Abstract
The origin and rapid diversification of the angiosperms (Darwin's “Abominable Mystery”) has engaged generations of researchers. Here, we examine the floral genetic programs of phylogenetically pivotal angiosperms (water lily, avocado, California poppy, and Arabidopsis) and a nonflowering seed plant (a cycad) to obtain insight into the origin and subsequent evolution of the flower. Transcriptional cascades with broadly overlapping spatial domains, resembling the hypothesized ancestral gymnosperm program, are deployed across morphologically intergrading organs in water lily and avocado flowers. In contrast, spatially discrete transcriptional programs in distinct floral organs characterize the more recently derived angiosperm lineages represented by California poppy and Arabidopsis. Deep evolutionary conservation in the genetic programs of putatively homologous floral organs traces to those operating in gymnosperm reproductive cones. Female gymnosperm cones and angiosperm carpels share conserved genetic features, which may be associated with the ovule developmental program common to both organs. However, male gymnosperm cones share genetic features with both perianth (sterile attractive and protective) organs and stamens, supporting the evolutionary origin of the floral perianth from the male genetic program of seed plants.
Keywords: ABCE model, fading borders, floral evolution, floral origin, transcriptional profiling
The evolutionary origin of flowering plants, or angiosperms, remains one of the greatest unsolved biological mysteries. The presence of a diverse assemblage of floral forms shortly after the sudden appearance of angiosperm fossils in early Cretaceous deposits ca. 130 Mya suggests that a rapid radiation established most of the modern lineages within a few million years (1). Famously declared an “abominable mystery” over a century ago by Charles Darwin (in a letter to J.D. Hooker in 1879) (2), the origin and subsequent diversification of flowering plants have captured the imagination of generations of researchers in wide-ranging botanical disciplines. Essential to any explanation has been the origin of the flower itself. Hypotheses on this topic, whether based on reconstructions from morphological features (3) or developmental genetics (4, 5), all attempt to resolve the evolution of floral organs from preexisting structures in nonflowering seed plants (gymnosperms).
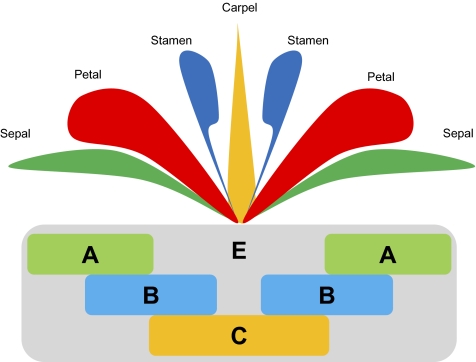
Flowers typically are composed of a perianth of leaf-like sepals and colorful petals surrounding stamens (the male reproductive organs) and carpels (the female reproductive organs). Angiosperm stamens and carpels are widely regarded as homologous with their functional counterparts in the simple strobili (cones) of gymnosperms, microsporophylls, and megasporophylls, respectively (see ref. 6 for alternative views), but sepals and petals are unique to flowers and therefore, lack clear evolutionary precursors. However, extensive research on the genetic control of flower development in Arabidopsis has demonstrated that floral organs are cross-transformable into one another and even can be modified into leaves through genetic manipulation of certain key developmental regulators (7–9). These insights are encapsulated in the ABCE model for the genetic control of floral organ identity (Fig. 1, reviewed in refs. 5, 10, and 11). Most of what we know about the regulation of floral development has been discovered through genetic manipulation of derived eudicot model systems, but comparative analyses suggest core components of the ABCE genetic program are conserved across angiosperms (12) and may trace to an original BC program that operated in the common ancestor of all seed plants (5). However, evolutionary dynamism in the spatial deployment of ABCE function, and of B function in particular, may underlie fundamental changes in floral organization during angiosperm diversification (13).
Fig. 1.
