Abstract
The Schizosaccharomyces pombe fbp1 gene is transcriptionally repressed by protein kinase A (PKA) that is activated by extracellular glucose via a cAMP-signaling pathway. We previously used an fbp1-ura4 reporter that places uracil biosynthesis under the control of the glucose-sensing pathway to identify mutations in genes of the cAMP pathway. More recently, this reporter has been used in high throughput screens for small molecule inhibitors of heterologously-expressed cyclic nucleotide phosphodiesterases (PDEs) that hydrolyse cAMP to 5’ AMP. Here we show that strains lacking the adenylyl cyclase gene respond to either exogenous cAMP or cGMP to activate PKA, thus regulating fbp1-ura4 expression and other PKA-regulated processes such as conjugation and the nuclear export of an Rst2-GFP fusion protein. Expression of cGMP-specific PDEs or ones that hydrolyse both cAMP and cGMP increase the amount of exogenous cGMP required to activate PKA in order to repress fbp1-ura4 expression, creating conditions that allow detection of inhibitors of these PDEs. As proof of this concept, we screened a collection of compounds previously identified as inhibitors of cAMP-specific PDE4 or PDE7 enzymes for their ability to inhibit the mammalian cGMP-specific PDE5A enzyme. We identified compound BC76, which inhibits PDE5A in an in vitro enzyme assay with an IC50 of 232 nM. Further yeast-based assays show that BC76 inhibits PDE1, PDE4, PDE5, PDE8, PDE10 and PDE11, thus demonstrating the utility of this system for detecting and characterising inhibitors of either cAMP- or cGMP-metabolising PDEs.
Keywords: Schizosaccharomyces pombe, cAMP, cGMP, PKA, phosphodiesterase, inhibitor
1. Introduction
The fission yeast Schizosaccharomyces pombe senses glucose through a G protein-mediated cAMP signaling pathway to repress the transcription of genes involved in gluconeogenesis and sexual development [1, 2]. Most of the genes of the glucose/cAMP pathway are represented in a collection of mutants that fail to repress transcription of an fbp1-ura4 translational fusion, in which the ura4 OMP decarboxylase gene of the uracil biosynthetic pathway is expressed from the fbp1 fructose-1,6-bisphosphatase promoter. These “git” mutants (git= glucose insensitive transcription) are readily identified by their ability to form colonies on medium containing high levels of glucose but lacking uracil [3]. Moreover, whereas wild type strains that repress fbp1-ura4 expression are resistant to the pyrimidine analog 5-fluoro-orotic acid (5FOA) in glucose-rich medium, these mutants are 5FOA-sensitive (5FOAS) due to the increased expression of the reporter. This 5FOAS phenotype has allowed for the cloning of the git genes or multicopy suppressors of git mutations by their ability to restore 5FOA-resistant (5FOAR) growth [4–9], as well as the isolation of suppressing mutations including loss of function alleles of the cgs1 PKA regulatory subunit gene and the cgs2 cyclic nucleotide phosphodiesterase (PDE) gene [10, 11].
Mammalian genomes possess 21 PDE genes encoding enzymes grouped into 11 pharmacologically-distinct families based on their substrate specificity (PDE4, PDE7, and PDE8 act on cAMP; PDE5, PDE6, and PDE9 act on cGMP; PDE1, PDE2, PDE3, PDE10, and PDE11 act on cAMP and cGMP) as well as their sensitivity to small molecules and conserved domains outside of the catalytic domains [12–15]. Although enzymes from this superfamily act on only two substrates, tissue-specific expression and subcellular localisation allow individual PDEs to control specific biological processes and to serve as unique therapeutic targets [16].
As mentioned above, mutations that reduce but do not eliminate cAMP signaling can be suppressed by mutations in the cgs2 PDE gene, by virtue of their ability to re-establish repression of fbp1-ura4 expression and confer 5FOAR growth [11]. Using this as a point of departure, we have employed this reporter and suppression phenotype to identify mammalian PDE inhibitors in high throughput screens (HTSs) of strains expressing cAMP-specific mammalian PDE4 and PDE7 enzymes [17, 18]. As a cell-based screen that detects compounds that stimulate growth in 5FOA medium, the compounds detected in this manner possess drug-like characteristics of being cell permeable, relatively non-toxic (suggesting that they do not promiscuously bind S. pombe proteins), and chemically stable, as it takes 48 hours for cells to reach saturated growth.
In the present study, taking advantage of the fact that neither adenylyl cyclase nor PKA activity are essential in S. pombe, we construct a collection of strains that lack the ability to synthesise cAMP, thus allowing us to regulate PKA activity by adding exogenous cAMP to the medium. Moreover, as expected from the sequence of the Cgs1 S. pombe PKA regulatory subunit [19], exogenous cGMP also activates PKA to control PKA-regulated processes including fbp1 transcription, sexual development, and the localisation of the PKA-regulated transcriptional activator Rst2 [20]. Not surprisingly, expression of cGMP-specific PDEs and cAMP/cGMP dual specificity PDEs in S. pombe increases the amount of exogenous cGMP required to confer 5FOAR growth, establishing conditions that allow us to detect inhibitors of these PDEs. From a collection of PDE4 and PDE7 inhibitors discovered in previous HTSs, we identify compound BC76 as an inhibitor of the cGMP-specific PDE5A enzyme and show that BC76 reduces the amount of exogenous cGMP required to activate PKA. We also confirm this activity by in vitro enzyme assays. Using strains expressing members of 10 of the 11 PDE families, we profile BC76 specificity. Thus, this study demonstrates that our screening platform can be used to identify and characterise inhibitors of both cAMP- and cGMP-specific PDEs.
2. Materials and methods
2.1 S. pombe strains and growth media
Yeast strains used in this study are listed in Supplementary Table 1. The fbp1-ura4 and ura4::fbp1-lacZ reporters are translational fusions integrated at the fbp1 and ura4 loci, respectively [3]. Yeast were grown and maintained using yeast extract agar (YEA) and yeast extract liquid (YEL) [21]. Defined medium EMM (MP Biochemicals) was supplemented with required nutrients at 75 mg/liter, except for L-leucine, which was at 150 mg/liter. Sensitivity to 5-fluoro-orotic acid (5FOA) was determined on SC solid medium containing 0.4 g/liter 5FOA and 8% glucose as previously described [3]. Strains were grown at 30°C.
