Abstract
Single molecule methods have given researchers the ability to investigate the structural dynamics of biomolecules at unprecedented resolution and sensitivity. One of the preferred methods of studying single biomolecules is single-molecule fluorescence resonance energy transfer (smFRET). The popularity of smFRET stems from its ability to report on dynamic, either intra- or intermolecular interactions in real-time. For example, smFRET has been successfully used to characterize the role of dynamics in functional RNAs and their protein complexes, including ribozymes, the ribosome, and more recently the spliceosome. Being able to reliably extract quantitative kinetic and conformational parameters from smFRET experiments is crucial for the interpretation of their results. The need for efficient, unbiased analysis routines becomes more evident as the systems studied become more complex. In this article we focus on the practical utility of statistical algorithms, particularly hidden Markov models, to aid in the objective quantification of complex smFRET trajectories with three or more discrete states, and to extract kinetic information from the trajectories. Additionally, we present a method for systematically eliminating transitions associated with uncorrelated fluorophore behavior that may occur due to dye anisotropy and quenching effects. We also highlight the importance of data condensation through the use of various transition density plots to fully understand the underlying conformational dynamics and kinetic behavior of the biological macromolecule of interest under varying conditions. Finally, the application of these techniques to studies of pre-mRNA conformational changes during eukaryotic splicing is discussed.
1. Introduction
One of the most significant advances in the single molecule field was the advent of measuring fluorescence resonance energy transfer (FRET) between a single FRET fluorophore pair (Ha et al., 1996). FRET relies on the distance dependent interaction between two fluorophores, a donor and acceptor, so termed because the former `donates' its energy through space to the latter, therefore decreasing in emission intensity while the acceptor increases. Due to the nature of the transition dipole interaction between the two fluorophores, energy transfer is more efficient when they are close in proximity than when they are further apart, allowing one to measure relative distances of up to ~10 nm between labeled sites on one of more biomolecules (Michalet and Weiss, 2002). A simple FRET ratio on a scale from 0 to 1 as a relative measure of the inter-fluorophore distance can be calculated as:
| (Eq. 1) |
where IA and ID are the fluorescence intensities of the acceptor and donor, respectively. Changes in this FRET ratio over time are then used as a measure of the conformational changes of the labeled biomolecule(s) over the course of an experiment. While absolute inter-fluorophore distances can also be estimated from this FRET ratio (Pereira et al., 2008), purely structural studies of this kind are not the focus of this article.
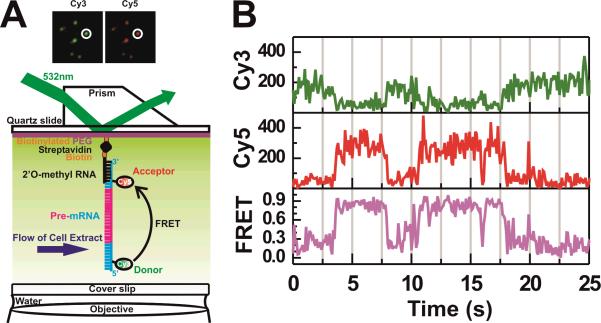
FRET measurements have long been performed in the ensemble (Stryer, 1978), however, advances in microscopy and sample preparation have allowed it to be performed at the single molecule level with relative ease and reliability (Roy et al., 2008; Walter et al., 2008). In a commonplace single molecule FRET (smFRET) experiment the emissions of the fluorophores attached to an immobilized biomolecule are monitored by wide-field video fluorescence microscopy in real-time (Fig. 1A; for a description of the necessary instrumentation the reader is referred to recent reviews (Roy et al., 2008; Walter et al., 2008)). Corresponding donor and acceptor spots are identified post-acquisition by mapping and pattern recognition of the two appropriately color-filtered video images, their signals integrated, and the FRET ratio calculated at the sampling rate of the detector used to collect the signal (Fig. 1B) (Roy et al., 2008). The most common fluorophores used for smFRET are Cy3 (donor) and Cy5 (acceptor) because of their relative brightness and photo-stability when compared to other fluorophores (Kapanidis and Weiss, 2002; Aitken et al., 2008). The Cy3–Cy5 FRET pair has a Förster radius of ~54 Å, allowing them to report on distance changes on the 20–100 Å scale (Ishii et al., 1999). Detection by smFRET has distinct advantages since it directly observes the kinetics of conformational changes of single molecules at (or away from) equilibrium without the need to synchronize and thus perturb them as ensemble FRET methods require; even highly heterogeneous biomolecular samples with rare and/or transient conformational states can thus be analyzed by smFRET in depth, revealing structural dynamics at unprecedented detail (Roy et al., 2008; Walter et al., 2008).
Figure 1. Capturing the conformational dynamics of single pre-mRNA molecules through smFRET in real-time.
(A) A pre-mRNA molecule is immobilized through a 2'-O-methylated capture oligonucleotide, and is bound to a PEG passivated quartz slide through a biotin-streptavidin interaction. The inset depicts a portion of a field of view captured by the I-CCD camera, and how the donor (Cy3) and acceptor (Cy5) fluorophores can be captured simultaneously. Peak finder algorithms are used to automatically find and match corresponding fluorophores from single molecules (white circles). (B) Exemplary fluorescence intensity and FRET changes of a single molecule.
The additional information gathered by smFRET over ensemble FRET approaches depends on the ability to detect subtle changes in inherently noisy data from individual molecules. The contribution of static and dynamic heterogeneity (or disorder) to biological systems, which is typically lost by ensemble-averaging, is being unraveled now by single molecule detection, contradicting the assumption that all molecules in a population are behaving exactly the same (Min et al., 2005; Ditzler et al., 2008; Fiore et al., 2008). In addition, rare and/or short lived intermediates often go undetected (or are ignored) in ensemble experiments because of their minimal contribution to the overall signal, yet in smFRET they are readily detectable as long as they are longer lived than the inverse of the rate of collection, usually on the order of 25–100 ms. Through the observation of a large population of molecules during wide-field video fluorescence microscopy sufficient statistics can be built up quickly to recapitulate the ensemble behavior of molecules, while not masking subpopulations. These features have made smFRET experiments popular, but the ability to extract all possible information from single molecule time trajectories in an unbiased fashion depends on (semi-)automated data analysis routines whose development so far lags behind that of the experimental techniques. We faced a particularly daunting task when analyzing complex smFRET trajectories from a pre-messenger RNA (pre-mRNA) during splicing in vitro (Abelson et al., 2010) and describe here the resulting, practical strategy for the largely unbiased extraction of kinetic information from smFRET trajectories with three or more states. Scripts for implementation of this strategy are available upon request.