The ABCE model of floral organ identity. Sepals are produced where A function acts alone, petals where A and B functions overlap, stamens where B and C functions combine, and carpels where C function acts alone. In the eudicot genetic model Arabidopsis thaliana, APETALA1 (AP1) and APETALA2 (AP2) are the A-function genes, APETALA3 (AP3) and PISTILLATA (PI) together specify B function, C function is specified by AGAMOUS (AG), and multiple SEPALLATA genes provide E function (7–9).
Here, we conduct comparative analyses of global transcriptome data which encompass the properties of whole developmental systems (e.g., 14, 15) to investigate floral developmental genetics beyond the action of candidate regulatory genes. Specifically, we analyze transcriptome data for the water lily Nuphar advena (Nymphaeales) representing the sister lineage of all extant flowering plants except Amborella (16), the magnoliid Persea americana (avocado; Laurales), the eudicots Eschscholzia californica (California poppy; Ranunculales) and Arabidopsis thaliana (Brassicales), and the gymnosperm Zamia vazquezii (Cycadales) to help reconstruct the origin and evolution of flowers. Our analyses extend previous comparisons of floral transcriptional programs (17–19) to several additional phylogenetically significant taxa, and we present insights into the genetic relationships among individual floral organs and gymnosperm reproductive cones.
Results
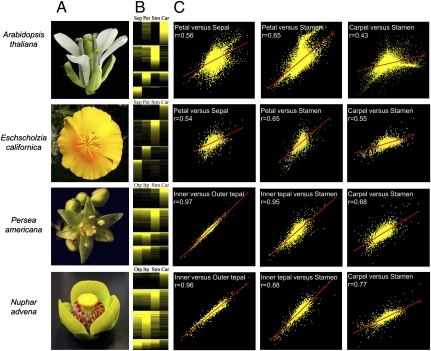
Global gene-expression data are rich in patterns of transcript abundances that we have explored through comparative cross-species analyses for evolutionary insights. We conducted a series of comparisons to examine the spatial distribution of florally biased expression in each species (Fig. 2). Manual ranking of positive log2 floral organ/leaf expression ratios by floral organ of primary expression indicates that genes participating in the transcriptional programs of Nuphar and Persea flowers are deployed in broad domains across adjacent whorls and beyond, whereas those in Arabidopsis and Eschscholzia are more tightly constrained to individual floral organ categories (Fig. 2B). Likewise, pairwise comparisons of the gene-expression profiles of adjacent floral organ categories translated into substantially greater correlations in Nuphar and Persea than in the two eudicots (Fig. 2C).
Fig. 2.
Canalization of floral organ transcriptional programs during angiosperm diversification. (A) Flowers of Nuphar and Persea bear an undifferentiated perianth of petaloid organs (tepals), whereas in Eschscholzia and Arabidopsis flowers the perianth is differentiated into leaf-like outer sepals and colorful inner petals. (B) Log2 floral organ/leaf gene expression ratios ranked by organs of peak expression contrast the blurred boundaries between adjacent floral organs in Nuphar and Persea versus the sharp boundaries in Arabidopsis and Eschscholzia. Stamen-preferential genes generally are more broadly expressed, but more so in Nuphar and Persea than in the eudicots, whereas carpel-preferential genes generally are more spatially restricted. Car, carpels; Itp, inner tepals; Otp, outer tepals; Pet, petals; Sep, sepals; Stm, stamens. The scale of log2 ratios ranges from saturated yellow (1 and higher = at least twofold up-regulated) to black (0 and lower = no change or down-regulated). (C) Scatter plots of log2 floral organ/leaf ratios and Pearson correlations indicate that transcriptional profiles of adjacent floral organs are more strongly correlated in Nuphar and Persea than in eudicots. Analyses are based on 4,588 Nuphar, 4,508 Persea, 5,468 Eschscholzia, and 12,785 Arabidopsis genes up-regulated in their respective flowers relative to leaves.
