Abstract
Positive-strand RNA viruses include a large number of human and animal pathogens whose essential RNA-dependent RNA polymerases (RdRPs) share a structurally homologous core with an encircled active site. RdRPs are targets for antiviral drug development, but these efforts are hindered by limited structural information about the RdRP catalytic cycle. To further our understanding of RdRP function, we assembled, purified, and then crystallized poliovirus elongation complexes after multiple rounds of nucleotide incorporation. Here we present structures capturing the active polymerase and its nucleotide triphosphate complexes in four distinct states, leading us to propose a six-state catalytic cycle involving residues that are highly conserved among positive-strand RNA virus RdRPs. The structures indicate that RdRPs use a fully prepositioned templating base for nucleotide recognition and close their active sites for catalysis using a novel structural rearrangement in the palm domain. The data also suggest that translocation by RDRPs may not be directly linked to the conformational changes responsible for active site closure and reopening.
Keywords: replication, picornavirus
Positive-strand RNA viruses cause diseases such as the common cold, acute hepatitis A, chronic hepatitis C, hemorrhagic fevers, meningitis, encephalitis, and paralytic poliomyelitis. The genome replication cycle of these viruses is entirely RNA-based, with replication being carried out by virally encoded RNA-dependent RNA polymerases that retain several hallmark polymerase sequence motifs within a ≈500-residue core structure composed of the typical polymerase palm, fingers, and thumb domains (1–5). A unique and conserved aspect of RNA-dependent RNA polymerase (RdRP) structures is that the fingers domain reaches across the palm to interact with the top of the thumb, encircling the active site.
Extensive biochemical and structural studies of polymerases in general have defined the catalytic cycle as a multistep process composed of initial NTP binding to an “open” conformation of the enzyme–template complex, a recognition event to ensure that only the correct nucleotide triggers the formation of the catalytically competent “closed” conformation, catalysis, and then an opening of the active site that is usually accompanied by translocation of the nucleic acid (6–8). RdRPs also use this multistep catalytic process, but the encircled active site structure makes it unlikely that major swinging motions of the fingers domain act to reposition the nascent base pair from an initial preinsertion site into the catalytic site, as is seen in other classes of single-subunit polymerases (9–12). Biochemical studies of poliovirus and foot-and-mouth disease virus (FMDV) RdRPs have shown that addition of the first one or two nucleotides onto a primer is the key initiation step on the pathway to forming a very stable and highly processive elongation complex (13, 14).
Despite the vast number of viral RdRP structures that have been solved over the past fifteen years, little is known about the structural interactions and conformational changes associated with nucleotide selection, active site closure for catalysis, and subsequent translocation of the RNA. The general path of RNA through the RdRPs is known for FMDV (15, 16) and Norwalk virus (17, 18) polymerases cocrystallized with short RNAs, resulting in initiation phase complexes that have at most incorporated one nucleotide. To gain further insights into the RdRP catalytic cycle, we assembled, purified, and then crystallized stable poliovirus polymerase elongation complexes (ECs) that had undergone multiple nucleotide addition cycles. The resulting structure was determined at 2.5 Å resolution and by soaking crystals in various NTP or NTP analogue solutions we were able to solve a series of structures that provide snapshots of the RdRP catalytic cycle.
Results
Elongation Complex Assembly and Crystallization.
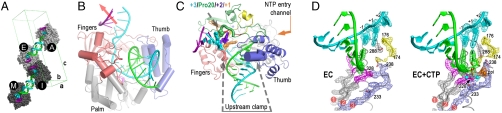
A library of primer-template RNAs was initially screened to identify sequences that could generate stalled polymerase elongation complexes stable enough to be purified by anion exchange chromatography. The purified complexes rapidly elongated through the remaining template sequence even in the presence of 400–600 mM NaCl, were unaffected by added competitor RNA, and retained activity for 2–3 weeks at 16 °C, indicating that the stalled ECs were very stable (Fig. S1). The EC whose structure was solved consisted of a 10-mer primer annealed to a 35-mer template strand that was stalled after incorporation of a G-A-G-A sequence, yielding a complex that contains both upstream and downstream RNA duplexes (Fig. S1). The complex crystallized with a unit cell containing four ECs arranged into two pairs via near coaxial stacking of their upstream template–product duplexes (Fig. 1A). The crystal packing interactions surrounding these two pairs of complexes are subtly different, which necessitated refinement in space group P1 with noncrystallographic symmetry applied within each EC pair, rather than in the closely related C2 space group.
