Abstract
In the budding yeast Saccharomyces cerevisiae, self-recognition and the thereby promoted aggregation of thousands of cells into protective flocs is mediated by a family of cell-surface adhesins, the flocculins (Flo). Based on this social behavior FLO genes fulfill the definition of “greenbeard” genes, which direct cooperation toward other carriers of the same gene. The process of flocculation plays an eminent role in the food industry for the production of beer and wine. However, the precise mode of flocculin-mediated surface recognition and the exact structure of cognate ligands have remained elusive. Here, we present structures of the adhesion domain of a flocculin complexed to its cognate ligands derived from yeast high-mannose oligosaccharides at resolutions up to 0.95 Å. Besides a PA14-like architecture, the Flo5A domain reveals a previously undescribed lectin fold that utilizes a unique DcisD calcium-binding motif for carbohydrate binding and that is widely spread among pro- and eukaryotes. Given the high abundance of high-mannose oligosaccharides in yeast cell walls, the Flo5A structure suggests a model for recognition, where social non-self- instead of unsocial self-interactions are favored.
Keywords: altruism, molecular recognition, fungal development, atomic resolution
Self-recognition is key to both microbial growth and tissue formation. The molecular basis of this phenomenon is well understood only for a few cases—e.g., during neural differentiation and immune recognition. In the microbial world, the formation of multicellular structures by self-recognition is mediated by specific, surface-exposed adhesins and provides protection and promotes long-term survival, substrate exploration, or host invasion (1, 2). In the budding yeast Saccharomyces cerevisiae, aggregation of vegetative cells is a paradigm for biofilm formation (3, 4) and the evolution of social behavior (5, 6). Self-recognition in this organism is mediated by flocculin proteins, which belong to a family of fungal adhesins that are present in pathogenic yeasts as well (7). The general architecture of flocculins includes an N-terminal A domain corresponding to a lectin-like adhesin domain, a stalk-like, repetitive and highly glycosylated B domain, and a C domain that carries a GPI anchoring site (8). The genome of S. cerevisiae contains at least four functional FLO genes (FLO1, FLO5, FLO9, and FLO10) whose N-terminal A domains are predicted to belong to the PA14-like protein family (9) first described in the anthrax-protective antigen (10). This protein family has been found to be widely distributed among all domains of life including human galactosyltransferases and fibrocystin (9, 11).
Flocculins confer dominant, calcium-dependent cell–cell adhesion that can be inhibited by hexoses like mannose (12, 13). This process is known as flocculation and has been exploited for centuries by the brewing industry (14). Flocculin genes also mediate social behavior of S. cerevisiae and have here been found to fulfill the definition of “greenbeard” genes (5, 6). Expression of flocculins causes cell aggregation, formation of multicellular flocs, and thereby protection against environmental stresses, while nonexpressing “cheater” cells are recruited preferentially at the nonprotective floc periphery (6). The nepotistic role exerted by such genes for social behavior obviously depends on therein encoded mechanisms for cell assortment, by which greenbeard gene carriers experience improved fitness compared to noncarriers. Such a greenbeard function, linking self-recognition with cooperative behavior, was postulated to be primarily implemented by cell-adhesion molecules (15).
Despite its significance in industry and theoretical biology, the precise mode of flocculin-mediated yeast surface recognition and the exact structure of the cognate ligands have remained elusive. The yeast cell wall has a complex architecture (16) exposing flocculins and O- and N-linked high-mannose oligosaccharides at the surface (17). Thus, an important question is how the A domains of flocculins preferentially interact with carbohydrate ligands in trans without being compromised by cis-interactions, which would prevent social behavior. Furthermore, the crucial role of calcium during flocculation has been speculated to depend either on the formation of rod-like superstructures of their heavily O-glycosylated B region (18) or a Ca2+-bridging mode for carbohydrate recognition as exemplified by C-type lectins (12). In this study, we have analyzed the A domain of the flocculin Flo5 (Flo5A). FLO5 plays a major role in industrial yeast flocculation (19) and has identical sequences in laboratory strains of the S288c and Σ1278b genetic background, which are both commonly used in adhesion studies (20). Furthermore, the Flo5A domain shares high similarity with the A domains of other flocculins including Flo1 (94% sequence identity on protein level), Flo9 (89%), and Flo10 (64%). We subjected Flo5A to a combined biochemical, structural and phenotypic analysis to get a concise view of the mechanisms that underlie A-domain-specific self-recognition in S. cerevisiae.