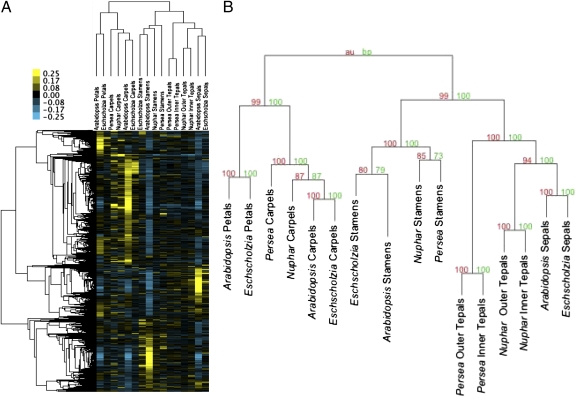
Next, comparisons of organwise transcriptional profiles based on relative abundance (RA) scores (20) of all putatively homologous genes were conducted to assess process homology (i.e., sharing a genetic program that is potentially, although not necessarily, inherited from a common ancestor (21) among floral organs. We found strong evidence that carpels are process homologous, as are stamens, across angiosperms (Fig. 3). Petals of Arabidopsis and Eschscholzia also appear to be process homologous, as are sepals of these two eudicots. In contrast to the eudicots, the outer and inner perianth organs of Nuphar and Persea are not differentiated into distinct sepals and petals [this traditional distinction in Nuphar may be spurious (22)] but are morphologically similar and are termed “tepals.” We found that Nuphar and Persea tepals are genetically most similar to each other, within their respective flowers, and collectively are more process homologous with eudicot sepals than with any other floral organs. Deeper in the cluster hierarchy, the sepals/tepals transcriptional program was closest to that operating in stamens, whereas eudicot petals were more similar to carpels. This hierarchy of organ relationships is strongly supported by random resampling analyses (Fig. 3) and also is robust to directed resampling in three modified data sets that exclude genes not sampled in all species, exclude data for Nuphar and Persea, or exclude data for Arabidopsis (Fig. S1).
Fig. 3.
Hierarchy of genetic relationships among floral organs supports the process homology of stamens, carpels, sepals, and petals, respectively, and places tepals with sepals in a group collectively closest to stamens. (A) RA scores of expression levels across floral organs within their respective flowers were clustered and subsequently mean centered for visual effect. The color scale ranges from 0.25 (yellow) to −0.25 (blue). (B) Bootstrap (red) and approximately unbiased (green) support values indicate strong stability for all clusters.
The potential biological significance of the gene clusters supporting these organ groupings was estimated through Gene Ontology (GO) annotation (Table S1 and Figs. S2–S7) and transcription factor binding sites (TFBS) enrichment analyses for their Arabidopsis members (Table S2). Genes coexpressed in sepals and tepals are involved in biological processes related to photosynthesis and defense (Fig. S2), and their promoters share various related G-box promoter elements associated with light responses (23–25), as well as the Unfolded Protein Response (UPR) motif associated with stress responses (26). Genes of the stamens cluster also participate in stress responses (Fig. S3) but additionally participate in cellular growth and differentiation and pollination (probably, pollen production). G-box and UPR motifs, as well the CArG binding site targeted by MADS-box genes, are enriched in their promoters. The cluster of genes coexpressed in sepals, tepals, and stamens is associated with stress responses, general energy-related processes (Fig. S4), and the G-box, UPR, and Low Temperature Responsive Element (LTRE) motifs. GO enrichment for petal genes highlights cellular growth and differentiation, morphogenesis, and response to stimuli (Fig. S5), and TFBS enrichment identifies the MYB1 binding site and two of the G-box elements that were found among the sepal, tepal, and stamen genes. Enrichment statistics for carpel genes indicate significant activity related to the regulation of gene expression, including both activation and silencing, along with several developmental processes (Fig. S6), and identify the TELO-box and E2F motifs that previously have been associated with rapid growth (27). Genes expressed in both petals and carpels share enrichment for the MYB1 binding site, the biological processes of petals, the cell cycle and floral development processes of carpels, and the stress responses of sepals, tepals, and stamens (Fig. S7).
