Abstract
Background
Copy number variants (CNVs) have recently been reported to be associated with several autoimmune conditions. Moreover, loci involved in immunity are enriched in CNVs. Therefore, we hypothesized that CNVs in immune genes associated with Graves' disease (GD) may contribute to the etiology of disease.
Methods
One hundred ninety-one North American Caucasian GD patients and 192 Caucasian controls were analyzed for CNVs in three major immune regulatory genes: CD40, PTPN22, and CTLA-4. Copy number was determined using quantitative-PCR (Q-PCR) assays specifically designed for determining copy numbers in genomic DNA. Additionally, a well-characterized CNV in the amylase gene was typed in a separate dataset of DNA samples that were derived from cell lines or blood.
Results
No CNVs could be confirmed in the CD40 and CTLA-4 genes, even though a CD40 CNV is cataloged in the Database of Genomic Variants. Only the PTPN22 CNV was confirmed in our cohort, but it was rare and appeared in only two individuals. A key finding was that the source of DNA has a significant effect on CNV typing. There was a statistically significant increase in amylase locus deletions in cell line-derived DNA compared to blood-derived DNA samples.
Conclusions
We conclude that CNV analysis should be performed only using blood-derived DNA Samples. Additionally, the CTLA-4, CD40, and PTPN22 loci do not harbor CNVs that play a role in the etiology of GD.
Introduction
Graves' disease (GD) represents one of the most common autoimmune diseases in the United States with a prevalence approaching 1% (1). GD is an organ-specific autoimmune disease characterized by the formation of thyroid-stimulating hormone receptor (TSHR) autoantibodies that stimulate the TSHR-inducing secretion of excess thyroid hormones leading to thyrotoxicosis (2,3). GD is a complex disease whose etiology involves an interaction of multiple inherent genetic components and various environmental triggers [reviewed in Ref. (4)]. Several susceptibility genes for GD have been identified, including tissue-specific genes, such as thyroglobulin and TSHR (1), and a set of immune regulatory genes. Of the immune regulatory genes associated with GD, the major one is HLA-DR (5–7). The key HLA-DR variant associated with GD is arginine at position 74 of the DRβ chain (8). Three non-HLA immune-regulatory genes also contribute significantly to the genetic risk for disease: CTLA-4, PTPN22, and CD40 [reviewed in Ref. (9)].
So far, variants that were the focus of genetic studies in GD included single-nucleotide polymorphisms (SNPs) and microsatellites. However, recently it has been shown that copy number variants (CNVs) are also associated with complex diseases, including autoimmune diseases [reviewed in Ref. (10)]. CNVs are large DNA segments, ranging from kilobases to megabases, that are altered within the genome as a result of duplications, deletions, insertions, inversions, or complex combinations of rearrangements [reviewed in Ref. (11,12)]. Immune regulatory genes have been found to be enriched with CNVs, suggesting that CNVs in immune-related genes may predispose to autoimmune diseases [reviewed in Ref. (12,13)]. We hypothesized that CNVs in the three major immune-regulatory genes associated with GD, CTLA-4, CD40, and PTPN22, may play a role in the etiology of GD. Therefore, the aim of this study was to test CNVs in these three genes for association with GD.
Subjects and Methods
Subjects
The project was approved by the Mount Sinai Institutional Review Board. One hundred ninety-one North American Caucasian GD patients were studied. GD was diagnosed by (i) clinical and biochemical primary hyperthyroidism, (ii) diffuse goiter, and (iii) the presence of TSHR antibodies and/or a diffusely increased I-131 uptake in the thyroid. Of the 191 GD patients 107 (56.0%) had ophthalmopathy. The average age of onset of GD was 40.4 years. Our controls consisted of 192 North American Caucasian individuals without personal or family history of thyroid disease. All controls had normal thyroid functions, and negative thyroid antibodies.
DNA purification
For the comparison of CNVs between blood- and cell line-derived DNA, 218 blood-derived DNA samples and 253 cell line-derived DNA samples were extracted using the Puregene kit (Gentra Systems, Minneapolis, MN). For the CNV analysis, 191 patient and 192 control blood-derived DNA samples were studied.