2.2 Construction of strains expressing mammalian PDE genes
Insertions of the murine PDE2A2 and PDE8A1 genes and the human PDE4A1, PDE4B2, PDE4D3, PDE7A1, and PDE7B1 genes into the S. pombe cgs2 (PDE) locus have been previously described [17, 18]. (Mammalian PDE nomenclature utilises a number to identify the PDE family, followed by a letter to indicate a particular gene within that family, followed by another number to indicate a splice variant when multiple splice variants are known.) Additional strains were constructed to insert the human PDE1B1 (RefSeq: NM_000924), murine PDE1C4 (RefSeq: L76947.1), human PDE3A1 (RefSeq: BC117371.1), murine PDE3B (RefSeq: AF547435.1), bovine PDE5A1 (RefSeq: NM_174417.2), human PDE9A5 (RefSeq: NM_001001570.1), and human PDE11A4 (RefSeq: NM_016953.3) genes into the cgs2 locus as previously described [17, 18]. A strain expressing the human PDE10A1 gene (RefSeq: AF127479.1) was created by PCR amplification of this open reading frame using oligonucleotides that target the integration of the PCR product into plasmid pRH1 [22], replacing the LEU2 gene and placing PDE10A1 under the control of the SV40 promoter. This plasmid was then stably integrated into the S. pombe lys2 locus by plasmid linearisation within lys2 prior to transforming strain CHP1239 to Lys+. All integrations were confirmed by PCR.
2.3 5FOA growth assays
Prior to the start of 5FOA growth assays, strains were grown to exponential phase under conditions that repress the fbp1-ura4 reporter (generally the presence of high levels of cAMP, cGMP, or a known inhibitor of the PDE expressed by the screening strain). Cells were collected by centrifugation and inoculated at a starting concentration of 0.5–2.0×105 cells/ml depending upon the strain into 50µl 5FOA medium in two to four replicate wells in a 384-well microtiter plate. The plates were incubated at 30°C for 48 hr in a container with moist paper towels to reduce evaporation in the wells. After vortexing the plates using a microtiter dish vortexer, optical densities (OD600) were measured using a microtiter plate reader. All assays were performed two or more times. In Fig. 1, values represent the averages from three independent assays. In the remaining figures, values are from a single representative experiment.
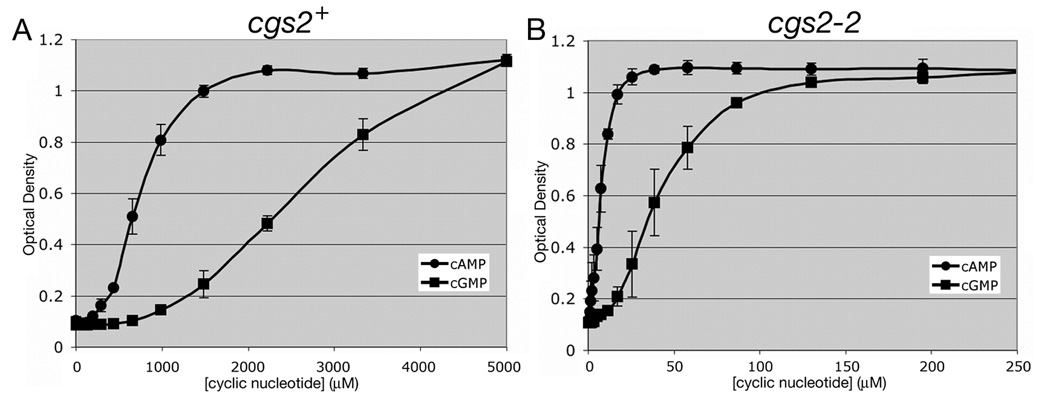
Fig. 1.
5FOA growth response to exogenous cAMP and cGMP in strains expressing or lacking the S. pombe Cgs2 PDE. Adenylyl cyclase deletion (git2Δ) strains CHP1197 (cgs2+; panel A) and CHP1207 (cgs2-2; panel B) were assayed for 5FOAR growth in a 384 well microtiter dish format (see Materials and Methods). Values represent the average OD600 ± standard deviations from three independent experiments after 48hr growth at 30°C. The values within each experiment represent an average from four replicate wells.
2.4 In vitro enzyme assays
In vitro enzyme assays were conducted to determine IC50 values for BC76 against PDE5A, PDE7A, and PDE8A. PDE5A assays were conducted via a two-step assay (PDE hydrolysis followed by snake venom conversion of 5’GMP to guanosine) as described by Weeks et al. [23] using recombinantly-expressed and purified His-tagged human PDE5A in the presence of 100nM cGMP. Parallel assays were carried out with sildenafil as a control. PDE7A and PDE8A assays were conducted via the Ba(OH)2 precipitation method of Wang et al. [24] in the presence of 35nM cAMP for PDE7A (BIOMOL International) and 10nM cAMP for PDE8A (BPS Bioscience Inc.). The concentrations of cyclic nucleotides used in these assays are at least three-fold below the experimentally-determine Km values, so that the IC50 values approximate the Ki for BC76. IC50 values represent the amount of compound required to inhibit 50% of the maximal PDE activity and are the averages of three independent experiments.
3. Results
3.1 Response of adenylyl cyclase deletion strains to exogenous cAMP and cGMP
Our initial efforts to construct S. pombe strains for small molecule PDE inhibitor screening focused on the cAMP-specific PDE4 and PDE7 families [17, 18]. The screen detects PDE inhibitors by establishing conditions under which cell growth in 5FOA medium is stimulated by elevating cAMP levels to activate PKA and repress an fbp1-ura4 reporter. To this end, these strains contain mutations in S. pombe genes that reduce glucose/cAMP signaling to a level that is insufficient to activate PKA unless the heterologously-expressed PDE is inactivated. While this approach worked well with PDE4 and PDE7 enzymes, many other PDEs tested hydrolyse so little cAMP when expressed in S. pombe that even gpa2− mutants that are severely defective for adenylyl cyclase activation are 5FOAR (data not shown). This led us to construct a series of strains lacking the git2/cyr1 adenylyl cyclase gene, allowing us to control PKA activity via exogenous cAMP. As seen in Fig. 1A, a strain that expresses the S. pombe cgs2 gene requires millimolar levels of cAMP or cGMP for growth in 5FOA medium. The amount of each nucleotide needed to promote growth is a function of the activity of the Cgs2 PDE against each nucleotide and of the ability of the Cgs1 PKA regulatory subunit to bind each nucleotide. By introducing a nonfunctional cgs2-2 frameshift allele [11], we show that 5FOAR growth can be achieved by <100µM of either cAMP or cGMP (Fig. 1B). As more cGMP than cAMP is required to stimulate PKA in a strain lacking PDE activity (Fig. 1B), it appears that the Cgs1 PKA regulatory subunit has a higher affinity for cAMP than for cGMP. The fact that this difference becomes ~2mM in a strain expressing the Cgs2 PDE (Fig. 1A) suggests that Cgs2 is more effective at hydrolysing cGMP than cAMP. However, we cannot rule out that a difference in the uptake or efflux of cGMP relative to cAMP partially accounts for these observations. The values in Fig. 1 represent averages ± standard deviations from three independent experiments. Not surprisingly, larger standard deviations are observed in the linear portion of the response curve (OD600 ranging from 0.3 to 0.8) than for cells growing in the presence of low or high cyclic nucleotide levels.