Fig. 1.
Poliovirus 3Dpol elongation complex. (A) Crystal packing showing staggered coaxial stacking of upstream template-product duplexes resulting in two nonequivalent pairs of ECs (A&E vs. I&M). Product strand is shown in green, template in cyan, and downstream nontemplate in purple. (B) EC structure showing up- and downstream RNA duplexes as the template strand (cyan) threads through the active site and the red arrow indicates trajectory of downstream RNA duplex. (C) Top view of EC showing the single stranded conformation of the +1, +2, and +3 downstream template nucleotides and the protein clamp of the upstream duplex. Palm domain is in gray, thumb is in blue, and the individual fingers (19) are colored with index in green, middle in orange, ring in yellow, and the pinky in pink. (D) 3,500 K composite simulated-annealing omit maps contoured at 1.5 σ showing quality of active site electron density for the native (EC) and Mg2+-CTP (EC + CTP) complexes. The pyrophosphate (ppi) in the CTP complex is shown in orange and the presence of metals ions was confirmed by an essentially identical Mn2+-CTP structure (see Figs. S3 and S4).
3Dpol Elongation Complex Structure.
The 2.5 Å resolution (Table S1) EC structure shows three base pairs in the downstream template RNA hairpin, three bases threading through the active site, and a well-defined 12-basepair upstream A-form duplex (Fig. 1B). The upstream duplex is held in place by a clamping structure composed of an extended loop from the pinky finger inserting into the major groove and an α-helix from the thumb domain packing into the opposite minor groove (Fig. 1C). This clamp is expanded compared to the apo 3Dpol structure (19) and there is no mechanism for strand separation of the newly synthesized RNA as it exits the 3Dpol EC (Fig. S2).
The three single-stranded nucleotides threading through the active site occupy distinct binding sites with different functions (Fig. 1C). The templating +1 base is fully stacked on the upstream duplex and is located directly above the catalytic site GDDC6–8 residues (superscripts indicate positions within conserved polymerase motifs A–F). The +2 base is bound in a pocket formed by the 3Dpol index finger, sits on top of the conserved RdRP signature motif F1 (4) within the ring finger, and is unstacked from the +3 base by Pro20 from the index finger. The +3 adenosine is found in an apparent strand-separation site where it cannot participate in base-pairing because residues 18 and 19 of the index finger block access for any base-pairing interactions with the nontemplate strand.
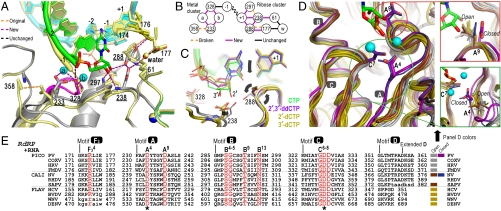
As is observed in RdRP structures without bound RNA, the central RRM-fold in the palm domain is not fully formed because the β-sheet interaction between motifs A and C is frayed at the end closest to the conserved active site GDDC6–8 residues. There are no Mg2+ ions present at the conserved active site Asp233A4 and Asp328C7 residues. This lack of a prebound metal at the catalytic site is likely due to the fact that the conserved Asp233A4 side chain that coordinates both catalytic metals in polymerases is pointing away from the active site and interacting with the backbone of residue 358D8 (Figs. 1D and 3A).
Fig. 3.