2. Analysis of Simple Trajectories
The practical value of smFRET experiments depends greatly on the ability to extract kinetic and conformational information about the biomolecule of interest. A priority for smFRET assays should be the design of a system (in terms of composition and fluorophore labeling sites) that leads to changes over time and differences between molecules that are significant enough to discern them in single molecule trajectories. Once established, such a system allows for the application of FRET distribution analyses and thresholding algorithms that, when incorporated with information about dye position, yield information regarding the structural dynamics of the molecule(s) of interest and how experimental conditions affect them. We will first describe a systematic set of analysis tools that are suitable for simple (two- or three-state) trajectories, and often can be used for the initial characterization of more complex trajectories.
2.1. Selection of trajectories for analysis
Even in the case of simple trajectories with few (two or three) FRET states the amount of information that is collected throughout a time series of images from an smFRET experiment requires the use of (semi-)automated scripts (often written for the programs MATLAB or IDL) to process the raw imaging data. First, peak-finder and signal integration algorithms (Roy et al., 2008) are usually performed in IDL to select molecules from a field of view and match peaks collected in different color-filtered channels (Cy3 and Cy5) (inset of Fig. 1A). For a wide-field microscope this is necessary since an average field of view of perhaps 50 × 100 μm2 often contain over 50 molecules, and over 5 fields of view may be observed per experiment. Second, the initial pool of candidate signals identified by the peak-finder algorithm is filtered to reject background noise and singly labeled molecules and select for bona fide single smFRET molecules. This selection is typically done by eye and may be subject to user bias; to minimize such bias it is helpful to establish a set of defined selection criteria. The more complex the trajectories the more relevant the establishment of objective selection criteria becomes since the behavior of molecules will often be heterogeneous and a single molecule may go from little or no (FRET ≈ 0) to high energy transfer efficiency (FRET ≈ 1). Perhaps the simplest two criteria to implement are the presence of photoactive fluorophores and some level of FRET between them. These criteria are satisfied when molecules exhibit significant anti-correlation in donor and acceptor fluorophore intensities (detected by eye or using mathematical correlation analysis), followed by single-step photobleaching of one or both fluorophores. For molecules locked in low-FRET states anti-correlation may not be easily observed, but the presence of an active acceptor fluorophore can be established upon direct acceptor excitation by a laser pulse at the end of the desired observation window (or intermittently). A photoactive acceptor is characterized by a sharp increase in emission upon such illumination (above a threshold to be established), regardless of the FRET state of the molecule; this feature helps distinguish molecules that are doubly labeled but reside in a low FRET state from those that are singly labeled. It is also helpful to define as a criterion an expected signal threshold based on the experimental background and signal-to-noise ratio, and to exclude molecules whose donor and acceptor trajectories are significantly positively correlated (Munro et al., 2007). The remaining time trajectories will provide the pool of molecules from which quantitative data can be extracted.
2.2. Analysis of FRET state distribution
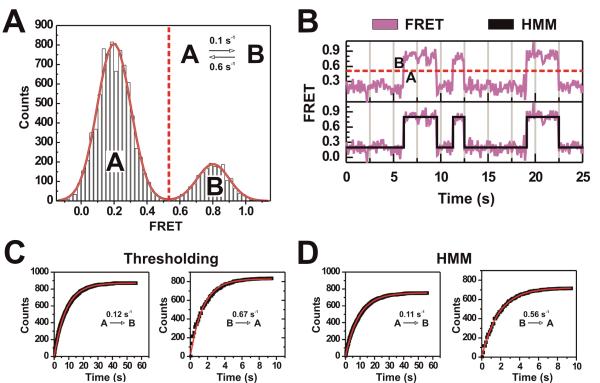
One of the first and most straightforward analysis steps is to create a FRET probability distribution by sampling the FRET values from an ensemble of molecules. Such analysis can be accomplished, for example, by binning all FRET values within a 10-s fragment of each molecule trajectory observed into a histogram (sometimes a single long molecule trajectory can be analyzed similarly). This simple procedure yields the relative occupancy of FRET states within the molecule population, as well as the associated relative inter-fluorophore distances. To this end, fitting with a sum of Gaussian functions as implemented in graphing software such as Microcal Origin is used to model the FRET distribution and obtain quantitative information on the mean FRET values, distribution widths (which is largely shot noise derived), and relative abundances of each molecule conformation (or state) detected (Fig. 2A). In Microcal Origin this is accomplished using the `Fit Multi-Peaks' analysis routine and selecting approximate peak centers and widths for initial guesses. For Fig. 2, a two-state system was simulated wherein 100 molecules reversibly interconvert between FRET states of 0.2 and 0.8 with rate constants of 0.1 s−1 and 0.6 s−1 (inset of Fig. 2A) over 1,000 data points each. The result in Fig. 2A shows that the multi-peak fitting procedure is able to identify both FRET states used in the simulation. Even though this procedure works reasonably well, it has been shown to be subject to bias from arbitrary values such as the chosen bin size (Okamoto and Terazima, 2008). Such inaccuracies become more problematic with an increasing number of states and a poor signal-to-noise ratio since the overlap between Gaussian distributions will become more severe. The utility of analyzing FRET distribution histograms is thus limited for complex, multi-state single molecule trajectories.
Figure 2. Histogram distribution analysis, thresholding, and hidden Markov modeling are well suited for simple trajectories.
(A) Simulated FRET probability distribution created by simulating 10 s at 100 ms repetition rate (1,000 data points) each of 100 molecules based on a two-state model (inset). The distribution was fit using two Gaussians whose sum models the distribution well (red outline). The centers of the Gaussians as determined by the fitting routine are situated at 0.203 and 0.802. (B) A sample trajectory of the simulated two-state system with discrete FRET states determined through either thresholiding (top) or hidden Markov modeling (HMM) (bottom). (C) Dwell time analysis of the simulated two-state system after determining the dwell times using the thresholding method or HMM. Measured dwell times were binned and plotted as a cumulative distribution and then fit with single-exponential functions using Microcal Origin.