To determine whether the 5FOAR growth is actually due to PKA activation, we investigated the effect of cAMP and cGMP on another PKA-regulated phenotype, sexual development. As seen in Fig. 2, a homothallic h90 strain lacking adenylyl cyclase and PDE activity displays a severe growth defect in that the cells conjugate and sporulate in the absence of a starvation signal. Growth of this h90 git2Δ strain is restored by levels of cAMP and cGMP (Fig. 2A) similar to those that promote 5FOAR growth in a strain expressing the fbp1-ura4 reporter (Fig. 1B). This is accomplished by shifting the h90 cells from conjugation and sporulation to mitotic growth (Fig. 2B). These results indicate that both exogenous cAMP and cGMP can be taken up by S. pombe at low concentrations and activate PKA when there is no PDE present to hydrolyse these cyclic nucleotides.
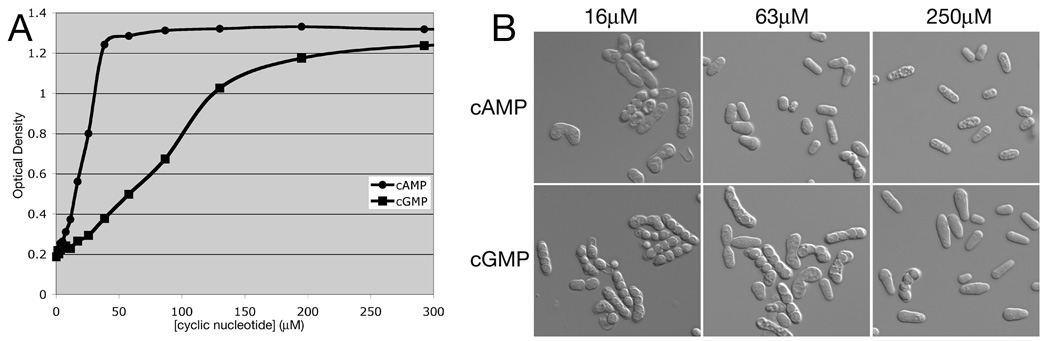
Fig. 2.
Suppression of unregulated mating by exogenous cAMP and cGMP restores vegetative growth in a homothallic strain lacking adenylyl cyclase and PDE activity. A) Exponential phase cells of strain CHP1440 (h90 git2Δ cgs2-2) were cultured in EMM containing various concentrations of exogenous cAMP or cGMP in four replicate wells and grown at 30°C for 48 hr before measuring OD600. B) Cells from the indicated cultures were examined by microscopy.
One way by which PKA represses transcription of genes regulated by glucose levels is to phosphorylate the Rst2 zinc finger transcriptional activator to mobilise it from the nucleus to the cytoplasm [20]. We observe the translocation of an Rst2-GFP fusion protein from the nucleus to the cytoplasm of cells lacking adenylyl cyclase and PDE activity within minutes of adding cAMP or cGMP to the growth medium, consistent with a rapid activation of PKA (Fig. 3). Thus, both exogenous cAMP and cGMP can be used to activate S. pombe PKA.
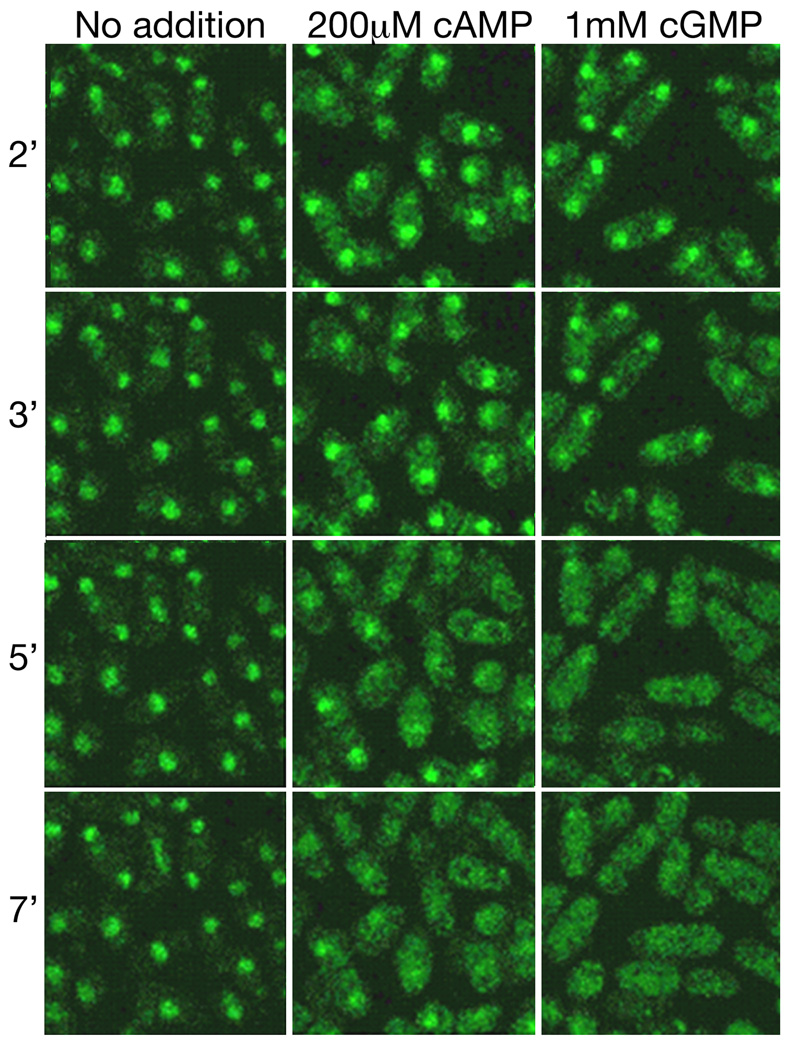
Fig. 3.
Localisation of Rst2-GFP upon PKA activation by exogenous cAMP or cGMP. Exponential phase cells of strain CHP1482 (git2Δ cgs2-2 rst2+-GFP) were examined by fluorescent microscopy using a Leica TCS-SP/2 confocal microscope with an objective heater to keep cells at 30°C. Time in minutes after addition of cAMP or cGMP are indicated.