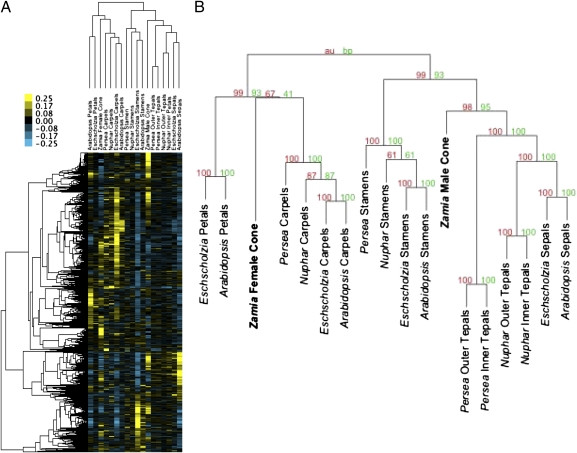
Because the reproductive cones of gymnosperms are the likely evolutionary precursors for flowers (5, 28, 29), we compared the expression profiles of male and female cones of the cycad Zamia vazquezii with those of angiosperm floral organs. Cluster analyses retrieved the organwise topology for floral organs described above with Zamia male cones placed next to the sepals/tepals cluster and female cones closest to angiosperm carpels (Fig. 4). Genes in the sepals/tepals cluster were unequally divided between expression primarily in male and female cones. Enrichment statistics identify photosynthesis and defense-related processes in both subsets (Table S1), but most enriched binding sites are associated with the subset expressed in male cones (Table S2). Genes coexpressed in stamens and male cones share significant enrichment for response to external stimuli and various metabolic processes (Table S1) and the suite of G-box, UPR and CArG binding sites of the stamen genes. Genes coexpressed in carpels and female cones regulate gene expression and share the TELO-box and E2F motifs (Table S2), as did carpels considered alone. Small gene clusters involving genes coexpressed in petals and either of the Zamia reproductive cones lack significant enrichment for GO annotation terms or TFBS.
Fig. 4.
(A) Expression profiles of Zamia reproductive cones share moderate similarity with angiosperm functional counterparts. Male cones are genetically closest to the angiosperm sepals/tepals cluster in the larger cluster with stamens, whereas female cones are genetically closest to carpels. Cluster analyses are based on previously analyzed RA expression data (Fig. 3) and an additional 825 Arabidopsis genes tagged as putative homologs in the Zamia data set. RA values for Zamia were mean centered across male and female cones and then halved to adjust their color range to that of the angiosperms; color scale ranges from 0.25 (yellow) to −0.25 (blue). (B) Bootstrap (red) and approximately unbiased (green) support values indicate strong support for the position of Zamia male cones, but the placement of female cones with carpels is not well supported.
Discussion
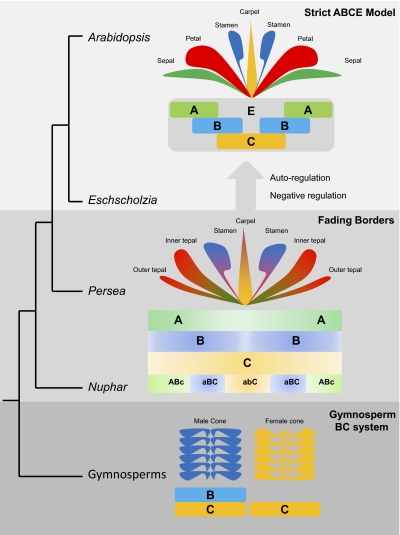
Evolutionary reconstructions of expression patterns of key floral transcription factors show progressively more spatially restricted deployment throughout angiosperm evolution, from across the floral meristem in Amborella, Nuphar, and Persea, for example, to specific organs in Arabidopsis and other eudicots (12, 30, 31). Likewise, the transcriptional cascades that are broadly deployed in Nuphar and Persea have been tightly constrained spatially within organ-specific boundaries in Eschscholzia and Arabidopsis. The molecular mechanisms responsible for this distinction likely lie within the machinery of the ABCE model itself. Especially relevant is the “fading borders” modification of the ABCE model (32), in which floral transcriptional regulators are broadly arrayed across the flower with gradually fading gradients of influence from focal to peripheral organ categories imparting intergrading morphologies across floral organs (Fig. 5).