Results and Discussion
The Flo5A Domain Is Required and Sufficient for Flocculation.
Sequence comparisons and secondary structure predictions of Flo5 and the related flocculins Flo1, Flo9, and Flo10 indicate that the Flo5A domain covers residues S23 to H271 and is essentially preserved in the other S. cerevisiae flocculins. To test whether the Flo5A domain is responsible for conferring calcium-dependent flocculation, several constructs (Fig. 1A and Table S1) were tested for functionality by expression in a nonflocculating yeast strain. The Flo5A domain was fused either to the BC region of Flo5 as in the natural protein or to the corresponding region of Flo11, an adhesin that comprises an unrelated type of A domain and mediates biofilm formation instead of flocculation (3, 4). To ensure comparable amounts of Flo5A-carrying flocculins on the cell surface, expression was driven by the FLO11 promoter (21) and monitored by fluorescence microscopy using Flo5A-specific antibodies (Fig. 1B and Fig. S1C). Subsequent flocculation assays demonstrated that the Flo5A domain was required and sufficient to cause calcium-dependent and mannose-inhibitable floc formation (Fig. 1C). Flocculation efficiency was similar with the BC regions of either Flo5 or Flo11. Unlike the Flo5A domain, the Flo11A domain conferred only marginal, mannose-insensitive flocculation but instead mediated adhesion to agar and polystyrene surfaces (Fig. S1 A and B).
Fig. 1.
The Flo5A domain is a surface-exposed Ca2+- and mannose-binding protein that mediates flocculation. (A) Diagram showing FLO5 and FLO11 constructs driven from the FLO11 promoter and expressed in a nonflocculating Σ1278b yeast strain in which FLO1, 5, 9, and 10 genes are transcriptionally silent and the FLO11 gene has been deleted. (B) Presence of the Flo5A domain at the surface of yeast strains described in A was visualized by differential interference contrast (DIC) and immunofluorescence microscopy using specific anti-Flo5A antibodies. Bars represent 10 μm. (C) Ca2+ dependence and mannose inhibition of FLO5A-mediated flocculation was determined by using constructs described in A and shows that the Flo5A domain is essential and sufficient for flocculation. (D) Overall structure of the Flo5A domain complexed to calcium (gold) and mannose (orange) at 0.95-Å resolution showing the primary and secondary ligand binding sites, the PA14 domain and the Flo5 subdomain (PDB ID code 2XJP).
Structure of the Flo5A Domain.
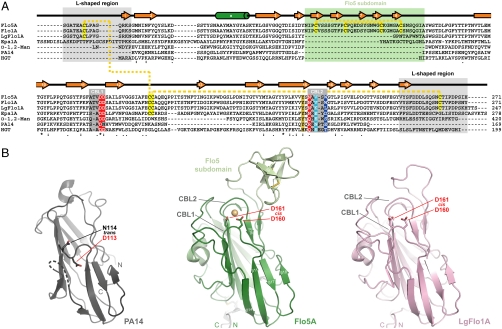
The crystal structure of the recombinant Flo5A domain was solved by SIRAS-phasing using a gadolinium derivative and refined either alone or in complexes with different sugars at atomic resolutions of up to 0.95 Å (Tables S2 and S3, PDB ID code: 2XJP). The overall structure of Flo5A shows a bipartite organization comprising a large β-sandwich domain that carries an insertion, the Flo5 subdomain, between β-strands 4 and 10 (Fig. 1D and Fig. S2A). The core of the β-sandwich domain is topologically related to the PA14 domain (9) that is part of the N-terminal propeptide fragment of the anthrax-protective antigen (10). This domain type is widely spread in bacteria and eukaryotes, where it has been hypothesized in functions related to carbohydrate binding. The superposition of both structures shows an rmsd of 1.4 Å that is restricted only to the central 10-stranded β-sandwich itself (78 Cα-positions) and is hence decorated by two additional regions (Fig. 2A). The inserted Flo5 subdomain (N84-A120) consists of five short β-strands that are stabilized by two disulfide bridges. A second region unique for Flo5A is mainly formed by its N and C termini. Together they extend as an L-shaped stretch from the core domain (Fig. 2 and Fig. S2C) and are fixed to it by disulfide bridges to two consecutive cysteines within the β12–β13 loop. This loop (G168-T189) is considerably longer than in the PA14 domain, and together with a second loop between β-strands 2 and 3 (Y46-T68) it seals the surface of the underlying β-sheet from solvent access.