2.3. Kinetic analysis with thresholding algorithms
FRET distribution histograms yield only the identity of FRET states in a population of molecules (or possibly a single long molecule trajectory), but do not extract any kinetic information on their dynamics. For this purpose, one needs to first reliably and efficiently identify the FRET states a molecule is sampling, then calculate the dwell times, or the amount of time spent in a state before transitioning to a specific other state, for all observed state transitions. In systems with FRET states that are easily distinguished by eye and/or histogram analysis it is possible to perform kinetic analysis with the use of simple thresholding. Thresholding is performed by implementing an algorithm wherein a FRET value that has little to no occupancy (as determined by state distribution analysis, Fig. 2A) is assigned as the threshold point. The algorithm then scans the FRET values within a trajectory and assigns each to a state based on its position relative to the threshold value (Fig. 2B, top panel). To avoid noise-induced assignment of artificial transitions, an additional requirement may be imposed that a new state be only assigned if there are two or more consecutive data points in a new position relative to the threshold value. After each data point within a trajectory is assigned a state, dwell times can be calculated, allowing for the determination of kinetic rate constants for state transitions. Rate constants are calculated directly by plotting dwell times as a cumulative probability distribution (or as a non-cumulative probability density function) that is then fit with a single- or, if necessary, multi-exponential growth curve (left side of Fig. 2C). An example where such analysis has been successful is the two- to three-state system of the hairpin ribozyme (Zhuang et al., 2002; Bokinsky et al., 2003; Rueda et al., 2004; Liu et al., 2007; Ditzler et al., 2008). Thresholding can be used reliably only for systems with three or fewer states since with more states threshold values become difficult to assign due to increasing overlap of the Gaussian distributions representing each state.
3. Analysis of Complex Trajectories
Due to the complex conformational behavior of many biomolecules, simple twoor three-states FRET systems may be rather the exception than the rule. Complex trajectories may arise from the presence of multiple interconverting structural or chemical states, and/or from trans-acting factors in solution that interact with the labeled molecule. Limited time resolution of the detector relative to the FRET state dwell times can also complicate trajectories (Lee, 2009), as well as photophysical effects such as blinking and dye anisotropy changes that lead to abrupt changes in FRET signal, often lacking anti-correlation of the donor-acceptor signal. Blinking can be detected with relative ease since the fluorescence intensity of the affected fluorophore drops to background level, and this portion of a trajectory can be ignored in (removed from) the analysis. Dye anisotropy changes due to local fluctuations in dye environment are more subtle effects that are harder to detect and are often disguised if relying solely on the FRET ratio for analysis. Distinction of true FRET changes from these photophysical artifacts becomes increasing difficult with an increasing number of states that occupy more of the limited FRET value range of 0–1. We therefore found that it is not sufficient to rely solely on the FRET ratio in the analysis of more complex trajectories, and that additional confidence can be gained from concomitantly analyzing the donor and acceptor signals (Abelson et al., 2010).
3.1. Hidden Markov analysis of complex FRET trajectories
Hidden Markov modeling (HMM) is a statistical algorithm that has been used for applications as varied as speech recognition, sequence alignment, and now smFRET analysis (Poritz, 1988; Eddy, 2004; McKinney et al., 2006). HMM is well suited for smFRET analysis because of its ability to find discrete states within noisy time series data and reliably find the most probable path through these states (Schuster-Böckler and Bateman, 2007). A hidden Markov model has three main parameter sets — the probability matrices of transition, emission, and initiation — that are optimized through an iterative process to find the set of parameters that best describes the data. The transition probability matrix contains the probabilities of any one FRET state changing to any other state in the subsequent time step. The emission probability matrix contains the probabilities of a specific FRET signal being emitted by each discrete FRET state. The initiation probability matrix gives the probabilities of starting at each of the possible discrete FRET states. Typically, a HMM is `trained' by using expectation maximization algorithms such as the Baum-Welch algorithm to iteratively modify these three parameter sets to better describe the data. The Viterbi algorithm, for example, can then be used to find the most likely sequence of states (MLSS) based on the trained Markov model parameters. For an in-depth discussion of the mathematical foundations underlying HMM we refer the reader to (Lawrence and Rabiner, 1989).
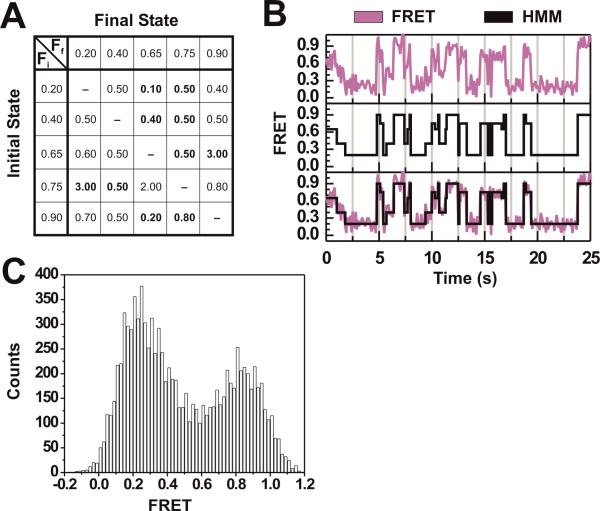
HMM has been successfully applied to smFRET analysis through the use of programs such as HaMMy (McKinney et al., 2006), QuB (Qin and Li, 2004), and vb-FRET (Bronson et al., 2009). These publicly available programs allow for the application of HMM algorithms to smFRET trajectories, and the extraction of FRET states and rate constants of their interconversion. These HMM programs present the most accessible form of data analysis that produces the most reliable results with minimal a priori assumptions required from the user. Additionally, they perform as well as if not better than thresholding algorithms. This point is demonstrated by applying HMM to the simulated system of Fig. 2A, resulting in the representative hidden Markov model shown in the lower panel of Fig. 2B. Even for this simple two-state system HMM achieves the best agreement with the raw data in terms of both kinetic rate constants and identity of the underlying FRET states, with less need for user defined (and possibly biased) input (compare Figs. 2C and 2D with inset of Fig. 2A). For more complex smFRET trajectories, such as those of the simulated five-state system of Fig. 3A (100 molecules, 1,000 data points per molecule) it is even clearer that thresholding and distribution analysis do not suffice. A sample trajectory in Fig. 3B (top panel) shows occupancy of nearly every point in the FRET range, making choosing a threshold value nearly impossible and largely arbitrary. The data are modeled well, however, with a five-state hidden Markov model that accurately identifies the underlying FRET states despite the data noise, and correctly detects when the states are changing (Fig. 3B, lower panels). The FRET probability distribution of these same data (Fig. 3C) shows what appears to be a simple two-state system, obscuring the three additional states used in the simulation due to the effects of binning and noise overlap between states.
Figure 3. Complex trajectories are not amenable to thresholding and distribution analysis, but require hidden Markov modeling.