Fig. 5.
The genetic regulation of floral development may have evolved from the BC system of gymnosperm cones into a “fading borders” program that later was shaped into the strict ABCE scheme. In the “fading borders” program, floral organs are influenced by transcriptional cascades regulated by “ABc,” “aBC,” and “abC” activities, where lowercase font indicates lower functional influence. These cascades promote the development of morphologically intergrading petaloid organs (tepals), stamens, and carpels, respectively. In the strict ABCE scheme, the sepal/petal distinction is maintained by strictly controlled B-function expression, although both perianth organs are influenced by A function. Strict regulation of the B-function domain also maintains a distinct boundary between stamens and carpels, but both are simultaneously influenced by C function. Similarly, repression of C function in petals would separate their developmental program from stamens, but both organs are influenced by B function. ABC functions extend beyond these organ boundaries and promote broadly overlapping transcriptional cascades in the “fading borders” program.
Our observations that transcriptional programs operate broadly across adjacent floral organs in Nuphar and Persea extend the “fading borders” model from the action of specific regulatory genes to downstream transcriptional cascades in floral development (see also ref. 17) and support the inference that this model is the ancestral regulatory program for flowers. A shared transcriptional program across the perianth organs and stamens of Aquilegia (Ranunculales) (33) suggests that some form of “fading borders” operates in some basal members of the eudicot clade as well. Moreover, extrapolating the trajectory of floral transcriptome evolution to its likely origin leads to an ancestrally uniform program in which separate components (if any) overlap fully, resembling the genetic program operating in unisexual gymnosperm cones (Fig. 5). This gymnosperm program would have been progressively compartmentalized during floral evolution, with flowers developing through the “fading borders” program before the strict ABCE scheme. Autoregulatory feed-back loops (5) and negative regulators that maintain strict spatial domains of ABCE function, at least in Arabidopsis (34; see ref. 35 for a comprehensive review), may have contributed to the sharpening of transcriptional boundaries, but the broadly overlapping floral transcriptional cascades evident in Nuphar and Persea suggest that adjacent organ identity functions are not separated fully from each other in these flowers (Fig. 5). Homologs of organ-identity genes also are broadly expressed during the initial stages of Nuphar and Persea floral development (12, 30), when organ identity is thought to be specified, suggesting that the transcriptional patterns we find in their late-stage flowers also may characterize their early developmental programs.
Despite broader spatial deployment of organwise transcriptional programs in Nuphar and Persea, we found substantial conservation in the genetic profiles of carpels and stamens, respectively, across angiosperms. The hierarchy of genetic relationships between these reproductive organs and sepals, petals, tepals, and gymnosperm reproductive cones, together with supporting data from GO annotation and TFBS enrichment analyses, provide unprecedented insights into the deep history of floral organ evolution.
The placement of Zamia female reproductive cones with carpels (Fig. 3), despite an evolutionary distance spanning perhaps 300 million years (36), may be a remarkable testament to the antiquity encapsulated in the ovule, the defining innovation of seed plants. Because both angiosperms and gymnosperms bear ovules, the transcriptional cascade contributed to carpels by ovules may be their most ancient component. Indeed, the regulation of gene expression and developmental processes that are dominant in both angiosperm carpels and female gymnosperm cones (Table S1) may relate to ovule development, which in carpels occurs during late floral development. The association of Arabidopsis and Eschscholzia petals with carpels is robust to random and directed resampling analyses (Fig. S1) but conflicts with the proposed recent origin of eudicot petals from stamens, sepals, or tepals (37). However, enrichment statistics indicate associations between carpel-specific genes and genes coexpressed in both petals and carpels (Table S1 and Figs. S6 and S7). Specifically, cell growth and differentiation and MYB1 binding sites are common to the two organs. Perhaps, in the evolution of petals from their ancestral organs, these components of the carpel development program were recruited for petal development, as has been envisioned previously on the basis of some key developmental regulators (38).