Analysis of copy number variants
To type CNVs in each DNA sample, we used the TaqMan Copy Number Assays (Applied Biosystems, Foster City, CA): for CD40, Hs04041560_cn and Hs99999100_s1; PTPN22, Hs07226371_cn; and CTLA-4, Hs04714283_cn. For the amylase 2A CNV we used the TaqMan assay; Hs04204136_cn. Quantitative-PCR (Q-PCRs) were performed using the applied biosystems universal genotyping Master Mix in a 20 μL duplex reaction that contained FAM-MGB target gene probes and VIC-TAMRA reference gene (RNaseP part number 4403326) probes and 20 ng of genomic DNA. The amplification reaction was performed using the Applied Biosystems 7300 real-time PCR machine using the following PCR program: 2 minutes of incubation at 50°C, initial DNA polymerase enzyme activation at 95°C for 10 minutes, followed by 40 cycles of: denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. Data were collected at the end of each 60°C step. All samples were run in quadruplicates and standard deviations were calculated. The 7300 software automatically excluded samples with large standard deviations based on a predesigned algorithm. The results were expressed as the threshold cycle (Ct), that is, the cycle number at which the PCR product crossed the threshold of detection. The Copy Caller program (Applied Biosystems) was used to determine copy numbers of the tested gene in the samples. This program is based on the ΔΔCt method and obtained copy number values as follows: Ct values were imported into the program and normalized to the reference gene (RNase P) generating a ΔCt value. To obtain the ΔΔCt value of the samples, since all samples had unknown copy numbers, the program found the ΔCt value obtained in the majority of the population and assigned this ΔCt value as that of a sample with two copies, as this is what would be expected since most normal individuals have two copies at all CNVs, having gotten one copy from the maternal side and one from the paternal side. Once found, this ΔCt value was then subtracted from the ΔCt values of all other samples, thus giving a ΔΔCt value. The ΔΔCt value was then used to find the relative quantification of the gene analyzed using the formula 2(−ΔΔCt). A relative quantification value of 1 is equivalent to two copies, and the program uses this to calculate all other sample copy numbers. This method has been tested and proven to provide reliable results.
Statistical analysis
The comparisons of the CNV typing between patients and controls were performed using the χ2 test. We used EpiInfo 3.4.2 software (CDC, Atlanta, GA) for the statistical analyses. A p-value of <0.05 was considered statistically significant.
Power calculations
Power calculations were performed using CDC simulation software (Epi Info, Version 3.3.2; CDC). We assumed the lowest population frequency of the susceptible CNV to be 10% since we were analyzing only common CNVs. Our power calculations indicated that our dataset of 191 patients and 192 controls would give our study 80% power to detect a difference between the patients and the controls resulting in an odds ratio of >2.37 with an α of 0.05. Thus, our dataset gave us enough power to detect biologically significant CNVs.
Results
Comparison of copy number variations in blood-versus cell line-derived DNA
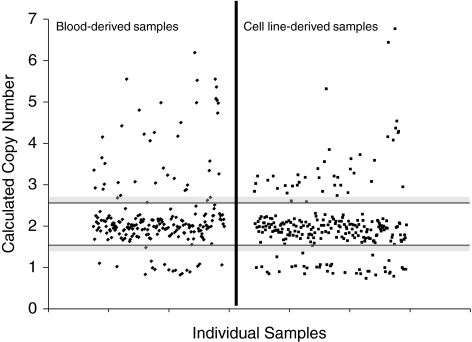
Since Epstein Barr Virus (EBV) immortalization of B-cells has been shown to introduce genetic changes in the DNA, we first compared cell line-derived DNA to whole blood-derived DNA. To test this potential artifact, we analyzed a well-characterized CNV in the amylase gene (chromosome 1, at base positions 103,911,245-104,109,897 [NCBI/hg18 Build 36; http://genome.ucsc.edu/]). We typed this CNV using the TaqMan assay Hs04204136_cn, and compared the frequency of deletions or duplications in DNA samples derived from whole blood versus cell lines (Fig. 1 and Table 1). DNA derived from cell lines consistently showed significantly higher number of deletions compared to blood-derived DNA (Table 1), suggesting that immortalization of B-cells introduces artificial copy number changes in numerous genetic loci. On the basis of this surprising and important finding, we limited our immune-gene CNV analysis only to blood-derived DNA.