(A) Parameters used for simulating a five-state system. FRET states and their transition rate constants are depicted as a transition matrix with the slowest and fastest rate constants (all in s−1) in bold. Fi = initial FRET state; Ff = final FRET state. (B) Raw FRET trajectory of one exemplary molecule simulated with the parameters in Fig. 3A (top). The idealized trajectory as determined by HMM executed in QuB (middle) and both trajectories overlaid (bottom). (C) FRET probability distribution of the simulated five-state system. This histogram shows that even in simulated data the five underlying states are obscured by noise, rendering distribution analysis less informative as in simple two-state systems.
It should be noted that Markov models generally assume that each stochastic transition is governed by a single rate constant with exponentially distributed waiting times. Molecular heterogeneity and limited observation windows often are, however, inherent to smFRET trajectories and cause non-exponentially distributed passage times and thus non-Markovian dynamics (Zhuang et al., 2002; Bokinsky et al., 2003; Rueda et al., 2004; Ditzler et al., 2008; Fiore et al., 2009). In situations where non-Markovian behavior is detected one can still apply HMM as a practical solution for the reliable identification of FRET states and quantification of the associated transition kinetics (Talaga, 2007).
The remainder of this article is dedicated to the practical aspects of using the programs HaMMy, QuB, and vb-FRET, and presents advances in their use derived from their application to the smFRET characterization of the conformational dynamics of a yeast pre-mRNA during splicing in vitro (Abelson et al., 2010).
3.2. Overview of HMM software available for smFRET analysis
The program HaMMy by Ha and coworkers presented the first readily available software specifically tailored for HMM analysis of smFRET trajectories (McKinney et al., 2006). The program employs HMM algorithms with a simple graphical user interface that processes the donor and acceptor intensities to determine the underlying discrete FRET states as well as the most likely sequence of states through a trajectory. HaMMy's user interface requires minimal input from the user, mainly guesses as to the number and mean FRET values of the states, and a choice of re-estimation algorithm. The simplicity of this program is perhaps its best attribute. One simply loads the single molecule trajectories formatted as *.dat files with a time column, followed by donor and acceptor intensity columns. The program will then analyze each trajectory and output three files that include the idealized FRET path, the dwell times, and a count of the total number of transitions between FRET states. This information can be utilized with the accompanying transition density plot (TDP) program to extract dwell times and rates for transitions of interest. We have successfully employed HaMMy, for example, to three-state smFRET trajectories of VS ribozyme folding (Pereira et al., 2008). The HaMMy and TDP programs and manuals are freely available at http://bio.physics.illinois.edu/.
The program QuB (available at http://www.qub.buffalo.edu/) was not originally designed for smFRET trajectories, but instead for single-ion channel measurements (Qin and Li, 2004). The electrophysiology field has been performing single molecule measurements on ion channels for over 30 years (Neher and Sakmann, 1976), and the signals acquired from such experiments have the same underlying features as smFRET trajectories with discrete states obscured by noise, and of interest are similarly the values of these states and their rate constants of interconversion. QuB has been successfully applied, for example, to smFRET experiments on the ribosome (Munro et al., 2007) and the spliceosome (Abelson et al., 2010). The graphical user interface is somewhat more challenging to master than that of HaMMy, but it provides the user with greater flexibility and control over the model parameters. For example, it provides the user with a variety of algorithms for re-estimation, MLSS calculations, and the ability to impose constraints on a model. One current major advantage of QuB is that it performs HMM much more rapidly than either HaMMy or vb-FRET.
The program vb-FRET by Gonzalez, Wiggins and coworkers (available at http://vbFRET.sourceforge.net) is yet another option for HMM analysis tailored for smFRET trajectories (Bronson et al., 2009). It provides an easy-to-use graphical interface similar to that of HaMMy, but with more customizable options similar to QuB. It is also a MATLAB executable file that for our use has been more stable. In addition to determining optimal parameters for the hidden Markov model, vb-FRET uses an approach known as “maximum evidence” to select the most likely model from a set of models being tested (Bronson et al., 2009). This approach alleviates the common problem of over-fitting data as a consequence of the use of a maximum likelihood approach, which often leads to the introduction of uninformative (superfluous) parameters to model the data, as well as to the need for post-processing efforts.
Table 1 lists the major differences between the three HMM programs. In the following we provide an example protocol for the use of each program, which may not necessarily be the best choice for all possible data sets but has worked effectively in our hands for both simple and more complex smFRET trajectories.
Table 1. Summary of features for HMM analysis programs.
A five-state system was modeled to simulate a series of complicated smFRET trajectories. The simulation included 105 total data points (100 molecules with each 1,000 frames of data collected) which is on par with what is expected from a series of sm-FRET experiments. This data set was independently analyzed with the various HMM analysis programs. Model selection was carried out with the indicated techniques, and the total time of analysis was recorded. The column “Data Range” indicates the acceptable input for the FRET, donor, and acceptor trajectories for each program. Because HaMMy and vb-FRET were designed to accept input of the range [0,1] the donor and acceptor trajectories need to be scaled accordingly.
| Program | Model Selection | Time to Analyze 5 state system with 105 data points | Data Range |
|---|---|---|---|
| HaMMy | Yes, BIC | ~3 h 50 min | [0,1] |
| QuB | No, Post-idealization BIC by user | ~1 h | [0, 32767] |
| vb-FRET | Yes, Maximum Evidence | ~15 min | [0,1], but can accept beyond this range |
3.3. Pre-processing trajectories for analysis by HMM
3.3.1. Removal of outliers
Often single molecule trajectories exhibit a state with a FRET value near the boundary of the range of 0–1 that will show transient excursions to FRET values either below 0 (when the acceptor intensity briefly dips into negative territory due to noise in the data) or above 1 (when the donor signal goes into negative territory). For trajectories dwelling extensively in low or high FRET states this phenomenon may lead to a significant occupancy of a virtual state below zero or above unity, which in turn may result in it being fit with a state by the HMM software. Consequently, the analysis will be complicated by introducing irrelevant states and by interrupting dwell times of more relevant states. The removal of outliers can be accomplished by the implementation of a simple algorithm that finds outliers and scales them back to within the FRET range of 0–1. Generally this algorithm involves identifying the points beyond the allowable FRET range and normalizing them to within the 0–1 range by taking an average of adjacent FRET values. A script containing this algorithm is available upon request.
3.3.2. Smoothing (noise reduction)
Single molecule trajectories are sometimes smoothed to help average out the inherent noise of the data collection process and emphasize the discrete states present. Although this is often useful for simple trajectories, complications arise if a larger number of states with rapid interconversion kinetics are present. One simple method of smoothing, rolling (point) averaging, may obscure transitions with dwell times that are shorter than the averaging window and introduce false FRET states in molecules where two or more states are rapidly interconverting. Rolling point averaging can work well with simple trajectories, but breaks down for more complex trajectories.