3.2 Cells expressing dual-specificity or cGMP-specific PDEs require additional cGMP to activate PKA
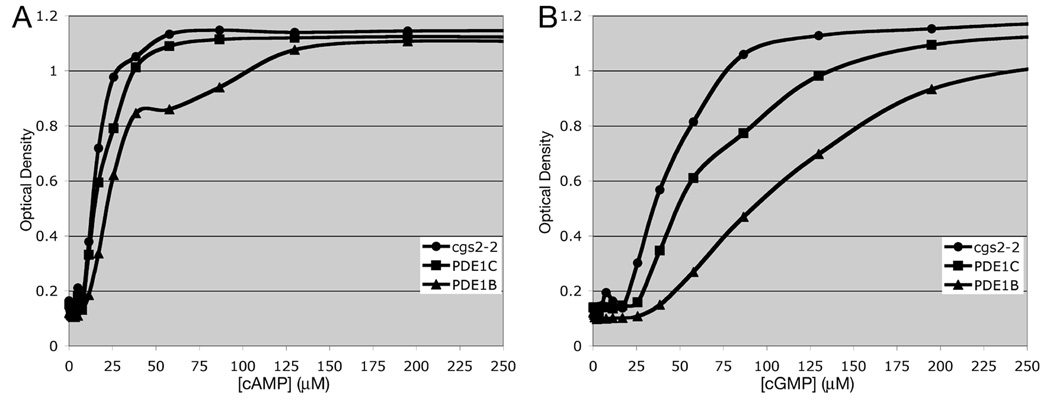
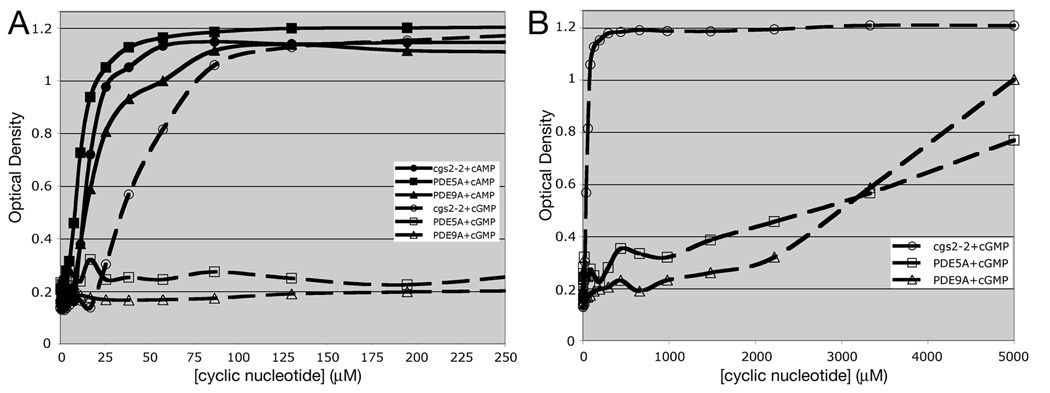
The ability to use exogenous cGMP to regulate PKA activity allows us to work with PDEs that are not amenable to study in strains that produce cAMP due to their limited activity against cAMP when expressed in S. pombe. This includes dual specificity PDEs such as members of the calcium-calmodulin activated PDE1 family (Fig. 4). Expression of PDE1B or PDE1C has a very small effect on the amount of cAMP required to promote 5FOAR growth (Fig. 4A), while creating a larger shift in the amount of cGMP required to promote growth (Fig. 4B). These limited effects of PDE1 expression may be due to the inability of S. pombe calmodulin to regulate mammalian PDE1 enzymes. However, the cGMP shift is sufficient to allow detection of PDE1 inhibitors that would increase the 5FOAR growth of the PDE1-expressing strain, raising it to that seen for the cgs2-2 strain that lacks PDE activity at a given cGMP concentration. We are also able to work with cGMP-specific PDEs (PDE5, PDE6, and PDE9). Fig. 5 shows that expression of PDE5A or PDE9A confers little or no change in the amount of cAMP needed to promote growth (Fig. 5A), but creates a requirement for an additional ~3.5mM cGMP to stimulate growth to an OD600 of 0.6 (approximately one-half the OD of a saturated culture; Fig. 5B). As such, it would be a simple matter to screen for PDE5A or PDE9A inhibitors by screening for 5FOAR growth of cells growing in medium containing 100µM to 1mM cGMP. Similar to screens using PDE4- and PDE7-expressing cells, this can be carried out in a 384 well format with 50µl cultures [17, 18].
Fig. 4.
5FOA growth response to exogenous cAMP and cGMP in strains lacking the S. pombe Cgs2 PDE or expressing mammalian PDE1B or PDE1C genes. 5FOA growth was measured as in Fig. 1 in strains expressing mammalian PDE1 genes in response to varying concentrations of A) exogenous cAMP or B) exogenous cGMP. Values represent the average OD600 from four replicate wells after 48hr growth at 30°C.
Fig. 5.
5FOA growth response to exogenous cAMP and cGMP in strains lacking the S. pombe Cgs2 PDE or expressing mammalian PDE5A or PDE9A genes. 5FOA growth was measured as in Fig. 1 in strains expressing mammalian cGMP-specific PDE5A or PDE9A genes in response to varying concentrations of exogenous cAMP or exogenous cGMP. A) Growth response at low concentrations of cyclic nucleotides. B) Growth response through the full range of cyclic nucleotide addition. Values represent the average OD600 from four replicate wells after 48hr growth at 30°C.