Molecular interactions involved in RdRP active site closure. (A) Superposition of 3Dpol active site in open state 1 (yellow) and closed state 4 (colors) showing how ribose hydroxyl recognition drives active site closure and divalent metal binding via the two distinct clusters of interactions diagrammed in panel (B). (C) Comparison of bound CTP, 2′-dCTP, 3′-dCTP, and 2′,3′-ddCTP conformations illustrating the NTP movement associated with active site closure. Only CTP with both ribose hydroxyls triggers the movements (arrows) of Asp238A9 and Ser288B4 that result in the closed conformation and catalysis. β and γ phosphates are omitted for clarity. (D) Conservation of motif A open conformation among all positive-strand RNA virus polymerases shown by superposition of four picornaviral (gray), two caliciviral (olive), and four flaviviral (brown) polymerases and a direct comparison with the closed conformation polio (purple) and Norwalk (blue) virus polymerase structures. Mg2+ ions and newly incorporated CMP are from the state 4 structure and the expanded views show the conformations of key side chains in all the structures. Note that a rabbit hemorrhagic disease virus polymerase structure adopting an intermediate conformation induced by Lu3+ binding is not shown. (E) Structure-based sequence alignment of RdRP motifs A–D and F3. Residues playing key roles in active site interactions and closure are colored red, the two invariant catalytic Asp residues are highlighted by asterisks, and residues in lower case letters either deviate from the consensus structure conformations or are not resolved in the crystal structures and are therefore included based only on sequence homology. See Fig. S8A for an extended alignment of all polymerase motifs across all classes of single-subunit polymerases and Fig. S8B for PDB codes.
NTP Complexes.
To further explore various structural states of the polymerase that occur during the catalytic cycle, we soaked EC crystals in solutions containing various NTPs and NTP analogs. We observed strong difference density allowing us to model the cognate CTP and its 2′-dCTP, 3′-dCTP, and 2′,3′-ddCTP deoxy derivatives (Figs. S3 and S4), but did not observe new density anywhere in the complex upon soaking with UTP, ATP, or GTP. Thus, the EC appears to be highly selective for cognate NTP binding, unlike apo 3Dpol structure that can bind all four NTPs in a common binding site (20) that is adjacent to the site seen in these EC structures (Fig. S3). A maximum likelihood based multiple structure superposition (21) of the complexes shows that addition of deoxy-CTPs results in only subtle structural shifts within the fingers domain, whereas addition of CTP results in a well-defined shift of the palm domain motif A that structures the active site for catalysis (Fig. 2A).
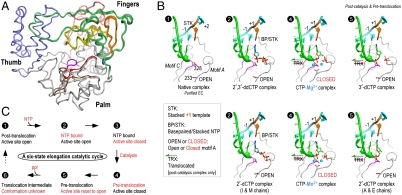
Fig. 2.
Structures of 3Dpol catalytic cycle states. (A) Superposition of the polymerase structures from all EC states showing small variations in the fingers domain structures and the significant CTP-induced movement of motifs A and D in the palm domain. This movement is highlighted by coloring motif A (red) and motif D (brown) of the closed conformation CTP complexes and showing spheres for the α-carbons of residues 233A4 and 355D5, and the remainder of the structures are colored as in Fig. 1C. (B) Structures of the EC and EC–NTP complexes arranged in order of the proposed catalytic cycle diagrammed in panel C. These include the open conformation state 1 in the absence of NTP, the open state 2 in the presence of bound deoxy-NTPs, the postcatalysis closed conformation state 4 upon CTP addition, and the postcatalysis and pretranslocation open state 5 with 3′-dCTP. Key residues and motifs are indicated in the state 1 panel and the presence of translocation, stacking, and base-pairing interactions involving the templating +1 base and bound nucleotide are indicated. See Figs. S3–S7 for electron density maps and additional analyses of the fingers and palm domain movements that take place upon NTP binding.
Deoxy CTP complexes.
Upon addition of 2′,3′-ddCTP we observe the incoming nucleotide base-paired with the templating base and stacked on the priming nucleotide (Fig. 2B). NTP binding in this site does not induce any significant structural changes in active site residues, Asp233A4 still points away from the catalytic center, no bound metal ions are observed, and there is no electron density evidence for catalysis in any of the four ECs in the P1 unit cell. Consistent with this, the triphosphate moiety is not fully loaded into the catalytic site, but instead sits ≈1 Å too high above the palm domain GDDC6–8 residues.