A non-linear forward-backward filter introduced by Haran has been used to smooth single molecule trajectories while minimizing their distortion, offering a clear advantage over rolling point averaging (Haran, 2004). We have applied this non-linear filter, for example, to trajectories of VS ribozyme folding where it reduces noise and emphasizes conformational changes between the three, clearly separated states (Pereira et al., 2008). One possible problem with this filter is the fact that the noise profile will no longer be Gaussian in shape, which is an assumption for analysis by HMM. In practice, this feature may lead to the fitting of small FRET changes that are within the noise of the raw data.
3.3.3. Formatting for HMM analysis
To perform HMM analysis it is necessary that smFRET trajectories be formatted in a manner amenable for manipulation with the desired analysis program. It should be noted that we are presenting only a limited, most commonly used set of input formats for each program, and for a more extensive list of input and output formats one should refer to the downloadable manuals for HaMMy (http://bio.physics.illinois.edu/HaMMy.html), QuB (http://www.qub.buffalo.edu/wiki/index.php/Main_Page), and vb-FRET (http://vbfret.sourceforge.net/). HaMMy loads files in ASCII format (e.g. *.dat) with the tabulated structure “time, donor intensity, acceptor intensity”. This structure is convenient as it is commonly the form in which the data are extracted from experiment. They can be loaded individually or in batch. HaMMy expects data values to fall within the range of [0, 1] and the data should be scaled to fit as needed. Based on the standard FRET ratio (eqn. 1) values outside of 0 and 1 do not have any physical meaning and should be considered outliers that can be corrected as described above.
Vb-FRET accepts data in ASCII format as does HaMMy, allowing one to load the same data into both programs for comparison. Although not formally supported, we have tested the program with data scaled outside of [0, 1] and found idealizations to work well.
QuB loads data in a variety of formats, including ASCII, without a need for normalization. QuB batch analysis can be performed by loading all molecules into a single *.txt file with each molecule separated from its neighbors by line breaks. QuB allows for the analysis of multiple channels in parallel, so one can visualize the donor, acceptor, and FRET ratio into the same file for observation and analysis. Scripts for the stitching and re-segmenting idealized trajectories are available upon request. We have also written a simple script that converts HaMMy formatted data into a *.txt file for input into QuB and comparison.
3.3.4. Stitching Trajectories
It is often the case that a single trajectory does not display all the possible transitions due to the limitations of photobleaching and the presence of some long-lived states that occupy much of the available time window. We have found that this scenario can limit the effectiveness of HMM programs in determining the discrete states for each molecule because there will be little to no occupancy of some states in some molecules. Additionally, the analysis of trajectories separately leads to HMM algorithms fitting the same conformational states with multiple idealized FRET values since subtle differences in background between trajectories can shift the value of the FRET state up or down. The variability of the values of discrete FRET states found when fitting trajectories separately requires that post-processing steps are carried out to group similar states before kinetic rates can be calculated for dwell time analysis. Such grouping of similar FRET states, however, requires a user-established criterion that is independent of the HMM analysis, which slows down the analysis routine and diminishes the objectivity of the analysis.
A better solution to this problem we found to be to `stitch' trajectories together so that a global analysis can be performed on an entire data set at once, which provides more data for the HMM algorithm to calculate reliable transition probabilities. This approach can be particularly helpful for shorter trajectories, but care should be taken to re-segment trajectories from one another after Markov modeling to prevent the introduction of false transitions at the molecule boundaries. Additionally, it is good practice to independently analyze a subset of molecules, large enough to recapitulate the behavior of the entire population, in both manners and compare the fitting results and HMM rates and states to ensure convergence. Scripts for the stitching and re-segmenting idealized trajectories are available upon request. HaMMy will only analyze segments containing up to 50,000 data points, whereas Vb-FRET has been tested up to 150,000 data points and the stability of the program is not affected. For both of these programs the idealization of such large trajectories will take a long time (Table 1), and in our hands has affected the stability of HaMMy. QuB has performed best with the stitched trajectories since it performs Markov modeling much more quickly than either of the currently available versions of HaMMy or vb-FRET (Table 1).
3.4. Selecting the appropriate number of FRET states
Perhaps the most difficult task in HMM analysis is deciding on the number of states to fit a data set with, a general problem when using statistical models to approximate an experimental distribution. For simple trajectories the number of states is often easily discernable by eye, but for complex trajectories this task becomes more difficult. Finding an objective and reproducible manner of choosing the appropriate number of states requires the use of a `rule'. The HaMMy manual suggests to allow for two more states than the total numbers of states one assumes to be present in the data set. The program then has the flexibility to sample higher order models, at the sacrifice of calculation speed. Inherently, a likelihood score based on the total probability calculated will continue to increasing with an increase in the number of model parameters (Bronson et al., 2009). Therefore, the goal should not be to maximize the likelihood score, but instead to strive for model parsimony, that is, to maintain the simplest model that best describes the data. HaMMy aims for model parsimony by calculating the total probability for each model tested and then using the Bayesian Information Criterion (BIC) to decide on the most appropriate model. BIC corrects for over-fitting, a common problem when using maximum likelihood approaches for determining model parameters, by introducing a penalty for complexity (Wasserman, 2000):
| (Eq. 2) |
where LL is the maximum likelihood reached by the model, k is the number of parameters, and N is the number of data points used in the analysis.
QuB in addition outputs a log-likelihood (LL) score for each model that can be used to compare the results of two independent idealizations and, when used to calculate the BIC, can help select the most appropriate model. The BIC calculation has to be performed outside of QuB as there is currently no nested model selection within the QuB algorithm, which speeds it up relative to the other analysis programs.
Vb-FRET takes a different approach to model selection, mainly the use of maximum evidence instead of maximum likelihood for idealization (Bronson et al., 2009). Maximum evidence aims to select the most likely model (and not only the most likely parameter values) through the assumption that the model with the correct number of states has the highest probability of reproducing the experimental data. Maximum evidence can be thought of as the probability of selecting the best parameterized model from the pool of all possible Markov models of a defined order (number of states). In a simple example, it is highly unlikely that a system with three distinct states will ever accurately be reproduced by a two-state Markov model. Conversely, a four-state model can reproduce the three-state system, but is overly complex, thus lowering the confidence in such a higher order model. This leaves the three-state model as the best choice since it can both reproduce the experimental data and is simple enough that the properly parameterized model is more likely to be chosen from the set of all possible outcomes.