FIG. 1.
Comparison of copy number variants between blood- and cell line-derived DNA. To compare the differences between blood- and cell line-derived DNA, we analyzed a well-characterized CNV in the amylase gene. Calculated copy numbers of the AMY2A gene in blood-derived (left) and cell line-derived (right) DNA samples are shown. Each dot represents the copy number of each individual sample. The area between the horizontal black bars represents the range of values that constitutes two copies. Gray-shaded regions signify borderline values that cannot be clearly assigned, either a one or two copies (lower region) or a two or three copies (upper region). Samples that fell in these gray areas were excluded from analysis. DNA samples derived from cell lines showed a significant increase in the number of samples showing deletions compared to those derived from whole blood (p = 0.008).
Table 1.
Comparison of the Frequencies of CNVs in the Amylase 2A Gene Between Blood- and Cell Line-Derived DNA Samples
| DNA source | <2 copies (%) | 2 copies (%) | >2 copies (%) |
|---|---|---|---|
| Blood-derived DNA (n = 218a) | 17 (8.0) | 157 (73.7) | 39 (18.3) |
| Cell line-derived DNA (n = 253b) | 42 (19.7) | 172 (68.8) | 36 (14.4) |
| p-Value | <2 vs. 2 copies: p = 0.007 | 2 vs. >2 copies: p = NS |
Of the 218 blood-derived DNA samples, 165 (75.7%) were healthy controls and 53 (24.3%) were patients with type 1 diabetes and thyroiditis.
Of the 253 cell line-derived DNA samples, 199 (78.7%) were healthy controls and 54 (21.3%) were patients with type 1 diabetes and thyroiditis.
NS, not significant.
Analysis of copy number variations in immune regulatory genes associated with GD
CNV assay selection
To identify CNVs in the CD40, PTPN22, and CTLA-4 genes, we searched the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation/?source = hg18) for cataloged CNVs. CD40 has one cataloged CNV in DGV that encompasses the entire gene and was analyzed by an assay specific for this CNV. PTPN22 has multiple listed CNVs in DGV, all overlapping and encompassing the entire gene and was analyzed by a CNV assay that covered the overlapping areas of its CNVs. CTLA-4 has no cataloged CNVs in the DGV, but many CNVs are still not deposited in DGV. Therefore, to test for potential disease-associated CNVs in the CTLA-4 gene, we used a CNV assay in the CTLA-4 gene locus.
CNV analysis of the CD40 gene
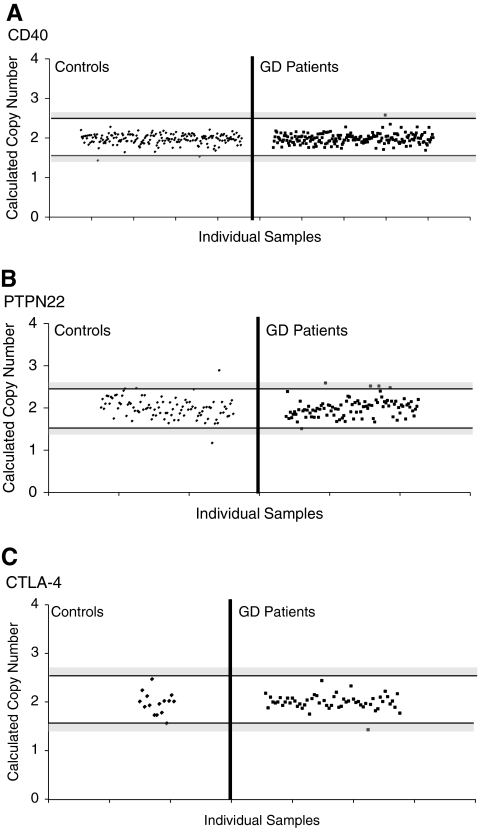
Even though a CD40 CNV is listed in the DGV, the CD40 gene showed no copy number variation in either the controls or GD patients (Fig. 2A). To confirm this surprising result, we used another CNV assay for the CD40 CNV (ABI assay No: Hs99999100_s1). This assay uses primers within exon 3 of the CD40 gene, and again showed 2 copies for all samples tested with no evidence for any deletions or duplications (data not shown).