Soaking crystals in 2′-dCTP has different effects in the A&E and I&M pairs of coaxially stacked ECs (Fig. 2B and Fig. S4). No catalysis is observed in the I&M pair and the resulting 2′-dCTP conformation resembles that described above for 2′,3′-ddCTP. In contrast, catalysis has taken place within the A&E pair and the active site has returned to the open conformation, resulting in 2′-dCMP addition and a complex similar to that discussed below for 3′-dCTP.
In the presence of 3′-dCTP it is clear that catalysis has occurred in the crystal as evidenced by defined density for the nascent phosphodiester bond in all four ECs (Fig. S4). However, the nucleotide is found in a very similar position as 2′,3′-ddCTP (Fig. 3C and Fig. S6C) and the polymerase active site has the same structure as in the 2′,3′-ddCTP complex, indicating that despite catalysis the polymerase has not been trapped in the catalytically competent closed conformation.
CTP complex.
Addition of the cognate CTP results in extensive conformational and hydrogen bonding changes in the complex that reflect active site closure and subsequent catalysis (Fig. 3A and Fig. S5). The heart of the conformational change is a 0.6–0.9 Å pivoting movement of the templating +1 base and its base-paired NTP down toward the active site (Fig. 3C). Active site closure is driven by extensive interactions with both 2′ and 3′ ribose hydroxyls that result in six new hydrogen bonding and charge interactions. As the ribose is pulled down into the active site, it collides with Asp238A9, repositioning that aspartate to establish interactions with Lys61 and Ser288B4. This in turn positions Ser288B4 to form a hydrogen bond with the NTP 2′-hydroxyl, as does Asn297B13 that was previously interacting with Asp238A9. The net result is a tight network of ribose hydroxyl interactions that not only position the NTP for catalysis, but also cause a realignment of the motif A backbone to form a complete 3-stranded β-sheet with motif C in the palm domain (Fig. 3D). Importantly, this realignment causes Asp233A4 to swing toward the RNA, where it now coordinates both Mg2+ ions necessary to complete the active site and enable catalysis. The binding of both divalent metal ions appears to be NTP dependent and associated with the Asp233A4 movement; there is no electron density evidence for bound metals when any of the noncognate NTPs were added with 5 mM excess MgCl2 nor when the EC crystals were soaked in 10 mM MgCl2 or MnCl2 solutions.
Closed Conformation Geometry.
The closed conformation CTP complex structure also allows us to identify the essential polymerase interactions that configure the active site for catalysis. Not surprisingly, the phosphoryl transfer reaction itself takes place by the same two-metal mechanism observed in other nucleic acid polymerases (8), using Mg2+ ions that are coordinated by residues Asp233A4, Asp328C7 and the NTP triphosphate (Fig. 3A). However, 3Dpol employs a unique closure step where a structural change in the palm domain, rather than the more commonly used fingers domain, triggers the transition from the open to the closed state. In all the EC structures there is also strong electron density for the Arg174F34 guanidinium group located directly above the NTP α and β phosphates where it may act as a proton donor (Fig. 1D, Fig. S4, and see discussion). The side chain is fairly flexible as judged by the weak density for its aliphatic chain, allowing the guanidinium group to maintain its position above the phosphates when the NTP descends into the catalytically competent closed conformation.
Discussion
From biochemical evidence it is known that viral RdRPs such as 3Dpol form the stable polymerase–RNA complex responsible for processive viral genome replication only after completing a few catalytic cycles (13, 14). Consequently, the approach of purifying 3Dpol-RNA complexes for crystallization after multiple rounds of nucleotide incorporation is a key element of being able to capture the structure of an authentic elongation complex and elucidate the structural changes associated with the RdRP catalytic cycle. The use of a long downstream template strand was also important for generating the specific interactions surrounding Pro20 and the +2 and +3 nucleotides that define the path of the template as it drops into the active site. The structure of the downstream duplex indicates that strand displacement occurs at a structurally conserved cleft between the pinky and index fingers with the nontemplate strand crossing the surface of the fingers domain.
A Six-State Catalytic Cycle.