4. Post-HMM processing and data visualization
Traditionally when analyzing smFRET trajectories the FRET ratio has been the only metric by which transitions have been detected, under the assumption that the donor and acceptor traces are always anti-correlated. When dealing with more complex trajectories, however, it becomes meaningful to maximize the observables to better understand the underlying dynamics of the molecules being studied. Incorporating the donor and acceptor trajectories in particular becomes necessary for systems with more than three states since uncorrelated changes between the dyes will often lead to a FRET value that exhibits some occupancy and therefore may appear as a bona fide FRET state. Such uncorrelated changes may arise from subtle variations in the local environment around the fluorophores such as changes in their rotational diffusion behavior (anisotropy) and fluorescence decay pathways.
4.1. Local detection of correlation based on HMM
Although the FRET ratio provides us with a bounded metric for conformational change, and can help correct for problems such as focal drift of the microscope, it is sensitive to unilateral changes in fluorescence intensity in either fluorophore. In complex systems, where cofactors may be acting on the molecule of interest in trans or the fluorophore is placed in a central position relevant for activity but exposed to a complex environment, the local environment around one fluorophore may not remain constant, which in turn may lead to changes in FRET ratio that are not caused by changes in FRET efficiency. Such apparent FRET changes uncorrelated in donor and acceptor signal may be intermixed with true changes in FRET efficiency, making their distinction within an smFRET trajectory nearly impossible. As a practical solution to this problem, we have adopted a strategy that analyzes by HMM the donor and acceptor trajectories alongside the corresponding smFRET trajectory, and subsequently scores each FRET change observed based on the presence or absence of corresponding donor and acceptor signal changes. Implementation of this algorithm requires that first the corresponding donor, acceptor and FRET trajectories are analyzed as follows.
HaMMy requires that the input data be scaled to fit between 0 and 1 so that the donor and acceptor intensities need to be normalized accordingly. In addition, HaMMy was designed to analyze FRET trajectories and therefore computes the FRET ratio (Eq. 1) from its input (“time, donor intensity, acceptor intensity”). Therefore, if one is interested in analyzing the donor channel, for example, the data should be formatted as: “time, 1-donor intensity, donor intensity”.
For Vb-FRET, the same input format as HaMMy can be used, except that no normalization is required.
QuB does not require that the data values lie between 0 and 1 so that no normalization is necessary. In addition, QuB's data input allows for flexibility so that the donor, acceptor, and FRET trajectories can all be loaded at once and analyzed independently by specifying how many channels are present in the data set. Alternatively, it also allows for each channel to be loaded separately.
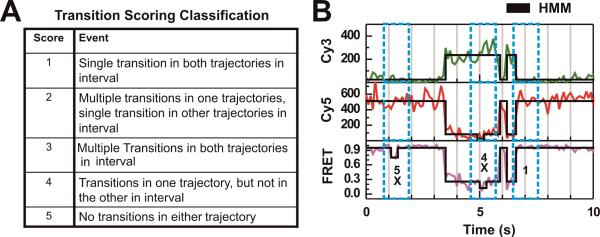
Once each channel has been idealized using HMM algorithms we employ a simple algorithm in MATLAB to detect donor-acceptor uncorrelated changes. The algorithm first scans the idealized FRET trajectory for each FRET change, then finds the corresponding time point in both the donor and acceptor trajectories to determine if there was a substantial change in idealized fluorescence intensity, and finally scores the transition using the simple scoring system outlined in Fig. 4. Each FRET transition is scored based using the same criteria, and the dwell times of FRET states are corrected after donor-acceptor uncorrelated FRET transitions have been identified and removed from consideration. Dwell time correction is important since uncorrelated changes in FRET otherwise distort (accelerate) the kinetics measured for a system since they shorten the apparent dwell times in specific conformational states.
Figure 4. Local correlation analysis utilizing HMM algorithms and transition scoring.
(A) Transition quality scoring used to exclude artificial FRET transitions caused by unilateral changes in one fluorophore. In our studies we are interested in only transitions with scores 1–3, since these transitions exhibit anti-correlated changes in the fluorescence intensity of both fluorophores simultaneously, as indicated. The time-window used to search for transitions in the donor and acceptor trajectories is set by examining the FRET trajectory. The size of the time window we chose to relate to the dwell time immediately before the FRET transition being scored. (B) Experimental trajectory and the scores of highlighted transitions after idealizing the donor signal, acceptor signal, and FRET using hidden Markov modeling (black lines). Transitions marked with an x are not characterized by donor-acceptor anti-correlation and not considered in any further analysis.
4.2. Data Condensation and Visualization
HMM techniques provide an efficient method for extracting quantitative measures of FRET states and rate constants of transitions between them from single molecule trajectories. With increasing complexity of smFRET trajectories the need to assess these parameters in a condensed and comprehensive representation becomes more evident. Of particular interest are transitions between pairs of sequential FRET states. Along with the release of HaMMy Ha and co-workers released a program that visualizes FRET transitions as so called transition density plots (TDPs), where the number of times a transition occurs is plotted as a heat map on a two-dimensional grid of final versus initial FRET state (Fig. 5) (McKinney et al., 2006). TDPs highlight the most prevalent transitions within a population of molecules and require that the transition rate constants be represent in a secondary plot (Joo et al., 2006; Pereira et al., 2008). The mirror symmetry relative to the main diagonal often observed in this type of plot is a result of reversible conformational changes (Fig. 5). It is important to note that the number of possible transitions that can be mapped onto a TDP is N*(N-1), where N is the number of discrete states found in the HMM analysis. For example, a trajectory that contains 5 states can have up to 20 distinct positions on the TDP, leading to a possibly quite complex plot. Importantly, transitions with slow kinetics will show up only infrequently in the trajectories due to their long dwell times relative to the limited observation window imposed by the photobleaching rate of the fluorophores.
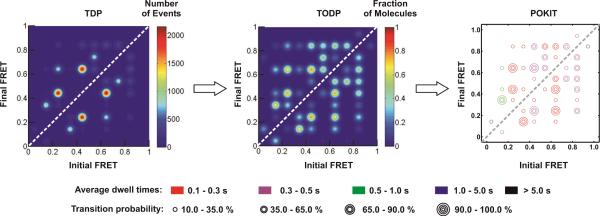
Figure 5. Data visualization for complex trajectories, including TDP and POKIT plot analysis.