FIG. 2.
Copy number variation analysis of CD40, PTPN22, and CTLA-4. TaqMan CNV assays were selected for CD40 and PTPN22 covering CNVs currently deposited in the Database of Genomic Variation. For CTLA-4, however, there was no cataloged CNV in the database, so a CNV assay encompassing the 5′UTR of the CTLA-4 gene was chosen. Each dot represents the copy number of each individual sample. The area between the horizontal black bars represents the range of values that constitutes two copies. Gray-shaded regions signify borderline values that cannot be clearly assigned, either a one or two copies (lower region) or a two or three copies (upper region). Samples that fell in these gray areas were excluded from analysis. Calculated copy numbers of CD40 (A), PTPN22 (B), and CTLA-4 (C) in control and Graves' disease (GD) patient samples are shown. CD40 and CTLA-4 showed no copy number variations in either the controls or the GD patients. PTPN22 showed no variation in the GD population, and only rare variation, one duplication and one deletion, in the control group.
CNV analysis of the PTPN22 gene
PTPN22 showed no copy variation in the GD patients, but in the control samples, there was one duplication and one deletion present (Fig. 2B), demonstrating that this is a rare CNV that is not associated with GD.
CNV analysis of the CTLA-4 gene
Since no CNV was described in the CTLA-4 gene we first typed a smaller cohort of 56 GD samples and 15 controls to detect a CNV in the CTLA-4 gene. In all 71 samples tested we found no copy variation in the CTLA-4 gene (Fig. 2C).
Discussion
Copy number variants (CNVs) are areas in the genome of duplications or deletions of large DNA sequences ranging from kilobases to megabases [reviewed in Ref. (11)]. CNVs have received much attention recently as they were suggested as a major source of human phenotypic variation specifically susceptibility to complex diseases. Moreover, recent data suggest that genes involved in immunity are particularly enriched in CNVs (13,14). These findings raised the possibility that immune-gene CNVs may predispose to autoimmune diseases, in a similar manner to the well-documented associations between immune-gene SNPs and autoimmunity (1). To date, several autoimmune diseases have been found to be associated with immune-regulatory gene CNVs (15–20). Therefore, we hypothesized that immune-regulatory genes associated with GD may harbor CNVs that influence susceptibility to disease. We tested the 3 major non-HLA immune-regulatory genes associated with GD: CTLA-4, CD40, and PTPN22.
CD40 is a surface receptor with diverse functions, including activation of B-cells and antigen presenting cells, immunoglobulin class switching, and IgG secretion. CD40 is a major susceptibility gene for GD (21); therefore, we tested whether a CD40 CNV, which is cataloged in the DGV, was associated with GD. However, we found no deletions or duplications in the locus of the DGV-listed CD40 CNV when testing 191 GD patients and 192 controls. All 383 individuals had two copies at this locus. This suggests that this CNV is very rare.
The PTPN22 gene, responsible for encoding a lymphoid tyrosine phosphatase, is a powerful inhibitor of T-cell activation. A C/T SNP in the PTPN22 gene causing an arginine to tryptophan change at position 620 was found to be associated with GD (22), as well as other autoimmune diseases (23). When we tested a DGV-deposited CNV in the PTPN22 gene in our large cohort, only two controls showed a copy number change at this locus, one deletion and duplication. None of the patients showed copy number changes at this locus, again suggesting that it is a rare CNV that does not contribute to the etiology of GD.