Fig. 2.
The PA14/Flo5-type protein family. (A) Multiple sequence alignment of representatives of the PA14/Flo5 family (Uniprot entries: Flo5, P38894; Flo1, P32768; LgFlo1, B3IUA8; Epa1 adhesin from C. glabrata, Q6VBJ0; a bacterial α-1,2-mannosidase, Q8A3K6; PA-14, P13423; human galactosaminyl transferase HGT, Q8N9V0). Important elements as the DcisD motif are highlighted (color gradient from red to blue corresponds to transition from conserved to variable). Disulfide bridges as present in the flocculins as well as predicted in Epa1 are marked in yellow. (B) Structural comparison between the PA14 domain (gray, 1ACC) of the protective antigen from Bacillus anthracis (10), the Flo5A domain (green), and the LgFlo1A domain (pink). The superposition of the Flo5A domain onto the PA14 domain gives an rmsd of 1.37 Å for 78 Cα-positions. Homology model of LgFlo1A was created using the Flo5A domain as template and MODELLER 9v7 (30). Color codes are equivalent to A. Calcium is represented by a golden sphere.
Calcium Is Directly Involved in Carbohydrate Binding.
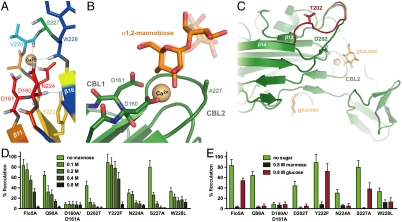
The role of calcium during flocculation has been discussed controversially. It has been speculated that calcium might enable the formation of rod-like superstructures by interaction with the glycosylated B regions (18) or provide a bridging mode for carbohydrate ligand recognition. Our biochemical and structural analyses of Flo5A revealed calcium-dependent binding specific for mannosides. The monosaccharide mannose that acts in vivo as an antiflocculant at concentrations greater than 0.2 M (Fig. 4D) exhibits in vitro weak binding to recombinant Flo5A as indicated by its rather high KD of 29 mM (Fig. 3E). Other hexoses like glucose or galactose failed to bind in fluorescence-titration experiments of Flo5A. The crystal structure of the mannose/Ca2+ complex of Flo5A shows a C-type lectin mode of carbohydrate binding via Ca2+-mediated recognition of the 2′- and 3′-hydroxyl groups, where the calcium ion is bound between sugar and protein in a distorted pentagonal bipyramidal coordination. The preference for mannosides instead of glucose and related sugars originates from a hydrogen bond between the axial 2′-hydroxyl group and the side-chain of Q98 (Fig. 3 B and C). Interestingly, Flo5A crystals soaked with 2 M glucose showed partial occupation of this sugar in the primary carbohydrate-binding site with concomitant displacement of Q98. Compared to mannose, glucose at high concentrations only weakly inhibits Flo5A-mediated flocculation in vivo (Fig. 4E).
Fig. 4.
Characterization of the Flo5A binding site and specificity. (A) Sequence conservation of the DcisD motif and other residues responsible for calcium-binding. The color gradient from red to blue corresponds to transition from conserved to variable and is based on a conservation analysis by the CONSURF server (31) using available sequences of PA14-related domains. (B) Structure of α1,2-mannobiose recognition by the S227A variant (2XJU). In this variant with increased affinity the ligand (orange) adopts a different binding mode than in the wild-type Flo5A domain (semitransparent). (C) Structure of the complex of the NewFlo-type variant D202T and glucose at 1.6-Å resolution showing that the D202T mutation exerts only an indirect effect on carbohydrate binding as it increases the distance of the variable β13–β14 loop from 15 to 19 Å (shown in red, 2XJV). (D) Flocculation efficiency and mannose inhibition of yeast strains expressing different FLO5A variants (Table S1). The presence of comparable amounts of Flo5A protein at the cell surface was confirmed by fluorescence microscopy. (E) Hexose-binding specificity of different Flo5A variants determined by inhibition assays for flocculation.