Based on structural snapshots obtained after soaking CTP, 2′-dCTP, 3′-dCTP, and 2′,3′-ddCTP into the EC crystals, we propose a six-state model for the 3Dpol RdRP elongation cycle (Fig. 2 B and C). In contrast to what has been observed in other polymerase classes, the 3Dpol catalytic cycle does not involve a major nucleotide repositioning step whereby the nascent template-NTP base pair is moved from a preinsertion site into the active site by a swinging motion of the fingers domain. Rather, 3Dpol active site closure is achieved via initial NTP base-pairing to a fully prepositioned templating nucleotide followed by a ribose hydroxyl recognition step that drives structural changes within the palm domain to enable Mg2+ binding and subsequent catalysis.
The catalytic cycle begins with state 1, the initial EC structure solved in the absence of a bound NTP where the +1 nucleotide is sitting above the active site and is fully stacked on the upstream duplex. This conformation appears to be unique to RdRPs as other classes of processive polymerases generally have the +1 templating nucleotide unstacked to allow for initial NTP binding in a preinsertion site. Next, state 2 is defined as loading of the NTP binding site observed in the 2′,3′-ddCTP and 2′-dCTP (I&M chains only) structures where an incoming nucleotide is bound in the open conformation active site via stacking and base-pairing interactions but catalysis has not taken place. This is followed by closure of the active site to generate a precatalysis state 3 and a postcatalysis state 4 that are likely to have essentially identical polymerase structures. We have captured the closed conformation state 4 in the CTP soaking experiments based on clear electron density for the phosphodiester bond (Fig. 1D), whereas a homologous Norwalk virus (NV) polymerase initiation complex has been captured in state 3 (discussed below). Following catalysis, the active site is reset to the open conformation in a postcatalysis and pretranslocation state 5 shown by the 3′-dCTP and 2′-dCTP (A&E chains) structures. Notably, the reset to the open conformation without translocation is unusual because these two events are usually tightly coupled in polymerases, and the state 5 structure thus suggests the existence of a not yet characterized state 6 that reflects the molecular interactions responsible for mediating RNA translocation.
Compared to prior FMDV (15, 16) and NV (17, 18) polymerase-RNA structures that represent initiation phase complexes after the incorporation of zero or one nucleotide and were solved using shorter RNAs with only two or four nucleotides in the downstream template (Fig. S3), the poliovirus EC shows well-defined density for template strand nucleotides, a highly selective NTP binding site, and catalytic activity. The FMDV complexes are all in the open conformation states 1 and 2 and show comparatively weak electron density for NTPs bound in sites that are intermediate between those seen in the poliovirus EC and apo 3Dpol structures (Figs. S3 and S4). They also exhibit structural heterogeneity, high B-factors, and reduced atomic occupancies of the RNA chains that are indicative of an initiation complex that is sampling multiple conformations in search of the processive elongation complex (15, 16). Unlike the poliovirus EC structures, the FMDV structures do show strong electron density for a single metal ion near the active site (Fig. S4), suggesting that a prebound ion may move into the active site as “metal a” in the catalytic step. Interestingly, NV polymerase has been captured in a precatalytic closed conformation (17) corresponding to state 3 and the lack of catalysis is likely due to a distortion of the upstream RNA that affects active site geometry (Fig. 3D and Fig. S2E).
Nucleotide Selection and Fidelity.
Nucleotide incorporation by 3Dpol can be summarized as a two-step process comprised of initial NTP binding followed by recognition of both the 2′ and 3′-ribose hydroxyl groups to trigger closure of the active site and subsequent catalysis. The structures with 2′,3′-ddCTP show the open conformation with the NTP sitting above the active site and those with 2′ and 3′-dCTP show a clear tendency for the remaining hydroxyl to be pulled down toward the palm domain, but only when both hydroxyls are present do we efficiently trap the catalytically competent closed conformation (Fig. 3C and Fig. S6C). This is consistent with the biochemical observation that 3Dpol does incorporate 2′- and 3′-deoxy NTPs (22), albeit at much lower efficiency than normal nucleotides due to a greater effect on kcat that reflects active site closure than on Km that reflects NTP binding. This leads us to suggest that nucleotide selection in RdRPs is a fairly simple process in which base-pairing interactions control the initial NTP binding geometry and the resulting positioning of the ribose hydroxyls becomes the major fidelity checkpoint for proper NTP selection. An incorrect nucleotide may bind, but its ribose hydroxyls will not be optimally poised for the rapid closure of the active site, thereby reducing incorporation efficiency.