CTLA-4 is a negative costimulatory molecule that suppresses the activation of T-cells. CTLA-4 is also expressed on T-reg cells and is important to their function. Several CTLA-4 polymorphisms are associated with GD, as well as with other autoimmune diseases (1). When we tested the CTLA-4 gene locus for copy number changes, we could not find a CNV in this gene, suggesting that CTLA-4 CNVs do not predispose to GD or other autoimmune diseases.
An important and surprising finding from our study is that the source of DNA, whether cell line-derived or blood-derived, significantly altered the CNV analysis. Our data clearly demonstrated that DNA derived from cell lines contained novel CNVs that have been introduced during transformation, but do not exist in blood-derived samples that represent the native unperturbed DNA. As we were concluding this study, the Wellcome Trust group published an article that reported the same phenomenon. They have shown increased presence of variation in cell line-derived samples compared to blood-derived samples (24). Typically, cell line-derived DNA is purified from EBV-transformed B-cells, resulting in immortalized B-cells that can serve as an unlimited source of DNA. However, the immortalization of B-cells can introduce duplications and deletions as a result of defects in cell cycle checkpoints. It has been shown that cells immortalized using viral oncogenes inactivate the p53 and p16INK4a/Rb pathways that extend cells life spans and allow them to override the normal cell cycle checkpoints. This can introduce genetic aberrations, including aneuploidy, and copy number changes [reviewed in Ref. (25)]. In addition to changes in large chromosome segments, the genomic instability in these cells can also involve subtle base substitutions and deletions or insertions of a few nucleotides [reviewed in Ref. (26)]. These findings may explain the presence of CNVs in the DGV (e.g., CD40) that could not be confirmed in our large cohort of patients and controls, as these CNVs may be cell line derived. Taken together, the findings from the Wellcome Trust study and our study demonstrate that the source of DNA for CNV analyses needs to be taken into serious consideration. Most likely CNV studies should be limited to blood-derived DNA samples. Therefore, in the current study all GD patients and control DNA samples were derived from whole blood.
In conclusion, our study showed that copy number variation in the immune regulatory genes CD40, PTPN22, and CTLA-4 do not play a role in the etiology of GD. It is possible that previous CNV studies in complex diseases may need to be re-analyzed if the DNA samples were derived from cell lines. For the 3 immune-regulatory genes we tested, the entire genetic risk is likely to be attributed to non-CNV genomic variations.
Disclosure Statement
The authors of this article have nothing to disclose.
References
- 1.Huber A. Menconi F. Corathers S. Jacobson EM. Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev. 2008;29:697–725. doi: 10.1210/er.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rapoport B. Chazenbalk GD. Jaume JC. McLachlan SM. The thyrotropin (TSH) receptor: interaction with TSH and autoantibodies. Endocr Rev. 1998;19:673–716. doi: 10.1210/edrv.19.6.0352. [DOI] [PubMed] [Google Scholar]
- 3.Prabhakar BS. Bahn RS. Smith TJ. Current perspective on the pathogenesis of Graves' disease and ophthalmopathy. Endocr Rev. 2003;24:802–835. doi: 10.1210/er.2002-0020. [DOI] [PubMed] [Google Scholar]