A side-by-side comparison of the same data set using three representations. Traditional TDPs are scaled by the number of times a transition is observed over all molecules, regardless of whether in only a small sub-population of molecules with rapid transitions or commonly in all molecules. TODP and POKIT plots are scaled by the fraction of all molecules within a population that exhibits a particular transition. POKIT plots additionally provide kinetic information encoded in the color of the concentric circles.
To help minimize this relative underrepresentation of slow compared to fast transitions in a TDP, we have developed complementary transitional occupancy density plots (TODPs) that scale transitions based on the fraction of molecules that exhibit them at least once, rather than the number of times they are observed over all molecules. This approach makes slow transitions that many molecules exhibit more visible on the heat map of a TODP than a TDP, and thus guards against a visual over-representation of unrepresentative fast transitions only few molecules exhibit (Fig. 5).
To also incorporate the kinetics of each transition into one and the same plot, we have further developed POpulation-weighted and Kinetically-Indexed Transition density (POKIT) plots. POKIT plots thus provide two additional, comprehensive pieces of information compared to TDPs. First, they present as a number of concentric circles the fraction of molecules in the entire data set that exhibits a specific FRET transition at least once (the information represented in TODPs as heat map). Transitions that are common in a majority of molecules can help identify, for example, conformational changes that are important in a reaction with intermediates leading to products, even if this reaction is irreversible and the transition occurs only once per molecule. Second, POKIT plots provide the average dwell time for each transition in the form of circle colors, facilitating the rapid visual comparison of the kinetics of various data sets (Fig. 5).
4.3. Applications to single-molecule studies of yeast pre-mRNA splicing
Eukaryotic pre-mRNA splicing is a dynamic process that involves the precise recognition and excision of intervening sequences from in between coding regions (exons) of transcribed pre-mRNAs. Splicing is carried out by a multi-mega Dalton ribonucleoprotein complex known as the spliceosome. Although rivaling the ribosome in size the spliceosome is unique in that it lacks a preformed catalytic core. Instead, assembly proceeds in a stepwise manner, and is influenced by the ATPase activity of several RNA-dependent ATPases. Some of the rearrangements involved include the disruption of RNA-protein, RNA-RNA interactions, and the binding of recognition sequences and accessory proteins that lead to the formation of the catalytically competent complex necessary for the two trans-esterification reactions of intron excision and exon ligation. Yeast genetics and the development of in vitro yeast splicing assays have allowed for the biochemical characterization of the components involved in splicing as well as the key assembly steps required for splicing.
The introduction of smFRET has now allowed us to directly observe the conformational dynamics of the pre-mRNA substrate during spliceosome assembly and catalysis in real-time (Fig 1A) (Abelson et al., 2010). The complexity of smFRET trajectories from these experiments (Fig. 1B) reflects the overall complexity of the rearrangements needed for splicing activity, as well as the asynchronous behavior of splicing in total yeast cell extract. These features lead to a diversity of FRET states, heterogeneous kinetics, and many occupied transitions between the states as a reflection of the range of conformations through which the pre-mRNA substrate is shuttling. To highlight those FRET transitions that are relevant to splicing it was necessary to compare the wildtype substrate with mutant substrates that are blocked at different stages of splicing (Abelson et al., 2010). Spliceosome assembly on a branchpoint mutant (BP) is impaired, leading to a complete lack of splicing activity. In the 3' splice site mutant (3'SS) the second step of splicing and thus exon-ligation is blocked. A detailed kinetic and conformational analysis of the substrates in ATP depleted or ATP supplement extract allowed us identify conformational states that are required for splicing activity (Abelson et al., 2010).
The analysis of the smFRET data went as follows: First, trajectories to be studied were pre-filtered by searching for the presence of any substantial anti-correlation by visual inspection. The raw donor, acceptor, and FRET trajectories were independently Markov modeled to determine transition boundaries, using a global fitting routine (stitched trajectories) that simultaneously analyzes all data points taken under a given experimental condition. The entire data set for each condition was analyzed by the iterative application of the Forward-Viterbi and Baum-Welch algorithms in the QuB program to generate idealized trajectories. The number of states assumed in the idealization was varied from 5 to 11, with all states initially being assigned equal probabilities and rate constants that then were iteratively optimized; the resulting fits were evaluated using the Bayesian Information Criterion (BIC). The model that resulted in the best BIC score was selected for further analysis. This process was performed on all three corresponding trajectories (donor, acceptor, and FRET). After idealization, a post-HMM processing algorithm (coded and executed in MATLAB) classified each FRET transition by counting the number and direction of transitions found in the donor and acceptor trajectories within a time window defined to begin one quarter of the immediately preceding dwell time before the FRET transition in question and ending one quarter of the following dwell time after this FRET transition, with a minimum set to 0.3 seconds in either direction from the transition. Each transition was scored based on the metric shown in Fig. 4A, and transitions with scores of one, two, or three were used for transition density plots. Additionally, FRET transitions with a FRET change smaller than 0.1 were not considered significant and consequently removed.
The idealized scored trajectories were used to create TDPs and POKIT plots to examine the conformational and kinetic differences between the mutants under the various experimental conditions. Both TDPs and POKIT plots show substantial differences between the mutants and various conditions, with the POKIT plots highlighting even more subtle differences. The POKIT plots show, for example, that after extended incubation of only the WT substrate in ATP-supplemented extract a population of molecules with a relatively stable high-FRET conformation arises that resembles that of the mature mRNA after splicing (Abelson et al., 2010).
4.3. Summary of a detailed strategy for analysis of complex trajectories with QuB
Molecule selection; requirements for an accepted trajectory are
-
1)
donor-acceptor anti-correlation; and
-
2)
presence of each a single donor and acceptor fluorophore as verified by the observation of: I) single step photobleaching and II) emission of Cy5 by direct excitation.
Preparing data for distribution analysis and HMM
-
1)
Background correction: A straightforward method of background correction involves subtracting the mean value of signal after photobleaching from each value in the trajectory.
-
2)
Removal of blinking events, and truncation of the trajectory at point of photobleaching.
-
3)
Output raw data in *.dat format (format “time, donor signal, acceptor signal”).
-
4)
Sample 10 s worth of signal from each molecule and create a histogram from binned data using Microcal Origin.
-
5)
Stitch (concatenate) all trajectories into a single trajectory, and use the frequency count function in Microcal Origin with a bin size of 0.05 or less.
Pre-processing
-
1)
Outlier Removal: Trajectories are inspected for data points outside of the FRET range of [0,1]. These points are corrected based on the values of the adjacent points in the trajectory. If the background correction is performed properly there should be few outliers. If the number of outliers is roughly ~10% or more of the trajectory this hints at incorrect background correction, or that blinking or photobleaching events have not been properly removed.