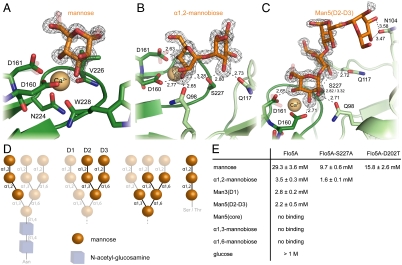
Fig. 3.
Crystallographic analysis of carbohydrate binding by the Flo5A domain. (A) The Flo5A•mannose complex shows alternative conformations of the mannose for its anomeric and 6-hydroxyl groups (semitransparent sticks, 2XJP). (B) The Flo5A•α1,2-mannobiose complex (1.30-Å resolution, 2XJS) shows a recognition of the second mannose moiety by Q117 and S227. (C) The 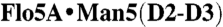 complex (1.35-Å resolution, 2XJR) harbors the mannopentaose mimicking the D2 and D3 arms in the binding site in such an orientation that only the D2-arm is recognized by the Flo5A domain. Distal mannoside moieties like the third and fourth have lowered occupancies (70% and 50%, respectively) due to increasing disorder. Alternative conformations are represented by semitransparent sticks, distances are given in Ångstrom. (D) Schematic diagram showing structures of the core N- and O-linked oligomannose modifications of budding yeast (24). Mannosidic substructures used for the characterization of the Flo5A domain are highlighted. (E) Table showing in vitro affinities of different carbohydrates toward the Flo5A domain and variants.
complex (1.35-Å resolution, 2XJR) harbors the mannopentaose mimicking the D2 and D3 arms in the binding site in such an orientation that only the D2-arm is recognized by the Flo5A domain. Distal mannoside moieties like the third and fourth have lowered occupancies (70% and 50%, respectively) due to increasing disorder. Alternative conformations are represented by semitransparent sticks, distances are given in Ångstrom. (D) Schematic diagram showing structures of the core N- and O-linked oligomannose modifications of budding yeast (24). Mannosidic substructures used for the characterization of the Flo5A domain are highlighted. (E) Table showing in vitro affinities of different carbohydrates toward the Flo5A domain and variants.
Flo5A Shows a Unique Calcium-Binding Site.
The calcium-binding site of Flo5A is a unique and characteristic hallmark of the PA14/Flo5-like protein family. The calcium ligands belong to two carbohydrate-binding loops, CBL1 and CBL2. CBL1 links the strands β11 and β12 and harbors a highly conserved Asp-Asp motif (D160, D161). CBL2 connecting β15 with β16 contributes to calcium-binding via the side chain of N224 and two carbonyl groups of its main chain, explaining the low conservation of V226 and W228 (Fig. 4A). CBL1 is unusual by bearing a rare cis-peptide between two nonproline residues, D160 and D161. This motif, in the following called DcisD, has so far only been found in the nucleotide-binding site of the ATP synthase (22) as well as in a Zn2+-dependent aminoprotease (23). In contrast to Flo5A, only the first of the aspartates is directly involved in metal binding in the active sites of these enzymes. In the Flo5A structure the unfavorable configuration of the cis peptide is stabilized by a hydrogen bond to the hydroxyl group of the likewise conserved Y222 (Fig. 4A and Fig. S3C). The DcisD motif is present throughout the PA14/Flo5-like family but is missing in the PA14 domain itself (Fig. 2B). In the latter, for which no carbohydrate-binding activity has been demonstrated, the loop corresponding to CBL1 shows major conformational differences including the replacement of one of the two crucial aspartates. Importantly, the Flo5A structure shows that the cis configuration allows a maximal spatial proximity of about 2.0–2.3 Å for the carboxylates of D160-D161 (Fig. S3). As such, the DcisD motif represents a unique structural type for calcium binding. A second consequence of the DcisD motif is the concomitant H-bonding between its carboxylates and the 3′- and 4′-hydroxyl groups of mannose ligands. The crucial role of the unique DcisD motif in the PA14/Flo5-like family of calcium- and carbohydrate-dependent adhesins is reflected by a complete lack of flocculation when this motif is mutated as exemplified by the Flo5AD160A,D161A variant (Fig. 4D). Similarly, flocculation was significantly reduced when another calcium ligand, N224, was exchanged by alanine.