As a result of the typically low fidelity of RdRPs, positive-strand RNA viruses are often described as quasispecies where individual genomes may contain one or two mutations compared to their parental virus (23). High mutation rates coupled with the ability of individual genomes to rapidly become dominant in a virus population allows viral genomes to rapidly evolve and thrive in different host cell environments, ensuring the efficient propagation of the virus by improving its fitness for survival (24). Because nucleotide selection and active site closure in the 3Dpol EC is a single step, one would expect that mutations altering the energetics and dynamics of the transition to the closed conformation could easily affect both elongation rate and nucleotide selection fidelity. This has indeed been observed in the form of a drug-resistant poliovirus wherein a Gly64 to serine mutation in 3Dpol conferred resistance to the antiviral nucleoside analog ribavirin by both increasing fidelity (25) and reducing elongation rate (26). Gly64 interacts with the buried N-terminus of 3Dpol that serves to position Asp238A9 in the active site (19), providing a direct connection between Gly64 and NTP-driven active site closure (Fig. S5).
Comparison with Other Polymerases.
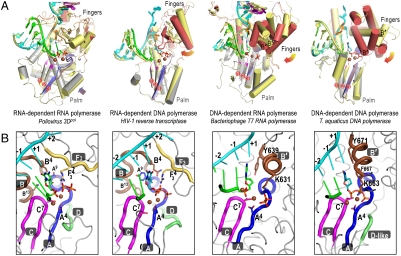
The mechanism for closing the 3Dpol RdRP active site via structural changes in the palm domain is fundamentally different from those of other polymerase classes. Processive DNA polymerases, single-subunit DNA-dependent RNA polymerases, and reverse transcriptases utilize large fingers domain movements to close their active sites by repositioning the nascent base pair from a preinsertion site into the catalytic site. The fingers domains of these polymerases are typically rich in α-helices to allow for structural fluidity as the domain swings down toward a preformed palm domain (Fig. 4A). In contrast, 3Dpol has a β-sheet rich fingers domain that remains essentially fixed during active site closure and instead the enzyme uses a conformational change in the palm domain to structure the active site for catalysis only upon correct NTP binding. The key element of 3Dpol active site closure is a subtle shift of motif A to position the essential Asp233A4 for metal binding and subsequent catalysis. In the open conformation, the 3-stranded β-sheet of the palm domain (motifs A and C) is only partially formed, but in the closed conformation it is structurally identical to the palm domains found in the other polymerase classes (Figs. 4A and Fig. S6B), allowing catalysis to take place by the common two-metal mechanism.
Fig. 4.
(A) A new mode of active site closure in positive-strand RNA virus polymerases. The viral RdRPs close their active site using a rearrangement of the palm domain that results in the complete formation of the 3-stranded palm domain β-sheet common to all polymerases. In contrast, representative structures from the other three classes of polymerases show major swinging motions of their fingers domains between the open (fingers yellow, motif A brown) and closed (fingers red, motif A blue) conformations that serve to reposition the +1 templating base (orange) and incoming nucleotide for catalysis. For clarity, only the closed conformation NTP, 6-bp of upstream duplex, and part of the protein structures are shown and spheres correspond to catalytic metals and the motif A AspA4 Cα atom. The motif B’ helix that plays a major role in translocation by the DNA-templated polymerases is labeled at its N-terminal end. (B) The common organization of the closed conformation active sites across all four classes of polymerases that is built on the 3-stranded antiparallel β-sheet of motifs A and C in the palm domain. In comparing the RNA- and DNA-templated polymerases, note the structural divergence of the B and B’ motifs and how the F3 and B’ motifs provides analogous charge interactions with the bound nucleotide. Conserved motifs A–D are indicated and key residues are labeled according to the alignment shown in Fig. 3E. See Figs. S6 and S8 for additional details.