3.3 Identification and characterisation of a PDE5A inhibitor
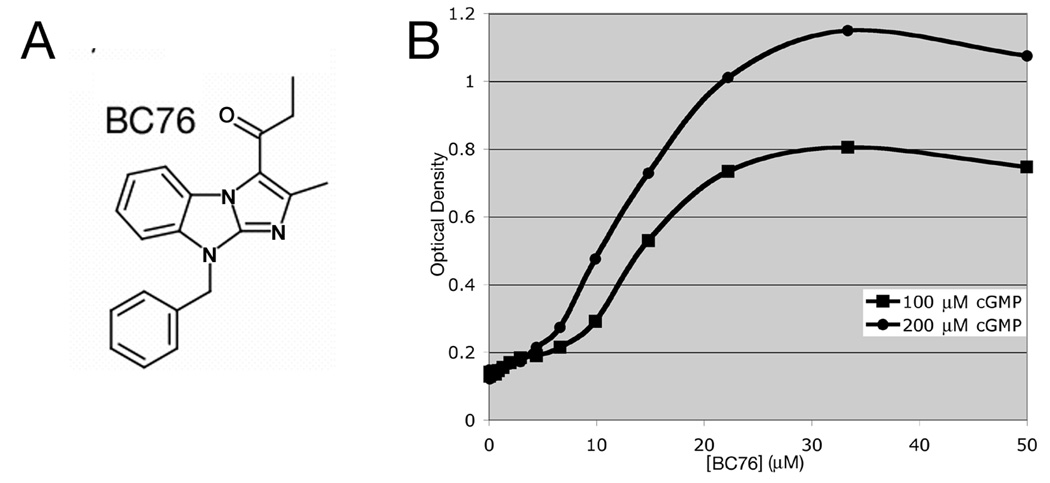
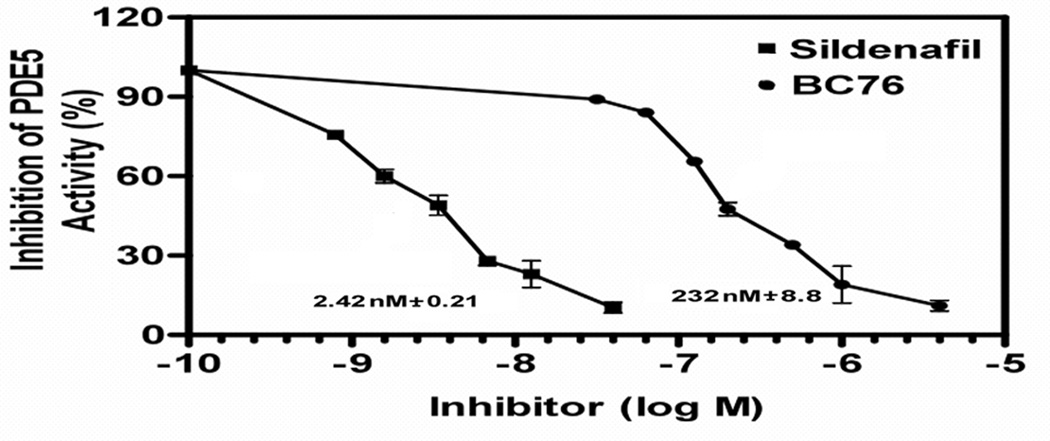
As a proof of principle that we can detect inhibitors of cGMP-specific PDEs, we screened a collection of compounds previously identified in PDE4 or PDE7 inhibitor screens [17, 18] for their ability to inhibit PDE5A. A few compounds display modest activity (data not shown), while BC76 (Fig. 6A) displays potent activity against PDE5A (Fig. 6B). 5FOAR growth is readily detected at 20µM, a concentration typically used in HTSs. To determine whether this 5FOAR growth is indeed due to PKA activation, we examined the effect of BC76 on Rst2-GFP localisation. BC76 addition to cells growing in 0.5mM cGMP causes the relocation of the reporter protein from the nucleus to the cytoplasm (Fig. 7). We also examined the long-term effect of BC76 exposure to cells expressing PDE5A. S. pombe strains lacking adenylyl cyclase or PKA divide at cell length nearly half that of wild type strains [7]. Restoration of wild type size control for cell division in strain CHP1223 (git2Δ cgs2Δ::PDE5A) requires both cGMP and BC76, and occurs within two rounds of cell division (Fig. 8). Finally, using an in vitro enzyme assay [23], we directly demonstrate that BC76 is an effective inhibitor of PDE5A displaying an IC50 of 232 nM, although it is ~100-fold less potent than sildenafil, which displays an IC50 of 2.42 nM in side-by-side assays (Fig. 9). Meanwhile, its activity as a modest PDE8A inhibitor (IC50 of 12.5 ± 1.6 µM) and a relatively poor PDE7A inhibitor (IC50 of 53.8 ± 5.3 µM) correlate well with our characterisation of this compound via yeast 5FOA growth assays (Fig. 10).
Fig. 6.
BC76 is a PDE5A inhibitor. A) Structure of compound BC76. B) 5FOA growth response by strain CHP1223 (git2Δ cgs2Δ::PDE5A) to varying concentrations of BC76 in 5FOA medium containing either 100µM cGMP or 200µM cGMP. Values represent the average OD600 from four replicate wells after 48hr growth at 30°C.
Fig. 7.
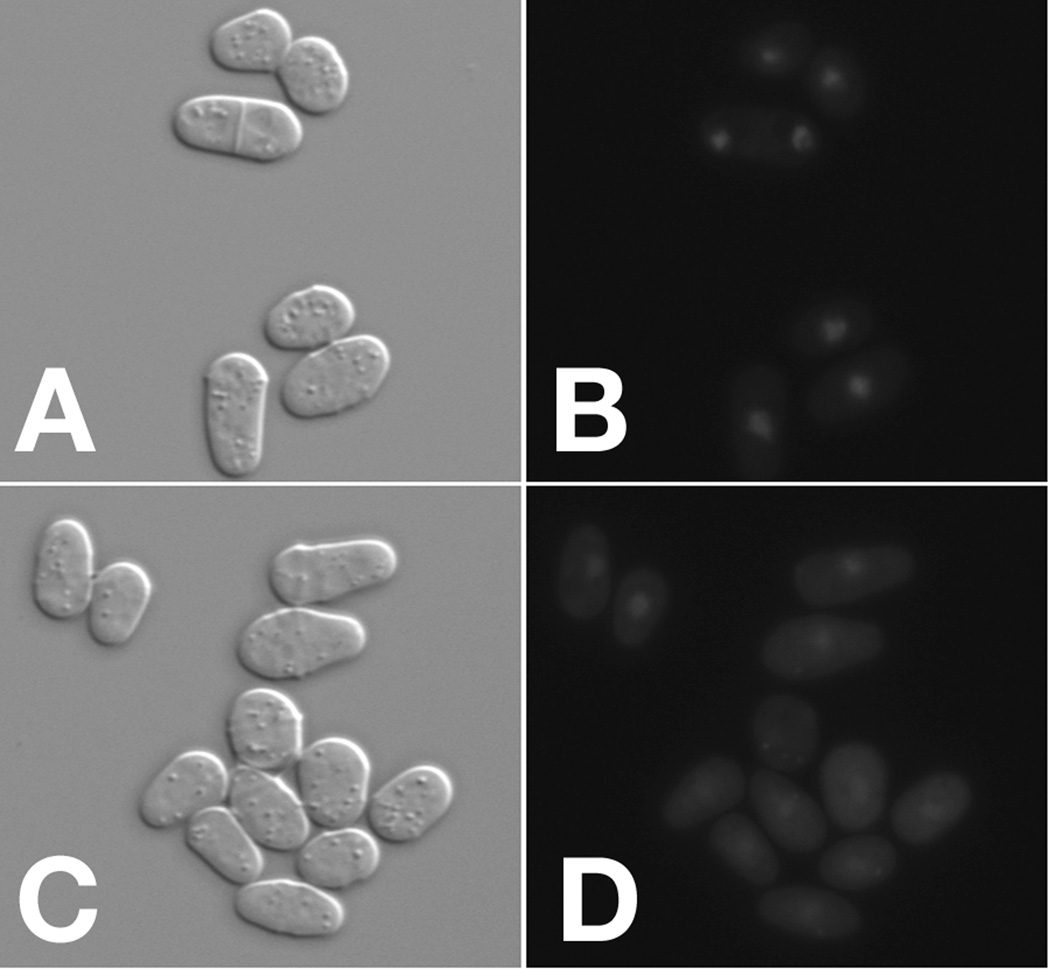
Translocation of the Rst2-GFP reporter from the nucleus to the cytoplasm upon treatment of PDE5A-expressing cells with BC76. Strain CHP1483 (git2Δ cgs2Δ::PDE5A rst2+-GFP) cells growing in the presence of 0.5mM cGMP were exposed to either DMSO (panels A and B as a negative control) or 30µM BC76 (panels C and D). Cells were incubated at 30°C for 20 minutes and imaged using an Axioplan 2 microscope with a 63X objective by DIC (panels A and C) or fluorescent (panels B and D) microscopy. BC76 treatment reduces the nuclear signal and increases the cytoplasmic signal by Rst2-GFP.