Terminal Arms of Yeast High-Mannose Oligosaccharides Act as Cognate Ligands.
The yeast cell wall has a complex architecture (16) and contains a large proportion of mannoproteins with either N- or O-linked oligosaccharides (17, 24). It has been speculated that these carbohydrates are involved in flocculation, but the exact structure of the ligands that are recognized by flocculins is not known. The open nature of the Flo5A carbohydrate binding site suggests the recognition of more complex mannosides. To identify the substructures of the known yeast core oligosaccharides, fluorescence-titration and crystal soaking experiments were performed with appropriate synthetic oligomannosides (Fig. S4). These experiments revealed that only α1,2-linked mannobiosides, but neither α1,3- nor α1,6-linked mannoses, are bound (Fig. 3 B, C, and E). Moreover, α1,2-linkage to a second mannose increased the affinity ninefold (KD = 3.5 mM) as compared to mannose (Fig. 3E). The recognition of the second mannose moiety is achieved by hydrogen bonds between its 3′-hydroxyl and S227 from CBL2 and, in addition, Q117 residing in the Flo5 subdomain. Mutation of S227 to alanine was found to increase the affinity toward mannose without affecting specificity by inducing a slightly altered conformation of the disaccharide within the binding site (Fig. 4B). The side chain of V226 is sandwiched between both sugar rings. Interestingly, mannose-α1,2-mannose-α1,2-mannose, a model for the D1 arm of N-linked or, alternatively, for O-linked core oligosaccharides of yeasts, shows the same affinity and binding mode as α1,2-mannobiose (Fig. 3E).
Synthetic mimics of mannopentaose fragments of the N-linked core oligosaccharide representing either the inner core or the outer core spanning the terminal D2 and D3 arms were tested for Flo5A binding (Fig. 3 D and E). Here, the inner core comprising only α1,3- and α1,6-linked mannose residues was not recognized. In contrast, the outer core mimic with its D2/D3 arms was bound with similar affinity as the α1,2-mannobiose. Both arms carry α1,2-linked mannobiose ends, but in the crystal structure only the D2 arm occupies Flo5A’s carbohydrate-binding site (Fig. 3C) with increased disorder observed for the third and fourth mannose moiety. Apparently, the Flo5 subdomain supports discrimination between the branches of the core oligosaccharide by steric hindrance or contributing a hydrogen bond between the fourth mannoside and N104.
A secondary carbohydrate-binding site was observed at the back of the Flo5A domain when crystals were soaked in molar concentrations of mannose or glucose (Fig. 1D and Fig. S2B). In this site, hexoses were recognized in a nonspecific and calcium-independent manner by making mostly hydrogen bonds with main chain atoms derived from the β3–β4 loop (Fig. S2B).
Structure-Derived Mutants Reveal NewFlo-Type Properties.
In the brewing industry, broadened sensitivity of flocculation to other sugars like glucose commonly depends on flocculins of the so called NewFlo type (13)—e.g., for the production of lager or pilsener (14). Previous work suggested that NewFlo-type variants like D202T and W228L are directly involved in carbohydrate binding (25). Interestingly, these variants as well as the N224A mutant in the calcium-binding site exhibit a loss of carbohydrate-binding specificity and flocculation efficiency (Fig. 4E and Fig. S5 B and C). However, these residues are not directly involved in carbohydrate recognition, because they are either distantly located from the primary binding site as D202 or only contribute to calcium-binding via their main chain carbonyl group like W228 (Fig. 4A). Instead, the D202T mutation changes the preferred conformation of the variable β13–β14 loop thereby indirectly relaxing the stringency of hexose binding, probably via alterations of the hydration network of the protein (Fig. 4C). The subsequent residue is one of four potential N-linked glycosylation sites present in the Flo5A domain (N135, N187, N203, N262) opening the possibility that glycosylation may also affect the sugar specificity of the primary binding site. Additionally, the D202T mutant with its slight increase of affinity by a factor of two improves the ability of free mannose to inhibit flocculation (Fig. 3E and Fig. S5B). Moreover, a variant of Flo5, LgFlo1 (19, 25), which is commonly found in bottom-fermenting yeast strains and promotes NewFlo-type flocculation, lacks the complete Flo5A-subdomain (Fig. 2B). This corroborates the finding that also structural alterations in greater distance to the primary binding site are capable to induce relaxed carbohydrate recognition, probably due to alterations in the hydration network. However, as already mentioned, residue Q98, which also resides in the Flo5 subdomain, is directly involved in carbohydrate binding specificity. Therefore, the observations might result from both direct and indirect effects.
