Abstract
The Epstein-Barr virus (EBV) is a γ-herpesvirus that infects B cells and epithelial cells and that has been linked to malignancies in both cell types in vivo. EBV, like other herpesviruses, has three glycoproteins, glycoprotein B (gB), gH, and gL, that form the core membrane fusion machinery mediating viral penetration into the cell. The gH and gL proteins associate to form a heterodimeric complex, which is necessary for efficient membrane fusion and also implicated in direct binding to epithelial cell receptors required for viral entry. To gain insight into the mechanistic role of gH/gL, we determined the crystal structure of the EBV gH/gL complex. The structure is comprised of four domains organized along the longest axis of the molecule. Comparisons with homologous HSV-2 gH/gL and partial pseudorabies virus gH structures support the domain boundaries determined for the EBV gH/gL structure and illustrate significant differences in interdomain packing angles. The gL subunit and N-terminal residues of gH form a globular domain at one end of the structure, implicated in interactions with gB and activation of membrane fusion. The C-terminal domain of gH, proximal to the viral membrane, is also implicated in membrane fusion. The gH/gL structure locates an integrin binding motif, implicated in epithelial cell entry, on a prominent loop in the central region of the structure. Multiple regions of gH/gL, including its two extreme ends, are functionally important, consistent with the multiple roles of gH/gL in EBV entry.
The Epstein-Barr virus (EBV) is a double-stranded DNA virus belonging to the Herpesviridae family, which is divided into three subfamilies (α, β, and γ) (1–3). EBV and Kaposi's Sarcoma herpesvirus (KSHV or HHV-8) are the two human host-specific viruses forming the γ-herpesviridae subfamily (2). EBV infects both B lymphocytes and epithelial cells and causes infectious mononucleosis. Nearly 95% of the population is infected by EBV by adulthood and carries EBV DNA throughout life. EBV is maintained in a latent state in infected B lymphocytes, with periodic reactivation of lytic replication. Numerous malignancies are associated with EBV, such as Burkitt's lymphoma, Hodgkin's lymphoma, and nasopharyngeal carcinoma (1, 3). In healthy individuals, the pathological effects of the virus are controlled by the immune system. However, in immunosuppressed individuals, such as transplant recipients and AIDS patients, EBV can cause tumor outgrowth and trigger fatal lymphoproliferative disease.
EBV uses different pathways for the infection of epithelial cells and B lymphocytes (4, 5). For both cell types, the minimal viral glycoprotein components that mediate membrane fusion have been identified (4, 5). As with other herpesviruses, EBV uses the core viral entry glycoproteins, glycoprotein B (gB) and the gH/gL complex. For the infection of B lymphocytes, EBV requires an additional protein, gp42, which binds to host HLA class II molecules, triggering the membrane fusion step (4, 5). gp42 has multiple functional sites for interaction with gH/gL, HLA class II, and potentially, another unknown binding ligand that could be engaged through a large surface-exposed hydrophobic pocket (6–12). The gp42 protein binds to the gH/gL complex with nanomolar affinity through its N-terminal region, and this interaction can be recapitulated with a synthetic peptide of ∼35 aa residues (10, 13, 14). EBV glycoprotein-mediated membrane fusion with epithelial cells does not require gp42 but only gB and gH/gL, and fusion can be completely blocked by saturating amounts of either gp42 or short gp42-derived peptides (13–16), consistent with the hypothesis that gp42 levels in the virion regulate the cellular tropism of the virus in vivo (16). Recent observations indicate that EBV gH/gL engages integrins αvβ6 and/or αvβ8 on epithelial cells to trigger membrane fusion and entry (17).