Fig. 8.
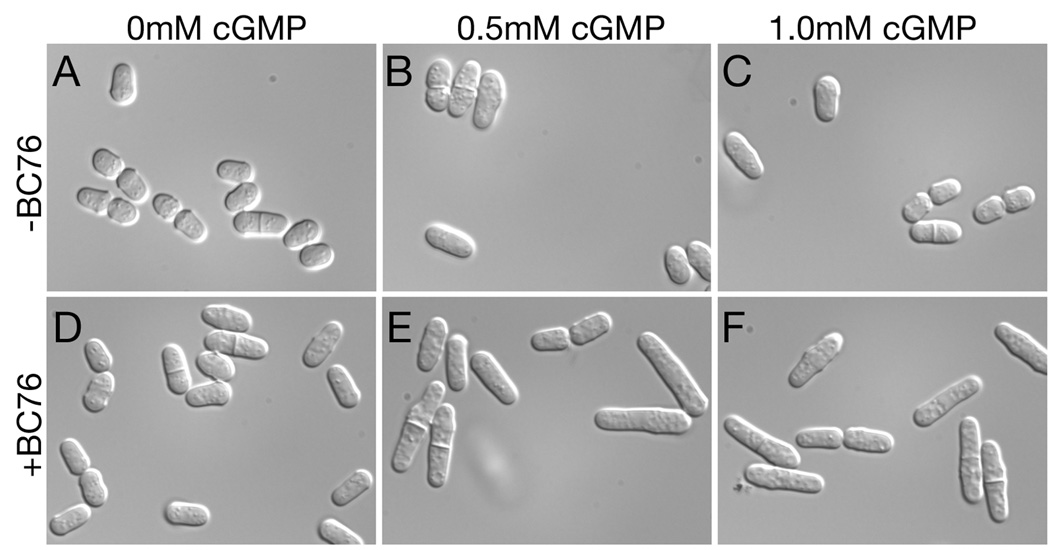
Exogenous cGMP and BC76 synergistically activate PKA in a PDE5A-expressing strain to alter cell length. Exponential phase cells of strain CHP1223 (git2Δ cgs2Δ::PDE5A) growing in EMM medium containing no cGMP (panels A and D), 0.5mM cGMP (panels B and E), or 1.0mM cGMP (panels C and F) were exposed to DMSO (vehicle control; panels A-C) or 30µM BC76 (panels D-F). Cells were imaged as in Fig. 7 after 4 hours growth at 30°C.
Fig. 9.
Inhibition of PDE5A by sildenafil and BC76 as measured by in vitro enzyme assays. Assays were conducted as described in the Materials and Methods. Values represent averages ± standard errors from three independent assays.
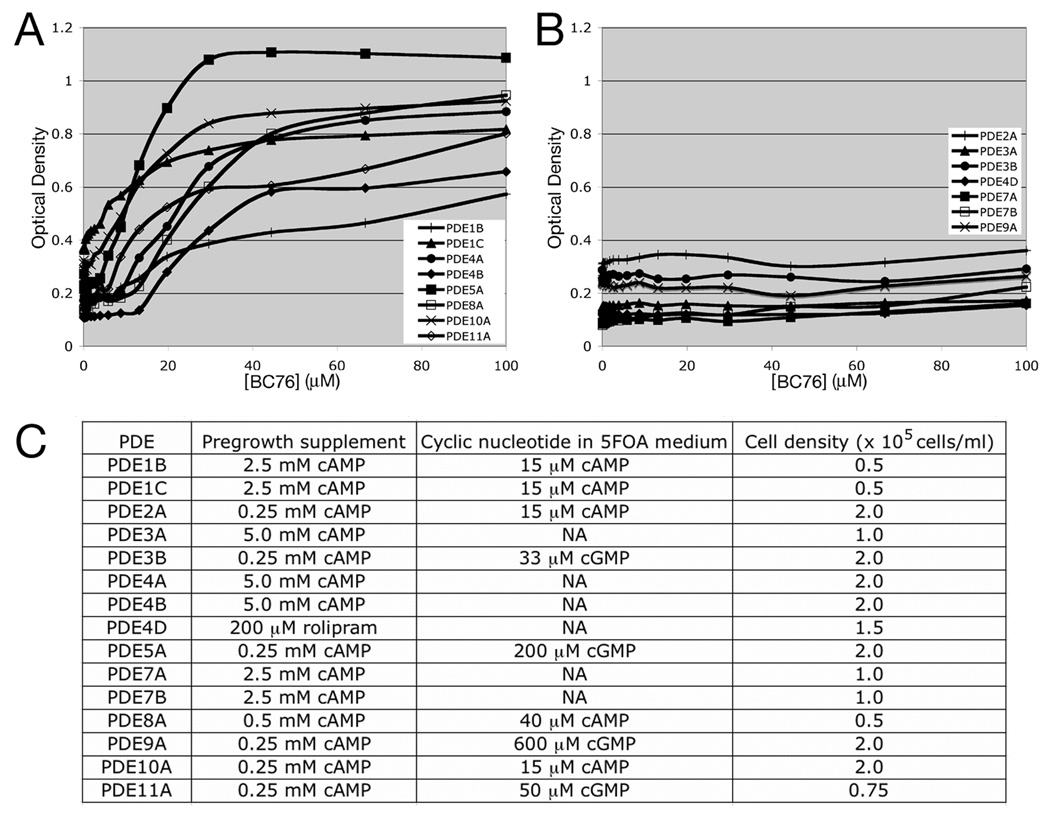
Fig. 10.