The Flo5A structure allowed the engineering of NewFlo-type behavior without a loss of flocculation efficiency as observed for indirect NewFlo mutants such as W228L. For example, for the Q98A variant a loss of interaction with the axial 2-hydroxyl group of mannose can be predicted to lessen specificity. Indeed, the Q98A variant shows wild-type-like flocculation efficiency but completely lacks discrimination between mannose and glucose (Fig. 4E and Fig. S5).
Flo5A Confers Social Behavior by Heterophilic Interactions.
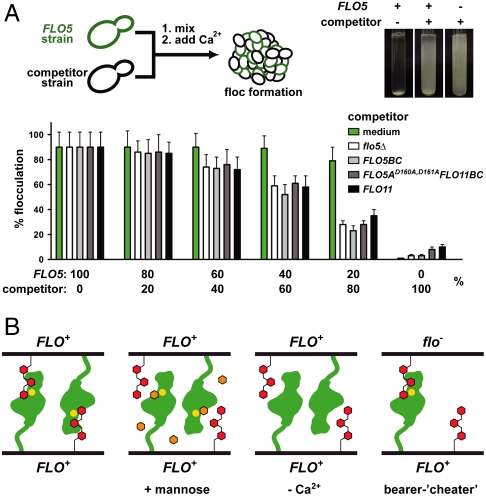
It has been suggested that flocculin-mediated self-recognition represents a heterotypic mode of social interaction (6). Our data on Flo5A support this view, because Flo5A binds to oligosaccharide structures that are ubiquitously present on yeast cell surfaces and are presented there by other, nonflocculin mannoproteins as well. Flocculation competition assays provide more direct evidence for heterotypic interactions because they show that different flocculation-deficient yeast strains efficiently compete with the flocculation of FLO5 bearing strains (Fig. 5A). Here, the flocculin-mediated heterophilic mode of self-recognition differs from homophilic interactions provided by other adhesins—e.g., the csA gene product of the social amoeba Dictyostelium discoideum (26) or the neural cell-adhesion molecule (NCAM) in higher eukaryotes (27). In addition to previous work (12), our data show that deletion of the Flo5A domain or disruption of the calcium-binding DcisD motif confers the same phenotype as deletion of the entire FLO5 gene.
Fig. 5.
Flo5A-mediated social behavior of budding yeast. (A) Inhibition of floc formation by different competitors determined by population mixing experiments using a flocculating FLO5 strain and different strains expressing either no flocculin (flo5∆), the FLO5BC domains, the nonfunctional FLO5AD160A,D161A variant, FLO11, or medium as a control. The inlay shows a typical flocculation-competition experiment using the competitor strain with the FLO5AD160A,D161A variant. (B) Model of cooperative cell–cell interactions underlying the greenbeard function as exemplified by FLO in budding yeast. Heterophilic Flo-type interactions are mediated by FloA domains (green) and terminal α1,2-mannobiose residues (red) present in trans at the surface of FLO+ bearer as well as in flo- cheater cells. Here, calcium-dependent interactions are inhibited by free mannose (orange hexagons; Ca2+: yellow dots).
Conclusions
Our data provide previously undescribed biochemical and structural characterization of a functional PA14/Flo5A-like domain. We showed that calcium is directly involved in carbohydrate binding and not only in an indirect manner, stabilizing the stalk-like B domain. Furthermore, its calcium-binding site, including the unique DcisD motif, is widely spread in all domains of life. Multiple sequence alignment based on PFAM 24.0 showed that 45% of the known PA14 domains, including human proteins like galactosyltransferases and fibrocystin, contain a double D at the equivalent position (11).