- 4.Tomer Y. Huber A. The etiology of autoimmune thyroid disease: a story of genes and environment. J Autoimmun. 2009;32:231–239. doi: 10.1016/j.jaut.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomer Y. Ban Y. Concepcion E. Barbesino G. Villanueva R. Greenberg DA. Davies TF. Common and unique susceptibility loci in Graves and Hashimoto diseases: results of whole-genome screening in a data set of 102 multiplex families. Am J Hum Genet. 2003;73:736–747. doi: 10.1086/378588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamani M. Spaepen M. Bex M. Bouillon R. Cassiman JJ. Primary role of the HLA class II DRB1*0301 allele in Graves disease. Am J Med Genet. 2000;95:432–437. doi: 10.1002/1096-8628(20001218)95:5<432::aid-ajmg5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 7.Ban Y. Concepcion ES. Villanueva R. Greenberg DA. Davies TF. Tomer Y. Analysis of immune regulatory genes in familial and sporadic Graves' disease. J Clin Endocrinol Metab. 2004;89:4562–4568. doi: 10.1210/jc.2003-031693. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson EM. Huber A. Tomer Y. The HLA gene complex in thyroid autoimmunity: from epidemiology to etiology. J Autoimmun. 2008;30:58–62. doi: 10.1016/j.jaut.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson EM. Tomer Y. The CD40, CTLA-4, thyroglobulin, TSH receptor, and PTPN22 gene quintet and its contribution to thyroid autoimmunity: back to the future. J Autoimmun. 2007;28:85–98. doi: 10.1016/j.jaut.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaschl H. Aitman TJ. Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feuk L. Carson AR. Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 12.Redon R. Ishikawa S. Fitch KR. Feuk L. Perry GH. Andrews TD. Fiegler H. Shapero MH. Carson AR. Chen W. Cho EK. Dallaire S. Freeman JL. Gonzalez JR. Gratacos M. Huang J. Kalaitzopoulos D. Komura D. MacDonald JR. Marshall CR. Mei R. Montgomery L. Nishimura K. Okamura K. Shen F. Somerville MJ. Tchinda J. Valsesia A. Woodwark C. Yang F. Zhang J. Zerjal T. Zhang J. Armengol L. Conrad DF. Estivill X. Tyler-Smith C. Carter NP. Aburatani H. Lee C. Jones KW. Scherer SW. Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey JA. Gu Z. Clark RA. Reinert K. Samonte RV. Schwartz S. Adams MD. Myers EW. Li PW. Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen DQ. Webber C. Ponting CP 2006 Bias of selection on human copy-number variants. PLoS Genet. 2:20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinney C. Merriman ME. Chapman PT. Gow PJ. Harrison AA. Highton J. Jones PB. McLean L. O'Donnell JL. Pokorny V. Spellerberg M. Stamp LK. Willis J. Steer S. Merriman TR. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 16.Mamtani M. Rovin B. Brey R. Camargo JF. Kulkarni H. Herrera M. Correa P. Holliday S. Anaya JM. Ahuja SK. CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythaematosus. Ann Rheum Dis. 2008;67:1076–1083. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitman TJ. Dong R. Vyse TJ. Norsworthy PJ. Johnson MD. Smith J. Mangion J. Roberton-Lowe C. Marshall AJ. Petretto E. Hodges MD. Bhangal G. Patel SG. Sheehan-Rooney K. Duda M. Cook PR. Evans DJ. Domin J. Flint J. Boyle JJ. Pusey CD. Cook HT. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 18.Willcocks LC. Lyons PA. Clatworthy MR. Robinson JI. Yang W. Newland SA. Plagnol V. McGovern NN. Condliffe AM. Chilvers ER. Adu D. Jolly EC. Watts R. Lau YL. Morgan AW. Nash G. Smith KG. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanciulli M. Norsworthy PJ. Petretto E. Dong R. Harper L. Kamesh L. Heward JM. Gough SC. de Smith A. Blakemore AI. Froguel P. Owen CJ. Pearce SH. Teixeira L. Guillevin L. Graham DS. Pusey CD. Cook HT. Vyse TJ. Aitman TJ. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y. Chung EK. Wu YL. Savelli SL. Nagaraja HN. Zhou B. Hebert M. Jones KN. Shu Y. Kitzmiller K. Blanchong CA. McBride KL. Higgins GC. Rennebohm RM. Rice RR. Hackshaw KV. Roubey RA. Grossman JM. Tsao BP. Birmingham DJ. Rovin BH. Hebert LA. Yu CY. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson EM. Concepcion E. Oashi T. Tomer Y. A Graves' disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684–2691. doi: 10.1210/en.2004-1617. [DOI] [PubMed] [Google Scholar]