Profiling the specificity of BC76 using the 5FOA growth assay. Strains expressing 15 of the 21 mammalian PDE genes were tested for growth stimulation by BC76 in the 5FOA growth assay. A) Data for strains that show growth stimulation in response to BC76. B) Data for strains that show no response to BC76. Values in panels A and B represent the average OD600 from duplicate wells after 48hr growth at 30°C. C) Conditions used for the 15 strains, including the pregrowth supplement to repress fbp1-ura4 transcription prior to inoculation of cells into 5FOA medium, the amount of cAMP or cGMP present in the 5FOA medium (NA= not applicable as the strain used can synthesise cAMP), and the initial cell density for a given strain.
As BC76 was originally detected as a PDE4A inhibitor, it inhibits PDEs from more than one of the 11 pharmacologically-defined families. We therefore used our panel of PDE-expressing strains to profile its activity against ten PDE families (excluding the PDE6 family). Strains expressing PDE1B, PDE1C, PDE4A, PDE4B, PDE5A, PDE8A, PDE10A, and PDE11A show varying degrees of growth stimulation in 5FOA medium upon exposure to BC76 at <40µM (Fig. 10A), indicating PDE inhibition. In contrast, strains expressing PDE2A, PDE3A, PDE3B, PDE4D, PDE7A, PDE7B, or PDE9A show no response to as much as 100 µM BC76 (Fig. 10B). Other compounds in our collection can stimulate 5FOAR growth of the strains that do not respond to BC76, suggesting that these PDEs are insensitive to BC76. Finally, we note that nine of the fifteen PDEs required the used of an adenylyl cyclase deletion strain, together with exogenous cAMP or cGMP in the medium to control PKA activation (Fig. 10C). These results demonstrate the flexibility of this screening platform to profile the activity of a PDE inhibitor against enzymes encoded by 15 of the 21 mammalian PDE genes. However, for detection to occur the compound must be taken up by S. pombe and must not be toxic at the concentrations used for testing.
4. Discussion
In this study, we show that the PKA-repressible S. pombe fbp1-ura4 reporter can be used to examine the activity of cGMP-hydrolysing PDEs and to detect small molecule inhibitors of these enzymes. cGMP-mediated activation of PKA suggests that the S. pombe Cgs1 PKA regulatory subunit can bind cGMP, as well as cAMP, in order to bring about Cgs1 release from the PKA catalytic subunit. PKA from two other species of yeast, Saccharomyces cerevisiae and Pichia pastoris, are likewise activated by either cyclic nucleotide [25, 26]. The ability to bind cAMP or cGMP by the PKA regulatory subunits from S. pombe (AAB20314), S. cerevisiae (NP_012231), and P. pastoris (XP_002492773) appears to be due to the presence of glutamine or asparagine residues at position 13 of the second phosphate-binding cassette (PBC B) consensus sequence. PKA regulatory subunits that are highly selective for cAMP over cGMP possess an alanine residue in this position [27].
Our platform for studying PDEs using S. pombe possesses a distinct advantage over the system for working with heterologously-expressed PDEs in S. cerevisiae that was used to clone mammalian PDE4A, PDE4B and PDE7A genes and to identify mutations in PDE4 that confer resistance to the PDE4 inhibitor rolipram [28–31]. This is due to the phenotype used to follow PDE activity in the two systems. The S. cerevisiae phenotype conferred by the absence of PDE activity is the loss of heat shock resistance in stationary phase cells. These stationary phase S. cerevisiae cells are likely to be less permeable to compounds in the medium than are the exponential phase S. pombe cells used in our screening platform. This may explain why the concentration of rolipram used in the S. cerevisiae system [31] is so much higher than the 5–20µM rolipram needed to fully stimulate 5FOAR growth in S. pombe strains expressing PDE4 isoforms [18]. It is also interesting to note that the PDE4 and PDE7 genes, along with PDE3A, were the only ones to express sufficient cAMP hydrolytic activity that we could work with them in strains that had a functional adenylyl cyclase gene (Fig. 10C). This may explain why the S. cerevisiae system was only able to identify clones of PDE4A, PDE4B and PDE7A in library screens.
The rapid translocation of the Rst2-GFP fusion protein from the nucleus to the cytoplasm upon the addition of cAMP or cGMP to the medium demonstrates that these nucleotides are taken up by cells within minutes to activate PKA (Fig. 3). Similarly, this assay shows that BC76 is taken up by cells within 20 minutes to inactivate PDE5A (Fig. 7), allowing exogenous cGMP to activate PKA. Any PDE inhibitor discovered by our HTS platform is likely taken up quickly as fbp1 transcription becomes highly derepressed in wild type cells within one hour of glucose starvation or in git− mutant cells upon removal of exogenous cAMP from the medium [32]. Still, this reporter should be a valuable tool for examining the relative rates by which PKA becomes activated in response to treatment with various PDE inhibitors.
To characterise the specificity of BC76, we used a panel of strains that express 15 of the 21 mammalian PDE genes, representing 10 of the 11 PDE families. BC76 inhibits enzymes from the PDE1, PDE4, PDE5, PDE8, PDE10, and PDE11 families (Fig. 10A), suggesting that it binds somewhere within the conserved active sites of these enzymes. With the exception of PDE4D, strains expressing genes within one family show a similar response to BC76 (Fig. 10). Thus PDE1B and PDE1C, as well as PDE4A and PDE4B, are inhibited by BC76, while PDE3A and PDE3B, as well as PDE7A and PDE7B, are unaffected by BC76. While most compounds are expected to display a similar effect on members of a family, we have seen subtype-selective inhibition similar to the failure of BC76 to promote 5FOAR growth in a strain expressing PDE4D (Fig. 10B). Previously we determined that the PDE7A inhibitor BRL50481 [33] is ~30-fold less effective against PDE7B in the 5FOA growth assay and ~80-fold less effective against PDE7B in an in vitro enzyme assay [17]. Therefore, our platform represents an effective and simple method for profiling the activity of PDE inhibitors producing results that generally agree with data from in vitro enzyme assays.
Finally, we cannot help but comment on the irony of the phenotype conferred by the PDE5A inhibitor BC76 upon S. pombe cells that express mammalian PDE5A (Fig. 8), the enzyme target of Viagra®, Levitra®, and Cialis® used to treat erectile dysfunction. This superficial similarity to the effect of PDE5A inhibitors in humans [34] should not be seen as an example of yeast serving as a model system to study a mammalian biological process. Rather, we propose that the use of the S. pombe fbp1-ura4 reporter system, with its ability to reflect the interactions between PDEs and their inhibitors, allows the detection of inhibitors of any PDE that displays activity when expressed in S. pombe. This system could also be used to detect PDE activators that stimulate Ura+ growth or reduce the ability of exogenous cyclic nucleotide to promote 5FOAR growth. In addition, this platform can be used to detect mutations in PDE genes that confer resistance or hypersensitivity to specific inhibitors, as well as hyperactive alleles that would confer increased mating in a homothallic (h90) strain background. Finally, one could screen cDNA libraries for biological regulators of target PDEs, which can be followed up by chemical genetic screens for small molecule regulators of these interactions. As such, the S. pombe fbp1-ura4 reporter can be used in many ways to study the function of heterologously-expressed PDEs via molecular and chemical genetic screens.