-
2)
Stitching: All FRET trajectories of a single experimental condition are stitched together, and formatted for QuB. This procedure is also performed for the donor and acceptor trajectories. The QuB input format is a *.txt file with the data columns time, donor intensity, acceptor intensity, and FRET that can be truncated to two columns such as time, donor intensity, to read our the donor or acceptor trajectories.
HMM analysis with QuB
-
1)
Import data into QuB: Specify the number of columns in the data file and whether or not there is a time column. We typically choose to analyze the donor signal, acceptor signal, and FRET ratio as separate files to allow for better visualization of idealized trajectories that are overlaid on the raw data.
-
2)
Create a hidden Markov model: In the modeling window of QuB begin by creating a model with the lowest number of states assumed to be possible (typically we start our models for complex trajectories with 5 states).
-
3)
Idealization: The amplitudes and standard deviation of the states is then estimated by using the `Amps' function in QuB. This will initiate the model, and then the Baum-Welch and Forward-Viterbi algorithms are used under the `Idl/Base' menu for idealization. There are several other algorithms available for idealization; SKM has been used successfully, for example, to model smFRET data from ribosomes (Munro et al., 2007) and performs idealizations faster than the Baum-Welch, Forward-Viterbi algorithms presented here. SKM did not fit our smFRET data well, perhaps because of the lower signal-to-noise ratio due to the use of crude yeast cell extract. After each idealization add a state to the model and repeat the amplitude initiation and idealization procedures. Save each idealization as a *.dwt file (QuB file format with FRET states and dwell times), and then copy the log-likelihood score for the model from the reports panel and paste it into a spreadsheet for the calculation of BIC.
Post Processing
-
1)
Model selection: The log-likelihood score output from QuB is used to calculate the BIC for each model tested, and the model with the lowest BIC score is selected for further analysis.
-
2)
Parsing idealized data: The *.dwt file for the selected model is read and the idealized FRET states are then matched with the raw data to create a path file, that has the format “time, donor signal, acceptor signal, FRET, idealized FRET”. This path file is then segmented back into the individual molecule trajectories that were initially used to generate the stitched data.
-
3)
Transition scoring: The transition scoring routine is run by loading the donor, acceptor, and FRET path files into a MATLAB script that then finds transitions in the FRET channel, and takes note of the directionality and number of transitions at the corresponding time point in the donor and acceptor trajectories. It then scores each transition based on the scale in Fig 4A, and a scored path file is created. Transitions of scores 1–3 are considered true FRET transitions and thus chosen for further analysis.
Data Visualization
The scored path file is then input into MATLAB scripts written to recognize the score of the transition, and create TDPs, TODPs, or POKIT plots based on this information. From the TDP and TODPs, dwell time information can be extracted from our scripts by boxing a region within MATLAB around the transition(s) of interest. For POKIT plots, the dwell times are automatically extracted for each transition, and an average is calculated and encoded by the color scheme in Fig. 5. The percent of molecules exhibiting a transition is calculated by counting how many molecules within the data set exhibit that particular transition at least once, and then dividing by the total number of molecules used in the analysis.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the work of Franklin Fuller in writing, and developing several of the MATLAB scripts used for transition scoring.
REFERENCES
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, Guthrie C, Walter NG. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat. Struct. Mol. Biol. 2010 doi: 10.1038/nsmb.1767. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr., Wiggins CH. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. What is a hidden Markov model? Nat. Biotechnol. 2004;22:1315–1316. doi: 10.1038/nbt1004-1315. [DOI] [PubMed] [Google Scholar]
- Fiore J, Kraemer B, Koberling F, Erdmann R, Nesbitt D. Enthalpy-Driven RNA Folding: Single-Molecule Thermodynamics of Tetraloop-Receptor Tertiary Interaction. Biochemistry. 2009;48:2550–2558. doi: 10.1021/bi8019788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore JL, Hodak JH, Piestert O, Downey CD, Nesbitt DJ. Monovalent and Divalent Promoted GAAA Tetraloop-Receptor Tertiary Interactions from Freely Diffusing Single-Molecule Studies. Biophys. J. 2008;95:3892–3905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran G. Noise reduction in single-molecule fluorescence trajectories of folding proteins. Chem. Phys. 2004;307:137–145. [Google Scholar]
- Ishii Y, Yoshida T, Funatsu T, Wazawa T, Yanagida T. Fluorescence resonance energy transfer between single fluorophores attached to a coiled-coil protein in aqueous solution. Chem. Phys. 1999;247:163–173. [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Weiss S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J. Chem. Phys. 2002;117:10953–10964. [Google Scholar]
- Lawrence R, Rabiner A. A tutorial on hidden markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Lee T. Extracting kinetics information from single-molecule fluorescence resonance energy transfer data using Hidden Markov Models. J. Phys. Chem. B. 2009;113:11535–11542. doi: 10.1021/jp903831z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc. Natl. Acad. Sci. USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Weiss S. Single-molecule spectroscopy and microscopy. Compt. Rend. Phys. 2002;3:619–644. [Google Scholar]
- Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Fluctuating enzymes: lessons from single-molecule studies. Acc. Chem. Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260:799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Terazima M. Distribution analysis for single molecule FRET measurement. J. Phys. Chem. B. 2008;112:7308–7314. doi: 10.1021/jp712104h. [DOI] [PubMed] [Google Scholar]
- Pereira MJB, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. Single VS Ribozyme Molecules Reveal Dynamic and Hierarchical Folding Toward Catalysis. J. Mol. Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz AB. Hidden Markov Models: A Guided Tour. International Conference on Acoustics, Speech, and Signal Processing; IEEE Press; 1988. pp. 7–13. ICASSP-88. [Google Scholar]
- Qin F, Li L. Model-Based Fitting of Single-Channel Dwell-Time Distributions. Biophys. J. 2004;87:1657–1671. doi: 10.1529/biophysj.103.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B, Bateman A. An introduction to hidden Markov models. Curr. Prot. Bioinform. 2007 doi: 10.1002/0471250953.bia03as18. Appendix 3A, A.3A.1–A.3A.9. [DOI] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- Talaga DS. COCIS: Markov processes in single molecule fluorescence. Curr. Opin. Colloid Interface Sci. 2007;12:285–296. doi: 10.1016/j.cocis.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG, Huang C-Y, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L. Bayesian Model Selection and Model Averaging. J. Math. Psychol. 2000;44:92–107. doi: 10.1006/jmps.1999.1278. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating Structural Dynamics and Function in Single Ribozyme Molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]