Among the core glycoproteins for EBV-induced membrane fusion, the crystal structures of gB (18), the gp42:HLA complex (6), and gp42 alone (10) have been determined. The EBV gB protein belongs to the recently identified class III viral fusion glycoproteins (19), which includes the herpesvirus gB proteins (18, 20), the Vesicular Stomatitis Virus G (VSV G) protein (21, 22), and the baculovirus gp64 protein (23). Both VSV G and baculovirus gp64 act alone as the fusogenic proteins for virus entry and both are activated by low pH changes during endocytosis of the virus (19, 21–23). The VSV G fusion protein undergoes reversible conformational changes that are pH-dependent in contrast to the irreversible transitions characterized for both class I and class II viral fusion proteins (24, 25). The VSV G protein structure has been solved in two conformational states, interpreted as representing the pre- and postfusion forms. The EBV and HSV-1 gB protein structures are most similar to the postfusion conformations of VSV G (19). A prefusion model of the EBV gB structure has been proposed (18), but there is no experimental evidence to date for this conformational transition in the herpesvirus glycoproteins. In contrast to the VSV and baculovirus fusion proteins, which act independently, herpesvirus gB requires gH/gL for the most efficient membrane fusion (15, 26, 27).
To better understand the role of gH/gL in virus entry and membrane fusion, we determined the crystal structure of the EBV gH/gL heterodimer. EBV gH and gL form a multidomain structure similar to the recently determined HSV-2 gH/gL (28) and partial pseudorabies virus (PRV) gH structures. The structure consists of four domains, forming a flat, elongated shape, with individual domains sequentially folded along the length of the protein. Each of the four domains adopts different tertiary folds and secondary structures, and the N-terminal domain seems more disordered in the crystal, representing a potentially mobile region of the heterodimer formed by both gH and gL proteins. Superpositions of the EBV, HSV-2 (28), and PRV structures reveal differences in the interdomain arrangements, with the largest difference observed for the first and second domains of the EBV and HSV-2 structures. Analysis of the gH/gL complex structure, based on previous mutational and functional experiments, points to specific regions in multiple domains that are critical for this protein complex to act as part of the core herpesvirus fusion machinery.
Results and Discussion
Determination of the EBV gH/gL Protein Structure.
The ectodomains of EBV gH and gL were coexpressed in insect cells. The two proteins form a stable complex and were purified by affinity chromatography with an antibody (E1D1) that recognizes an epitope dependent on both gH and gL (13, 29). The complex elutes in gel filtration chromatography with the expected molecular weight of the heterodimer (13).
EBV gH/gL crystallized in the P21 space group and a native dataset to 3.58-Å resolution was collected (Table S1). The structure was determined by heavy-atom methods, and the asymmetric unit was expected to contain 10–20 copies of gH/gL. Initial phases were obtained using a Ta6Br12 cluster and further improved with a second (K2IrCl6) derivative (Table S1). A low-resolution selenomethionine dataset was used to validate the chain tracing and model building but not in initial phasing. Sixteen copies of gH/gL were identified in the asymmetric unit, and phases were extended to 3.58 Å using noncrystallographic symmetry averaging. Side-chain electron density was significantly improved using B-factor sharpened electron density maps (Fig. S1). The final Rfree is 31.3%, and the refinement and model statistics are collected in Table S1. The structure includes the majority of the gH and gL residues included in the expression construct.
Overall Description of the Structure of EBV gH/gL.
The EBV gH/gL structure reveals an elongated rod-like shape ∼100 Å in length and 30–60 Å in width, with the middle of the molecule being the widest (Fig. 1). The overall shape agrees with the rod-like shapes observed in negatively stained EM images (Fig. S2). gH/gL is divided into four major domains laid out along the length of the molecule, whose folds involve only immediately adjacent polypeptide segments. There are no domain insertions or long interdomain excursions in the structure.
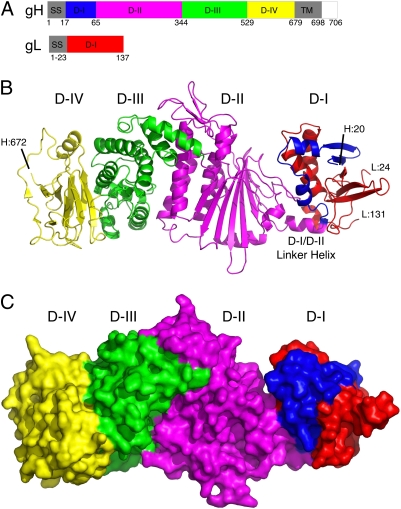
Fig. 1.