- 22.Velaga MR. Wilson V. Jennings CE. Owen CJ. Herington S. Donaldson PT. Ball SG. James RA. Quinton R. Perros P. Pearce SH. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J Clin Endocrinol Metab. 2004;89:5862–5865. doi: 10.1210/jc.2004-1108. [DOI] [PubMed] [Google Scholar]
- 23.Begovich AB. Carlton VE. Honigberg LA. Schrodi SJ. Chokkalingam AP. Alexander HC. Ardlie KG. Huang Q. Smith AM. Spoerke JM. Conn MT. Chang M. Chang SY. Saiki RK. Catanese JJ. Leong DU. Garcia VE. McAllister LB. Jeffery DA. Lee AT. Batliwalla F. Remmers E. Criswell LA. Seldin MF. Kastner DL. Amos CI. Sninsky JJ. Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craddock N. Hurles ME. Cardin N. Pearson RD. Plagnol V. Robson S. Vukcevic D. Barnes C. Conrad DF. Giannoulatou E. Holmes C. Marchini JL. Stirrups K. Tobin MD. Wain LV. Yau C. Aerts J. Ahmad T. Andrews TD. Arbury H. Attwood A. Auton A. Ball SG. Balmforth AJ. Barrett JC. Barroso I. Barton A. Bennett AJ. Bhaskar S. Blaszczyk K. Bowes J. Brand OJ. Braund PS. Bredin F. Breen G. Brown MJ. Bruce IN. Bull J. Burren OS. Burton J. Byrnes J. Caesar S. Clee CM. Coffey AJ. Connell JM. Cooper JD. Dominiczak AF. Downes K. Drummond HE. Dudakia D. Dunham A. Ebbs B. Eccles D. Edkins S. Edwards C. Elliot A. Emery P. Evans DM. Evans G. Eyre S. Farmer A. Ferrier IN. Feuk L. Fitzgerald T. Flynn E. Forbes A. Forty L. Franklyn JA. Freathy RM. Gibbs P. Gilbert P. Gokumen O. Gordon-Smith K. Gray E. Green E. Groves CJ. Grozeva D. Gwilliam R. Hall A. Hammond N. Hardy M. Harrison P. Hassanali N. Hebaishi H. Hines S. Hinks A. Hitman GA. Hocking L. Howard E. Howard P. Howson JM. Hughes D. Hunt S. Isaacs JD. Jain M. Jewell DP. Johnson T. Jolley JD. Jones IR. Jones LA. Kirov G. Langford CF. Lango-Allen H. Lathrop GM. Lee J. Lee KL. Lees C. Lewis K. Lindgren CM. Maisuria-Armer M. Maller J. Mansfield J. Martin P. Massey DC. McArdle WL. McGuffin P. McLay KE. Mentzer A. Mimmack ML. Morgan AE. Morris AP. Mowat C. Myers S. Newman W. Nimmo ER. O'Donovan MC. Onipinla A. Onyiah I. Ovington NR. Owen MJ. Palin K. Parnell K. Pernet D. Perry JR. Phillips A. Pinto D. Prescott NJ. Prokopenko I. Quail MA. Rafelt S. Rayner NW. Redon R. Reid DM. Renwick Ring SM. Robertson N. Russell E. St. Clair D. Sambrook JG. Sanderson JD. Schuilenburg H. Scott CE. Scott R. Seal S. Shaw-Hawkins S. Shields BM. Simmonds MJ. Smyth DJ. Somaskantharajah E. Spanova K. Steer S. Stephens J. Stevens HE. Stone MA. Su Z. Symmons DP. Thompson JR. Thomson W. Travers ME. Turnbull C. Valsesia A. Walker M. Walker NM. Wallace C. Warren-Perry M. Watkins NA. Webster J. Weedon MN. Wilson AG. Woodburn M. Wordsworth BP. Young AH. Zeggini E. Carter NP. Frayling TM. Lee C. McVean G. Munroe PB. Palotie A. Sawcer SJ. Scherer SW. Strachan DP. Tyler-Smith C. Brown MA. Burton PR. Caulfield MJ. Compston A. Farrall M. Gough SC. Hall AS. Hattersley AT. Hill AV. Mathew CG. Pembrey M. Satsangi J. Stratton MR. Worthington J. Deloukas P. Duncanson A. Kwiatkowski DP. McCarthy MI. Ouwehand W. Parkes M. Rahman N. Todd JA. Samani NJ. Donnelly P. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AL. Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer C. Kinzler KW. Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]