Less evolutionary conservation in male reproductive genetics is suggested by the position of Zamia male cones next to sepals/tepals rather than stamens (Fig. 4). However, this genetic relationship could be related to the pollen-producing program of the developmentally mature stamens in our data set, as GO annotations indicate, separating the stamens from the effectively sterile premeiotic male cones. The origin of the floral perianth from sterile male organs, an implicit corollary of evolutionary scenarios for the origin of the flower, also is consistent with their genetic relationships with premeiotic gymnosperm male cones. Notably, homologs of both B- and C-function genes are expressed in the tepals of Persea (17, 30), Nuphar (18), and other Nymphaeales (31), supporting the link between male reproductive genetics and perianth organs in the flowers of these species. The appearance of the evolutionarily derived eudicot sepals within the tepals cluster is compatible with an “intercalation” hypothesis for the origin of “true” leafy sepals via restriction of petaloidy from the outer perianth in core eudicot flowers (38, 39). Accordingly, the ancestral, tepaloid, angiosperm perianth may have been genetically programmed to function both as a protective envelope for the reproductive organs and to attract floral visitors for pollination purposes.
TFBS add to the commonalities across the genetic profiles of angiosperm sepals, tepals, and stamens. Whether organ-specific or shared across organs, the sets of genes expressed in these organs are significantly enriched for a common suite of binding sites, some of which are shared with the petal genes (Table S2). Shared binding sites are anticipated features of coexpressed genes, indicating their likely coregulation (e.g., 40). Therefore, these genomic features of genes with distinct expression patterns in floral organs may represent vestiges of an ancestral set of coexpressed genes. This scenario gains credibility in the context of hypotheses of floral origin from a single gymnosperm reproductive cone, in which case the preponderance of binding sites shared between Zamia male cones and angiosperm sepals, tepals, petals, and stamens (Table S2) supports the “Mostly Male” (29) and “Out of Male” (5) hypotheses. An alternative scenario in which these binding sites evolved de novo in the distinct sets of genes expressed in sepals/tepals/male cones, in stamens, and in petals is less parsimonious.
Our results establish connections between organwise transcriptome evolution and the morphological evolution of flowers. Ancestral female genetic programs inherited from gymnosperms are moderately conserved in angiosperm carpels, whereas male gymnosperm reproductive programs are dispersed among stamens and perianth organs. Canalization of ancestrally overlapping “fading borders” transcriptional cascades to produce organ-specific patterns of expression in angiosperm flowers may trace to the origin of the strict ABCE scheme characteristic of the eudicot angiosperms, although an earlier canalization that includes the monocots and/or multiple independent canalizations cannot be discounted.