Structure of EBV gH and gL heterodimer. (A) Schematic diagram of the EBV gH and gL domain structures. Colored domains correspond to the crystal structure shown in B. Regions not in the construct are colored gray or white. Sequence numbers are indicated below. (B) Ribbon diagram of the EBV gH/gL structure. gH is colored blue in D-I, magenta in D-II, green in D-III, and yellow in D-IV. gL is colored red. The D-I/D-II linker helix and the gH and gL N- and C-terminal residues are indicated, with L:24 and L:131 referring to gL residues 24 and 131 and H:20 and H:672 referring to gH residues 20 and 672. At the N terminus of gH and not visible in the structure were residues AMT that were added to the soluble protein as a result of cloning. Similarly, for gL, N-terminal residues AMD (from cloning) were not observed in the structure. At the C terminus, gH residues 673–679 and gL residues 132–137 were not observed. (C) Surface of the EBV gH/gL structure colored by domain as in B.
The gL protein forms intimate contacts with the N-terminal residues of gH, which together define the first domain (D-I) of the heterodimer structure (Fig. 1B). The central two domains of gH (D-II and D-III) create a core region of predominantly helical structure, surrounded on either side by β-sheet. The exclusive involvement of gL with the N-terminal residues of gH in D-I is consistent with previous mapping experiments (30) and the HSV-2 gH/gL structure (28).
There are 11 cysteine residues in gH and 4 in gL. Five disulfide bonds were observed within the gH structure between residues 120 and 312, 278 and 335, 454 and 478, 534 and 587, and 612 and 615. C153 does not form a disulfide bond with any other cysteine residue and bound Hg in heavy atom-soaked crystals. gL contains two disulfide bonds between residues 28 and 56 and 29 and 79. Extra density consistent with N-linked glycosylation was observed at gH residues 60, 435, 549, 604, and 664 and gL residues 44, 53, and 69. We have defined the gH/gL domain boundaries differently from those previously reported for HSV-2 gH/gL (28). The EBV gH/gL domains seem more consistent with interdomain rearrangements evident in structural comparisons, and these domains are, therefore, described in more detail below.
N-Terminal Residues of gH and gL Form D-I.
The gL subunit is important for gH folding (28, 30, 31) and is implicated in the specificity of EBV gB activation (32). In EBV gH/gL, gL (residues 24–131) plays a key structural role in D-I (Fig. 1), constructing an α-/β-domain that is intimately intertwined with the N-terminal residues of gH (20–65).
D-I is a flat, semiglobular domain formed by a layer of five-stranded mixed parallel/antiparallel β-sheets supported by a layer of three α-helices (Fig. S3A). The secondary structure and hydrophobic core of D-I seems dependent on both gH and gL residues and is similar at its core to the HSV-2 gH/gL structure (28). The five-stranded β-sheet contains three strands from gL and two strands from gH, whereas the underlying three-helix layer is formed by two helices from gL and one from gH. A long protrusion from the surface of D-I is formed by strands Lβ-2 and Lβ-3 (Fig. 1B and Fig. S3A). Two neighboring cysteine residues in gL (residues 28 and 29) form disulfide bonds with residues 56 and 79, located in the loop between strands Lβ-1 and Lβ-2 and in Lβ-3, respectively. The N terminus of gL forms a long loop leading into the central Lβ-1 strand beginning at residue 47. In HSV-2 gL, the N-terminal residues form additional structure on the face of the D-I β-sheet (28).
The three-helix layer separates D-I from D-II, forming a wall that includes a number of charged and polar residues (Fig. 1B). The polypeptide link between D-I and the rest of gH is mediated by a single α-helix (residues 66–74) at the base of the interdomain interface (Fig. 1B), which might allow repositioning of the two domains, with the helix acting as an internal hinge or lever. Superposition of the D-I domain from the EBV and HSV-2 structures reveals a common core tertiary structure, with the HSV-2 domain exhibiting significant variations at the N terminus of gL and in extended loop structures throughout the domain.
gH Residues 66–672 Fold into Three Sequential Globular Domains.