Moreover, our study offers a detailed view on an adhesion domain derived from a greenbeard gene because such a function has been recently assigned to flocculin genes of the Flo5 family (6). As a greenbeard, the expression of a Flo5 family member causes not only simple aggregation to yeast flocs and hence protection against environmental stresses but also recruitment of non-FLO-expressing cheater cells at the nonprotective floc periphery (6). Whereas homophilic interactions as exerted by Dictyostelium CsA are beneficial for segregating the bearers of the adhesion gene (26), the observed heterophilic interaction between Flo5-like flocculins and the α1,2-linked mannosides present at the outer yeast cell wall (Fig. 5B) provides an additional function by utilizing cheater cells for protection (6). In this context, the relatively low affinity of 3.5 mM for the recognition of α1,2-linked mannosides might be crucial for the greenbeard function of Flo5 family members, because it allows flocculin-expressing survivors to return from being trapped in flocs to the planktonic lifestyle.
On the yeast cell surface, the number of oligosaccharide ligands largely exceeds that of flocculin molecules. Therefore, mechanisms are required to ensure that cis-interactions of the flocculins’ A domains with the α1,2-mannoside ligands are not ablating the binding potential provided for trans-ligands. Homophilic cell-adhesion molecules of higher eukaryotes like NCAM resolve this issue by providing a second, specific binding mode with other adhesion molecules in cis, while retaining at the same time their trans-binding capability (27). The secondary carbohydrate-binding site of the Flo5A domain may fulfill the analogous function by preferentially immobilizing the Flo5A domain at the yeast surface. Covalent fixation of the N and C termini of the Flo5A domain via disulfide bridges to the protein’s core domain could further inhibit free rotation relative to the adjacent BC region. However, the BC region may further contribute to cell–cell adhesion. Length variation of repetitive tandem repeats within the B region was reported to alter flocculation and biofilm formation of yeast (28). Furthermore, amyloid-forming sequences within the B regions are postulated to confer clustering of A domains on yeast cell surfaces and thereby improve intercellular binding (29). Accordingly, the combination of secondary carbohydrate binding and stiffened domain presentation could be a further clue that enables Flo5-like adhesion molecules to preferentially recognize and bind to ligands in trans (Fig. S6) while being immersed in a pool of cis-ligands.
Methods
Protein Production and Purification.
Recombinant Flo5A was generated in a thioredoxin- and glutathione-reductase deficient Escherichia coli strain using a low-temperature protocol for slow expression and maturation. Flo5A was purified using Ni-affinity and subsequent size exclusion chromatography.
Crystallization and Structure Determination.
Crystals of Flo5A belonging to space group P212121 diffracted to atomic resolution. Accordingly, the structure of Flo5A was solved at 0.95-Å resolution by single anomalous dispersion with isomorphous replacement (SIRAS) using a single gadolinium derivative. Ligand soakings of preobtained crystals were performed using different hexoses, mannobioses, and three oligomannosides. Data collection statistics and refinement statistics are given in Tables S2 and S3, respectively.
Flocculation Assays.
Flocculation assays were performed using a nonflocculent and nonadhering yeast strain carrying appropriate FLO5 and FLO11 gene constructs. Methods for agar and plastic adhesion tests are detailed in SI Text. Polyclonal rabbit antibodies were generated using purified Flo5A and a standard immunization procedure.
Microscopy.
Fluorescence microscopy was carried out to verify comparable amounts of Flo5A carrying proteins on cell surfaces using Flo5A specific antibodies via (i) differential interference microscopy (DIC) and (ii) fluorescence microscopy using a GFP filter set.
In Vitro Assays.
Fluorescence titration was performed to determine binding constants of Flo5A by exciting the intrinsic tryptophane fluorescence at a wavelength of 295 nm and recording the fluorescence quench during titration with different saccharides.
Extended Information.
Yeast strain and plasmid construction, synthesis of oligomannosides, and detailed information on other methods used in this study are described in SI Text.
Supplementary Material
Acknowledgments.