Methods
Microarray expression data for leaves and mature floral organs were extracted from data sets for Eschscholzia [Gene Expressio Omnibus (GEO) accession no. GSE24237], Arabidopsis (GEO accession no. GSE5632), Persea (GEO accession no. GSE13737), and Nuphar (GEO accession no. GSE23082). Cross-species analyses were conducted on RA measures of gene expression among floral organs within species to remove systemic biases (i.e., normalized) across the species-specific datasets (20). RA scores were assembled into a multispecies expression matrix composed of 6,584 Arabidopsis, 4,006 Nuphar, 3,725 Persea, and 4,568 Eschscholzia genes identified as likely homologs through best reciprocal tBLASTx (National Center for Biotechnology Information) E-scores <10–5. A total of 125,502 transcripts was collected by 454 transcriptome sequencing of nonnormalized cDNA libraries made from immature (likely premeiotic) male and female Zamia cones [Sequence Read Archive (SRA) accession nos. SRX019097 and SRX019098, respectively]; 78,843 Zamia transcripts assembled into unigenes potentially homologous with 6,920 Arabidopsis genes (tBLASTx E-score <10–5). Tag counts per unigene per library were summarized on the basis of putative homology with Arabidopsis genes and normalized to the total number of tags in the respective libraries. RA measures of Zamia gene expression across cones were calculated by dividing normalized tag counts by their sum across cones and were appended to the angiosperm data (together with data for 825 additional Arabidopsis genes not tagged in Nuphar, Persea, or Eschscholzia) on the basis of putative homology. Cluster analyses are based on Pearson correlation scores with the average-linkage clustering algorithm implemented in Cluster 3.0 (41), and the results were visualized with Java TreeView (42). Support values for organwise clusters were estimated with the same clustering parameters using the R package Pvclust (43) with 10,000 replicate runs. GO annotation enrichment analyses were conducted with Benjamini and Hochberg false discovery rate correction and significance set at P < 0.05 using the Cytoscape (44) plug-in BiNGO (45). TBFS enrichment analyses were conducted with Bonferroni correction and significance set at P < 0.05 using Athena (46).
Supplementary Material
Acknowledgments
We thank Dennis Stevenson and Christian Schultz (New York Botanical Garden, Bronx, NY) for Zamia tissues. Oligonucleotide probes for the Persea, Nuphar, and Eschscholzia arrays were designed by Raad Gharaibeh and Cynthia Gibas (University of North Carolina-Charlotte). This study was supported by National Science Foundation Grants PRG-0115684 for the Floral Genome Project and PGR-0638595 for the Ancestral Angiosperm Genome Project. Publication of this article was funded in part by the University of Florida Open Access Publishing Fund.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013395108/-/DCSupplemental.
References
- 1.Friis EM, Pedersen KR, Crane PR. Diversity in obscurity: Fossil flowers and the early history of angiosperms. Philos Trans R Soc Lond B Biol Sci. 2010;365:369–382. doi: 10.1098/rstb.2009.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman WE. The meaning of Darwin's 'abominable mystery'. Am J Bot. 2009;96:5–21. doi: 10.3732/ajb.0800150. [DOI] [PubMed] [Google Scholar]
- 3.Endress PK, Doyle JA. Reconstructing the ancestral angiosperm flower and its initial specializations. Am J Bot. 2009;96:22–66. doi: 10.3732/ajb.0800047. [DOI] [PubMed] [Google Scholar]
- 4.Frohlich MW, Chase MW. After a dozen years of progress the origin of angiosperms is still a great mystery. Nature. 2007;450:1184–1189. doi: 10.1038/nature06393. [DOI] [PubMed] [Google Scholar]
- 5.Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot (Lond) 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudall PJ, Bateman RM. Defining the limits of flowers: The challenge of distinguishing between the evolutionary products of simple versus compound strobili. Philos Trans R Soc Lond B Biol Sci. 2010;365:397–409. doi: 10.1098/rstb.2009.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 8.Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 9.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Soltis PS, Soltis DE, Kim S, Chanderbali AS, Buzgo M. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC function. Adv Bot Res. 2006;44:385–402. [Google Scholar]