The structural mechanism we have described for NTP recognition and catalysis in poliovirus 3Dpol appears to be a universal feature of positive-strand RNA virus polymerases. The partial β-strand conformation for motif A in the palm is a common feature of apo RdRP structures (3) and all structures solved thus far show conservation of the residues that are important for NTP ribose recognition and active site closure; i.e., Asp233A4, Asp238A9, Ser288B4, Gly289B5, Asn297B13, Asp328C7. This includes rhino, coxsackie, Norwalk, dengue fever, West Nile, hepatitis C, and bovine viral diarrhea viruses representing a diverse array of positive-strand RNA viruses (Fig. 3D). In contrast, RdRPs from double-stranded RNA viruses contain both a fully structured palm domain and a β-sheet rich fingers domain and appear to close their active sites by more subtle changes in sidechain conformations and backbone orientation (27, 28).
Solving the closed conformation structure of the poliovirus EC also enables us to directly compare the closed conformations of all four classes of single-subunit polymerases (Fig. 4B). The active site geometry is always built on a 3-stranded β-sheet scaffold in the palm that is surrounded by functionally and structurally homologous residues from the various conserved polymerase sequence motifs. RNA-templated polymerases establish ribose selectivity using the cluster of palm domain residues A9, B4, and B13; RdRPs utilize polar residues that form hydrogen bonds with each other and with the 2′-hydroxyl (22), whereas RdDPs use nonpolar amino acids to create a hydrophobic site for 2′-deoxy ribose binding (29–32). The DNA-templated polymerases, on the other hand, establish ribose selection from above the active site via fingers domain residues from their helical B′ motif (often termed “O-helix”); DdRPs utilize a tyrosine hydroxyl at position B′9 for rNTP selection (10, 33) whereas DdDPs use an aromatic ring at position B′5 for dNTP selection (34, 35). All the polymerases also contain a fingers domain lysine or arginine residue positioned directly above the α–β phosphate linkage. In DNA-templated polymerases this is a conserved lysine at B′1 and in RNA-dependent polymerases it is a conserved arginine at 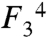 within the RdRP ring finger. Poliovirus 3Dpol Arg174F34 is structurally equivalent to Arg73 of HIV-1 RT (31), Arg194 of Tribolium castaneum telomerase (32), and His1085 from the yeast RNA polymerase II trigger loop that has been implicated as a proton donor for the pyrophosphate leaving group (36).
within the RdRP ring finger. Poliovirus 3Dpol Arg174F34 is structurally equivalent to Arg73 of HIV-1 RT (31), Arg194 of Tribolium castaneum telomerase (32), and His1085 from the yeast RNA polymerase II trigger loop that has been implicated as a proton donor for the pyrophosphate leaving group (36).
Finally, the postcatalysis state 5 structures show a clear reset of the active site to the open conformation without translocation of the nucleic acid, and in light of comparisons with other polymerase structures this suggests that translocation in RNA-templated polymerases may occur via a different mechanism. The lack of translocation itself in the poliovirus ECs is most certainly due to crystal packing constraints exacerbated by the coaxial stacking of the upstream duplexes from two complexes. However, the fact that the active site can open back up without translocating and/or distorting the RNA structure suggests that translocation is a distinct step that occurs after opening of the active site. This would be fundamentally different from what is observed in DNA-templated single-subunit polymerases where these two events appear to be tightly coupled to the swinging motions of the fingers domain. The structures of Taq DNA (35) and T7 RNA (10, 11) polymerases have shown that the origin of this coupling is a tyrosine residue at the C-terminal end of the fingers domain B′ helix that is placed immediately downstream of the nascent base pair upon active site closure. When the polymerase returns to the open conformation, the rotation of this helix (Fig. 4A) pushes the tyrosine toward the nascent base pair, leading to translocation. Viral RdRPs, however, do not have a B′ helix and instead contain conserved B and F3 motifs that diverge from the B′ motif in both sequence and structure (Fig. 4B). It is therefore quite possible that they could use a different mechanism to drive translocation via a new state 6 whose structure we do not yet know. Furthermore, the B and F3 motifs are conserved among RdRPs, reverse transcriptases, and telomerases, implying that RNA-templated polymerases in general may share this mechanism for translocation.