The authors thank Edward Mitchell and Tobias Klar for support at European Synchrotron Radiation Facility, Grenoble (beamlines ID14-2 and ID29), Petra Gnau, Diana Kruhl and Sandra Benthin for technical support, Uwe Linne for mass-spectrometric analyses, and Michael Kock for the analysis of the S227A mutant. M.V. thanks International Max Planck Research School for Environmental, Cellular and Molecular Microbiology for funding. This research is supported by grants of the Deutsche Forschungsgemeinschaft (H.-U.M. and L.-O.E.) and the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz center of Synthetic Microbiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2XJP, 2XJQ, 2XJR, 2XJS, 2XJT, 2XJU, and 2XJV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013210108/-/DCSupplemental.
References
- 1.Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Virji M. In: Glycoscience and Microbial Adhesion. Lindhorst TK, Oscarson S, editors. Heidelberg: Springer; 2009. pp. 1–15. (Topics in Current Chemistry). [DOI] [PubMed] [Google Scholar]
- 3.Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 4.Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 5.Dawkins R. The Selfish Gene. Oxford, U.K.: Oxford Univ. Press; 1976. [Google Scholar]
- 6.Smukalla S, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linder T, Gustafsson CM. Molecular phylogenetics of ascomycotal adhesins—a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet Biol. 2008;45:485–497. doi: 10.1016/j.fgb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol Mol Biol R. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigden DJ, Mello LV, Galperin MY. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem Sci. 2004;29:335–339. doi: 10.1016/j.tibs.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the Anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 11.Sonnhammer ELL, Eddy SR, Durbin R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Miki BLA, Poon NH, James AP, Seligy VL. Possible mechanism for flocculation interactions governed by gene FLO1 in Saccharomyces cerevisiae. J Bacteriol. 1982;150:878–889. doi: 10.1128/jb.150.2.878-889.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratford M, Assinder S. Yeast flocculation: Flo1 and NewFlo phenotypes and receptor structure. Yeast. 1991;7:559–574. doi: 10.1002/yea.320070604. [DOI] [PubMed] [Google Scholar]
- 14.Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. Yeast flocculation: What brewers should know. Appl Microbiol Biotechnol. 2003;61:197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- 15.Haig D. Gestational drive and the green-bearded placenta. Proc Natl Acad Sci USA. 1996;93:6547–6551. doi: 10.1073/pnas.93.13.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol R. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lommel M, Strahl S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology. 2009;19:816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 18.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 19.Ogata T, Izumikawa M, Kohno K, Shibata K. Chromosomal location of Lg-FLO1 in bottom-fermenting yeast and the FLO5 locus of industrial yeast. J Appl Microbiol. 2008;105:1186–1198. doi: 10.1111/j.1365-2672.2008.03852.x. [DOI] [PubMed] [Google Scholar]
- 20.Dowell RD, et al. Genotype to phenotype: A complex problem. Science. 2010;328:469–469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupp S, Summers E, Lo H-J, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 23.Chevrier B, Schalk C, D’Orchymont H, Rondeau JM, Moras D. Crystal structure of Aeromonas proteolytica aminopeptidase: A prototypical member of the co-catalytic zinc enzyme family. Structure. 1994;2:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 24.Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man. A model organism helps elucidating congenital human diseases. Angew Chem, Int Edit. 2006;45:6802–6818. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi O, Hayashi N, Kuroki R, Sone H. Region of Flo1 proteins responsible for sugar recognition. J Bacteriol. 1998;180:6503–6510. doi: 10.1128/jb.180.24.6503-6510.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queller DC, Ponte E, Bozzaro S, Strassmann JE. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science. 2003;299:105–106. doi: 10.1126/science.1077742. [DOI] [PubMed] [Google Scholar]
- 27.Kiselyov VV, Soroka V, Berezin V, Bock E. Structural biology of NCAM homophilic binding and activation of FGFR. J Neurochem. 2005;94:1169–1179. doi: 10.1111/j.1471-4159.2005.03284.x. [DOI] [PubMed] [Google Scholar]
- 28.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsook CB, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eswar N, et al. In: Current Protocols in Bioinformatics. Suppl. 15. Baxevanis AD, editor. Malden, MA: John Wiley & Sons; 2006. pp. 5.6.1–5.6.30. [Google Scholar]
- 31.Landau M, et al. ConSurf: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.