- 11.Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. The ABC model and its applicability to basal angiosperms. Ann Bot (Lond) 2007;100:155–163. doi: 10.1093/aob/mcm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, et al. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- 13.Soltis PS, et al. Floral variation and floral genetics in basal angiosperms. Am J Bot. 2009;96:110–128. doi: 10.3732/ajb.0800182. [DOI] [PubMed] [Google Scholar]
- 14.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 15.Parikh A, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanderbali AS, et al. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae) Proc Natl Acad Sci USA. 2009;106:8929–8934. doi: 10.1073/pnas.0811476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo MJ, Chanderbali AS, Altman NS, Soltis PS, Soltis DE. Evolutionary trends in the floral transcriptome: Insights from one of the basalmost angiosperms, the water lily Nuphar advena (Nymphaeaceae) Plant J. 2010;64:687–698. doi: 10.1111/j.1365-313X.2010.04357.x. 10.1111/j.1365-313X.2010.04357.x. [DOI] [PubMed] [Google Scholar]
- 19.Zahn LM, et al. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica: A reference for comparison with core eudicots and basal angiosperms. Genome Biol. 2010;11:R101. doi: 10.1186/gb-2010-11-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 21.Hall BK. Descent with modification: The unity underlying homology and homoplasy as seen through an analysis of development and evolution. Biol Rev Camb Philos Soc. 2003;78:409–433. doi: 10.1017/s1464793102006097. [DOI] [PubMed] [Google Scholar]
- 22.Warner KA, Rudall PJ, Frohlich MW. Environmental control of sepalness and petalness in perianth organs of waterlilies: A new Mosaic theory for the evolutionary origin of a differentiated perianth. J Exp Bot. 2009;60:3559–3574. doi: 10.1093/jxb/erp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 24.Jiao Y, Ma L, Strickland E, Deng XW. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giuliano G, et al. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frohlich MW. An evolutionary scenario for the origin of flowers. Nat Rev Genet. 2003;4:559–566. doi: 10.1038/nrg1114. [DOI] [PubMed] [Google Scholar]
- 29.Frohlich MW, Parker DS. The mostly male theory of flower evolutionary origins: From genes to fossils. Syst Bot. 2000;25:155–170. [Google Scholar]
- 30.Chanderbali AS, et al. Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae) Int J Plant Sci. 2006;167:1075–1089. [Google Scholar]
- 31.Yoo M, Soltis PS, Soltis DE. Expression of floral MADS-Box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int J Plant Sci. 2010;171:121–146. [Google Scholar]
- 32.Buzgo M, Soltis PS, Soltis DE. Floral developmental morphology of Amborella trichopoda (Amborellaceae) Int J Plant Sci. 2004;165:925–947. [Google Scholar]
- 33.Voelckel C, Borevitz JO, Kramer EM, Hodges SA. Within and between whorls: Comparative transcriptional profiling of Aquilegia and Arabidopsis. PLoS ONE. 2010;5:e9735. doi: 10.1371/journal.pone.0009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irish VF. The Arabidopsis petal: A model for plant organogenesis. Trends Plant Sci. 2008;13:430–436. doi: 10.1016/j.tplants.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Zahn LM, Feng B, Ma H. Beyond the ABC-model: Regulation of floral homeotic genes. Adv Bot Res. 2006;44:163–204. [Google Scholar]
- 36.Savard L, et al. Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc Natl Acad Sci USA. 1994;91:5163–5167. doi: 10.1073/pnas.91.11.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronse De Craene LP. Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Ann Bot (Lond) 2007;100:621–630. doi: 10.1093/aob/mcm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albert VA, Gustafsson M, Di Laurenzio L. In: Molecular Systematics of Plants II. Soltis DE, Soltis PS, Doyle JJ, editors. Boston: Kluwer; 1998. pp. 349–374. [Google Scholar]
- 39.Ainsworth C, Crossley S, Buchanan-Wollaston V, Thangavelu M, Parker J. Male and female flowers of the dioecious plant sorrel show different patterns of MADS box gene expression. Plant Cell. 1995;7:1583–1598. doi: 10.1105/tpc.7.10.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Creux NM, Ranik M, Berger DK, Myburg AA. Comparative analysis of orthologous cellulose synthase promoters from Arabidopsis, Populus and Eucalyptus: Evidence of conserved regulatory elements in angiosperms. New Phytol. 2008;179:722–737. doi: 10.1111/j.1469-8137.2008.02517.x. [DOI] [PubMed] [Google Scholar]
- 41.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 42.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 46.O'Connor TR, Dyreson C, Wyrick JJ. Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.