In summary, the crystallization of prepurified active poliovirus 3Dpol elongation complexes has revealed the molecular details and conformational changes associate with NTP recognition, active site closure, and catalysis in RdRPs from positive-strand RNA viruses. These enzymes feature a fully prepositioned templating nucleotide and close their active sites via changes in the palm domain, utilizing a method that is fundamentally different from that of other polymerases. The structures increase our understanding of RdRP mechanism and will open new avenues for the development of antiviral pharmaceuticals targeting the unique open state of the RdRP active site.
Materials and Methods
Please see SI Text for detailed methods. Briefly, 3Dpol L446D (19) was expressed in Escherichia coli BL21 (DE3) pCG1 cells using a ubiquitin fusion construct system where in vivo cleavage results in full length protein with a native Gly1 residue at the N-terminus (37). The RNA template strand was produced by in vitro T7 transcription and annealed to a chemically synthesized primer strand. Elongation complexes were generated by incubating 3Dpol with annealed primer-template RNA, GTP, and ATP. The resulting stalled complexes were purified by anion exchange chromatography and concentrated.
Crystals grew in 2–4 weeks by sitting drop vapor diffusion at 10 °C using 8–9 mg/mL EC with precipitant solution containing 95 mM trisodium citrate, pH 5.6–5.9, 6.5–7.0% v/v isopropanol, 5% v/v glycerol, 16.5–18.0% (w/v) PEG 4000. Crystals were gradually buffer exchanged into a cryostabilizer solution prior to flash freezing in liquid nitrogen or soaked for 15–20 h at 16 °C in the cryostablizer with 10 mM MgCl2 and 5 mM CTP, 2′-dCTP, 3′-dCTP, or 2′,3′-ddCTP.
Diffraction data were collected at beamline 4.2.2 (Advanced Light Source) and integrated, merged, and scaled using d*Trek v9.9 with resulting statistics being listed in Table S1. The initial structure solution was obtained by molecular replacement with the apo-3Dpol structure (1RA6) and refined used Coot and PHENIX. Composite simulated-annealing omit maps were calculated by CNS and figures were generated with PyMOL.
Supplementary Material
Acknowledgments.
We thank P. S. Ho and R. W. Woody for helpful discussions and Jay Nix for a steady stream of X-rays. This work was supported by National Institutes of Health Grant AI-059130 (O.B.P.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3OL6, 3OL7, 3OL8, 3OL9, 3OLA, and 3OLB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007626107/-/DCSupplemental.
References
- 1.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Lescar J, Canard B. RNA-dependent RNA polymerases from flaviviruses and Picornaviridae. Curr Opin Struct Biol. 2009;19:759–767. doi: 10.1016/j.sbi.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svetlov V, Nudler E. Macromolecular micromovements: How RNA polymerase translocates. Curr Opin Struct Biol. 2009;19:701–707. doi: 10.1016/j.sbi.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramer P. Common structural features of nucleic acid polymerases. Bioessays. 2002;24:724–729. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- 10.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 11.Temiakov D, et al. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 12.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 14.Arias A, et al. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J Virol. 2008;82:12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrer-Orta C, et al. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 16.Ferrer-Orta C, et al. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci USA. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamyatkin DF, et al. Structural insights into mechanisms of catalysis and inhibition in norwalk virus polymerase. J Biol Chem. 2008;283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 18.Zamyatkin DF, Parra F, Machin A, Grochulski P, Ng KK. Binding of 2′-amino-2′-deoxycytidine-5′-triphosphate to norovirus polymerase induces rearrangement of the active site. J Mol Biol. 2009;390:10–16. doi: 10.1016/j.jmb.2009.04.069. [DOI] [PubMed] [Google Scholar]
- 19.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by NTP binding and fingers–thumb interactions. J Mol Biol. 2007;366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theobald DL, Wuttke DS. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gohara DW, Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): Kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry. 2004;43:5149–5158. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domingo E, et al. Viruses as quasispecies: Biological implications. Curr Top Microbiol Immunol. 2006;299:51–82. doi: 10.1007/3-540-26397-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 28.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage—Structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 29.Sarafianos SG, Pandey VN, Kaushik N, Modak MJ. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 30.Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:19759–19767. doi: 10.1074/jbc.M910361199. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 32.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 33.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gohara DW, et al. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expres Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.