Supplementary Material
Acknowledgments
We are extremely grateful to Joseph Beavo, Miles Houslay and Vincent Manganiello for providing clones of some of the PDE genes used in this study, and Masayuki Yamamoto for a strain expressing the Rst2-GFP fusion protein. We thank Josh Rosenberg for expert assistance with the microscopy. This work was supported by a grant from Boston College and by NIH grant R21 GM079662 to C.S.H.. We thank Nicola Tolliday, Lynn VerPlank, Jason Burbank, and Stephanie Norton of the Broad Institute’s Screening facility for guidance on high throughput screening that led to the discovery of BC76. The project has been funded in part with Federal funds from the National Cancer Institute's Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400 and has been performed with the assistance of the Chemical Biology Platform of the Broad Institute of Harvard and MIT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Didem Demirbas, Email: demirbas@bc.edu.
Ozge Ceyhan, Email: ceyhan@bc.edu.
Arlene R. Wyman, Email: arlene.wyman@bc.edu.
F. Douglas Ivey, Email: iveyfr@bc.edu.
Christina Allain, Email: chrijala@gmail.com.
Lili Wang, Email: liliwang@broadinstitute.org.
Maia N. Sharuk, Email: maia.sharuk@umassmed.edu.
Sharron H. Francis, Email: sharron.francis@Vanderbilt.Edu.
Charles S. Hoffman, Email: hoffmacs@bc.edu.
REFERENCES
- 1.Hoffman CS. Eukaryotic Cell. 2005;4:495–503. doi: 10.1128/EC.4.3.495-503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman CS. Biochem Soc Trans. 2005;33(Pt 1):257–260. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman CS, Winston F. Genetics. 1990;124(4):807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alaamery MA, Hoffman CS. Genetics. 2008;178(4):1927–1936. doi: 10.1534/genetics.107.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dal Santo P, Blanchard B, Hoffman CS. J Cell Sci. 1996;109(Pt 7):1919–1925. doi: 10.1242/jcs.109.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman CS, Winston F. Genes Dev. 1991;5(4):561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- 7.Jin M, Fujita M, Culley BM, Apolinario E, Yamamoto M, Maundrell K, Hoffman CS. Genetics. 1995;140(2):457–467. doi: 10.1093/genetics/140.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landry S, Pettit MT, Apolinario E, Hoffman CS. Genetics. 2000;154(4):1463–1471. doi: 10.1093/genetics/154.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schadick K, Fourcade HM, Boumenot P, Seitz JJ, Morrell JL, Chang L, Gould KL, Partridge JF, Allshire RC, Kitagawa K, Hieter P, Hoffman CS. Eukaryot Cell. 2002;1(4):558–567. doi: 10.1128/EC.1.4.558-567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stiefel J, Wang L, Kelly DA, Janoo RTK, Seitz J, Whitehall SK, Hoffman CS. Eukaryotic Cell. 2004;3(3):610–619. doi: 10.1128/EC.3.3.610-619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Griffiths K, Zhang YH, Ivey FD, Hoffman CS. Genetics. 2005;171:1523–1533. doi: 10.1534/genetics.105.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender AT, Beavo JA. Pharmacol Rev. 2006;58(3):488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 13.Conti M, Beavo J. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 14.Lerner A, Epstein PM. Biochem J. 2006;393(Pt 1):21–41. doi: 10.1042/BJ20051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugnier C. Pharmacol Ther. 2006;109(3):366–398. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Houslay MD. Trends Biochem Sci. 2010;35(2):91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Alaamery MA, Wyman AR, Ivey FD, Allain C, Demirbas D, Wang L, Ceyhan O, Hoffman CS. J Biomol Screen. 2010;15(4):359–367. doi: 10.1177/1087057110362100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivey FD, Wang L, Demirbas D, Allain C, Hoffman CS. J Biomol Screen. 2008;13(1):62–71. doi: 10.1177/1087057107312127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVoti J, Seydoux G, Beach D, McLeod M. Embo J. 1991;10(12):3759–3768. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higuchi T, Watanabe Y, Yamamoto M. Mol Cell Biol. 2002;22(1):1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutz H, Heslot H, Leupold U, Loprieno N. In: Handbook of Genetics. King RC, editor. New York: Plenum Press; 1974. [Google Scholar]
- 22.Hoffman RL, Hoffman CS. Curr Genet. 2006;49(6):414–420. doi: 10.1007/s00294-006-0065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks JL, 2nd, Corbin JD, Francis SH. J Pharmacol Exp Ther. 2009;331(1):133–141. doi: 10.1124/jpet.109.156935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Liu Y, Chen Y, Robinson H, Ke H. J Biol Chem. 2005;280(35):30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 25.Cytrynska M, Wojda I, Frajnt M, Jakubowicz T. Can J Microbiol. 1999;45(1):31–37. [PubMed] [Google Scholar]
- 26.Frajnt M, Cytrynska M, Jakubowicz T. Acta Biochim Pol. 2003;50(4):1111–1118. [PubMed] [Google Scholar]
- 27.Canaves JM, Taylor SS. J Mol Evol. 2002;54(1):17–29. doi: 10.1007/s00239-001-0013-1. [DOI] [PubMed] [Google Scholar]
- 28.Colicelli J, Birchmeier C, Michaeli T, O'Neill K, Riggs M, Wigler M. Proc Natl Acad Sci U S A. 1989;86(10):3599–3603. doi: 10.1073/pnas.86.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colicelli J, Nicolette C, Birchmeier C, Rodgers L, Riggs M, Wigler M. Proc Natl Acad Sci U S A. 1991;88(7):2913–2917. doi: 10.1073/pnas.88.7.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaeli T, Bloom TJ, Martins T, Loughney K, Ferguson K, Riggs M, Rodgers L, Beavo JA, Wigler M. J Biol Chem. 1993;268(17):12925–12932. [PubMed] [Google Scholar]
- 31.Pillai R, Kytle K, Reyes A, Colicelli J. Proc Natl Acad Sci U S A. 1993;90(24):11970–11974. doi: 10.1073/pnas.90.24.11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Nature. 2008;456(7218):130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 33.Smith SJ, Cieslinski LB, Newton R, Donnelly LE, Fenwick PS, Nicholson AG, Barnes PJ, Barnette MS, Giembycz MA. Mol Pharmacol. 2004;66(6):1679–1689. doi: 10.1124/mol.104.002246. [DOI] [PubMed] [Google Scholar]
- 34.Corbin JD, Francis SH. Int J Clin Pract. 2002;56(6):453–459. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.