, on one hand, RAUL is capable of catalyzing polyubiquitination constitutively, an ability that is required for regulating the resting forms of the IRF7 and IRF3 proteins; whereas on the other hand, RAUL activity can also itself be regulated through post-translational mechanisms under pathogenic settings, which is important for regulating the active forms of both IRF proteins.
Several recent studies have indicated that some Ub E3 ligases are physically associated with DUBs that directly reverse self-ubiquitination of the ligase, thus regulating its activity. However, only one HECT domain ligase Rsp5 has been previously shown to be regulated by a DUB Ubp2, and no effect on its E3 ligase activity was observed (Kee and Huibregtse, 2007; Shembade et al., 2010). Our findings suggest that RAUL can be rescued from undergoing self-ubiquitination-mediated degradation by Usp7 via direct deubiquitination, which represents an important post-translational regulatory mechanism for RAUL. Its activity is increased through deceleration of the self-induced degradation and decreased via acceleration of the degradation. Thus it appears that Usp7 is capable of cleaving Ub chains with both lysine 48- and 63-linkage from both RING finger and HECT domain E3 Ub ligases.
Much of our knowledge with regard to immunity comes from understanding how cells deal with viral infection, from smallpox to human immunodeficiency virus (HIV). The observation that KSHV RTA activates RAUL activity through Usp7 embodies a striking example of virus-mediated manipulation of a cellular ubiquitination pathway. This is reminiscent of another HECT domain Ub E3 ligase E6AP, which mediates the ubiquitination of p53 in cells that express the human papillomavirus (HPV) E6 oncoprotein. Evidently Sendai virus is also able to take advantage of RAUL signaling during infection, suggesting that this scheme may be generally relevant to other viral infections. A common strategy adopted by viruses to counteract innate immunity is to mimic or seize existing cellular pathways and to utilize them in their favor (McCormick and Ganem, 2005; Scheffner et al., 1990). Therefore exploring these “commandeered pathways” unveils not only the pathogenesis involved but also uncharted cellular pathways
Methods
Reagents
pCMV-MyC-RAUL (WT, 1083 amino acids) and the pCMV-MyC-RAUL HECT domain point mutant (C1051A), as well as pET3a-RAULWT, pET3a-RAUL-ND (1-655) and pET3a-RAUL-CD (655-1083) for in vitro translation were provided by Cecile M. Pickart (Bloomberg School of Hygiene, Johns Hopkins University). RAULΔ132 was provided Ming Wang. pIRF7A (1-503), pFLAG-IRF7A (1-503), pGST-IRF7 (1-503) and pGST-IRF7 (255-503) and reporter plasmids encoding firefly luciferase (LUC) under the control of human IFN-α1 (−140 to +9) and IFN-β (−280 to +20) promoter elements were provided by Yan Yuan (Zhu et al., 2002). pIRF7DN (aa1-12 and 103-503) was provided by Tom Maniatis (Wathelet et al., 1998). pIRF7C was provided by Joseph S. Pagano (University of North Carolina) (Zhang and Pagano, 1997). IRF7S477D and S479D) was provided by John Hiscott. pHA-Ub was provided by Ken Watanabe (Sasaki et al., 2002). pFLAG-Ub, pHA-IRF3, pHA-IRF3 (1-128), pHA-IRF3 (1-374), pGST-IRF3 (58-427), pGST-IRF3 (175-427) were provided by Takashi Fuita (Wathelet et al., 1998). pGST-IRF3 was provided by Michele E. Hardy (Veterinary Molecular Biology Laboratory) (Graff et al., 2002), 293T-TLR3 cell lines was provided by Kate Fitzgerald(Fitzgerald et al., 2003), HCT116 Usp7+/+ and Usp7−/− cell lines were provided by Bert Vogelstein(Cummins et al., 2004). Purified Usp7 protein was provided by Roger Everett (Everett et al., 1997), pcDNA3-Pin1 was provided by Akihide Ryo(Saitoh et al., 2006), pSR-Pin1 (siRNA-Pin1), pSR-Rli (control siRNA) and pEFHA-IRF3S339A were provided by Shoji Yamaoka(Saitoh et al., 2006). Rag2 −/− 5c.c7 TCR transgenic cells were provided by Yan Zheng and Jonathan D. Powell(Zheng et al., 2007).
Monoclonal antibodies against β-actin, ubiquitin, FLAG, HA and Myc-epitopes were purchased from Sigma. Rabbit polyclonal antibody against GST, Usp7, Ro52, RBCK1, IRF7 and IRF3 were purchased from Santa Cruz. Control non-immune antibodies are indicated as IgG. RAUL antibody was generated in rabbits using a peptide corresponding to the near N-terminal sequence of RAUL (Y-KTRPKVSLGGASRKEEK-SC). shRNA-Usp7 was provided by Roger Everett.
Highlights.
▶ Ub ligase RAUL limits type I IFN by catalyzing K48 ubiquitination of IRF7 and IRF3
▶ Suppression of RAUL augments production of type I IFN and reduces viral replication
▶ RAUL activity is positively regulated by deubiquitinase Usp7
▶ Virus KSHV RTA recruits the Usp7-RAUL mechanism to antagonize antiviral responses
Supplementary Material
Acknowledgements
These studies were funded by National Cancer Institute research grants R01 CA73585 to Gary S. Hayward from the National Institute of Health and American Cancer Society Institutional Research Grant IRG-58-005-43 to Yanxing Yu. We thank Drew M. Pardoll, Frank Housseau, Cecile M. Pickart, Min Wang, Bert Vogelstein, Yan Zheng, Jonathan D. Powell, Ken Watanabe, Takashi Fuita, Yan Yuan, Shoji Yamaoka, Kate Fitzgerald, Akihide Ryo, Michele E. Hardy, Tom Maniatis, Roger Everett, John Hiscott, Joseph S. Pagano, S. Diane Hayward, Prashant Desai, Shaoxian Jiang for gifts of plasmids or cell lines or technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrington RE, Subler MA, Rands E, Omer CA, Miller PJ, Hundley JE, Koester SK, Troyer DA, Bearss DJ, Conner MW, et al. A farnesyltransferase inhibitor induces tumor regression in transgenic mice harboring multiple oncogenic mutations by mediating alterations in both cell cycle control and apoptosis. Mol Cell Biol. 1998;18:85–92. doi: 10.1128/mcb.18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro M, Patton JT. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J Virol. 2007;81:4473–4481. doi: 10.1128/JVI.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomvarin C, Siripornmongcolchai T, Chaicumpar K, Limpaiboon T, Wongkham C, Yutanawiboonchai W. Evaluation of polymerase chain reaction, conventional and MRSA screen latex agglutination methods for detection of methicillin-resistant, - borderline and -susceptible Staphylococcus aureus. Southeast Asian J Trop Med Public Health. 2004;35:879–885. [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. following 486. [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. Embo J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Rosa JL. The HERC proteins: functional and evolutionary insights. Cell Mol Life Sci. 2005;62:1826–1838. doi: 10.1007/s00018-005-5119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JW, Mitzel DN, Weisend CM, Flenniken ML, Hardy ME. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J Virol. 2002;76:9545–9550. doi: 10.1128/JVI.76.18.9545-9550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Kaisho T, Tanaka T. Turning NF-kappaB and IRFs on and off in DC. Trends Immunol. 2008;29:329–336. doi: 10.1016/j.it.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodadek T, Sikder D, Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- McCormick C, Ganem D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science. 2005;307:739–741. doi: 10.1126/science.1105779. [DOI] [PubMed] [Google Scholar]
- Nakagawa K, Yokosawa H. Degradation of transcription factor IRF-1 by the ubiquitin-proteasome pathway. The C-terminal region governs the protein stability. Eur J Biochem. 2000;267:1680–1686. doi: 10.1046/j.1432-1327.2000.01163.x. [DOI] [PubMed] [Google Scholar]
- Okumura A, Alce T, Lubyova B, Ezelle H, Strebel K, Pitha PM. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J Biol Chem. 2002;277:22541–22546. doi: 10.1074/jbc.M109728200. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SY, Otsuji M, Gorski K, Huang X, Slansky JE, Pai SI, Shalabi A, Shin T, Pardoll DM, Tsuchiya H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. Embo J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhou W, Jeon MS, Demydenko D, Harada Y, Zhou H, Liu YC. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol Cell. 2006;21:135–141. doi: 10.1016/j.molcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- You J, Pickart CM. A HECT domain E3 enzyme assembles novel polyubiquitin chains. J Biol Chem. 2001;276:19871–19878. doi: 10.1074/jbc.M100034200. [DOI] [PubMed] [Google Scholar]
- You J, Wang M, Aoki T, Tamura TA, Pickart CM. Proteolytic targeting of transcriptional regulator TIP120B by a HECT domain E3 ligase. J Biol Chem. 2003;278:23369–23375. doi: 10.1074/jbc.M212887200. [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity. 2005;22:59–70. doi: 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Zhang L, Pagano JS. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. J Immunol. 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
- Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci U S A. 2002;99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.