The remainder of gH folds into three additional and potentially, semiautonomous domains with extensive interdomain interfaces, similar to the structures of the HSV-2 gH/gL (28) and PRV gH fragment.
D-II starts with the C-terminal end of the D-I/D-II linker α-helix (Fig. S3B), followed by a long extended chain that wraps around the outside surface of D-II. After the linker from D-I, residues 79–97 climb ∼60 Å diagonally across D-II to the opposite corner, forming flaps that cover the core helical bundle of the domain. Electron density of this segment is more disordered, and it is likely less constrained and flexible. D-II can be split into two major secondary structure sections (Fig. 1 and Fig. S3B), with a prominent eight-stranded antiparallel β-sheet (β-sheet II), followed by an antiparallel five-helix bundle (helical bundle II). β-sheet II forms a picket fence separating D-I from helical bundle II (Fig. 1B). The β-sheet II fence facing D-I is covered by two extended polypeptide loops (residues 133–154 and 201–217) that include a short helix (2α-3) at the tip of the first loop (residues 143–148), forming the interface with D-I (Fig. 1B).
The three main α-helices of helical bundle II lie parallel to the long axes of β-sheet II, forming the central and widest region of the entire gH/gL molecule. Residues 245–336 form the majority of this helical region that is mainly helix–loop–helix structure. The core three helices consist of residues 258–274, 285–307, and 315–335 (2α-6–2α-8), and two shorter α-helices from residues 123–130 and 246–254 (2α-2 and 2α-5) add to the outside of this three-helix core, with five helices total. A final helix forms the transition between D-II and D-III.
D-III (Fig. S3C), starting from S345 and extending to L529, is a mainly α-helical structure, with its helices lying nearly perpendicular to the length of helical bundle II. The domain begins with five helices arranged in four layers of a helix–loop–helix structure that spirals down the side of D-II, involving residues 345–454 (Fig. 1B and Fig. S3C). The final four helices form a distinct subdomain bundle, making a total of nine helices in D-III. There is one disulfide bond near the center of D-III (C454–C478).
D-IV (Fig. S3D), beginning at P530 and ending at G672, forms a β-sandwich domain, with two antiparallel β-sheets (β-sheet IIIA and IIIB). Both β-sheets consist of four core antiparallel β-strands (4β-1–4β-4 and 4β-7–4β-10), which are linked by a single cross-over loop (residues 584–617). The β-sandwich is aligned with one edge abutting D-III and the axis of the β-strands aligned with the helix and sheet axis of D-II (Fig. 1B). Extensive loops connect the β-strands within each sheet, and the two disulfide bonds within this domain are located on the exterior chains decorating the core β-sandwich.
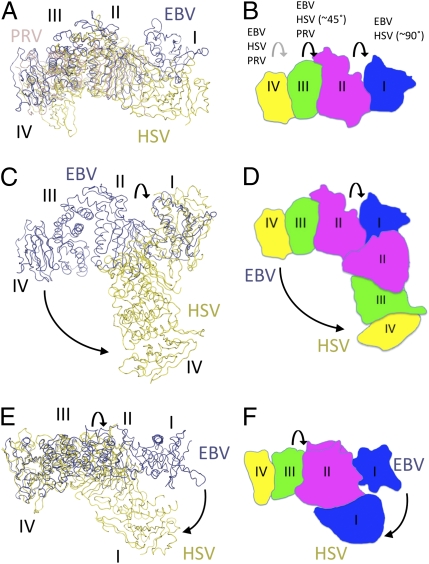
Comparison of the EBV, HSV-2, and PRV Structures.
The recent structure determinations of HSV-2 gH/gL (28) and a three-domain PRV gH fragment allow a structural comparison across the α- and γ-herpesviruses (33). The core secondary structures of the domains, as defined here for EBV gH/gL, are very similar in the three structures. However, the interdomain packing arrangements differ significantly, particularly in comparisons of the EBV and HSV-2 structures (Fig. 2). The PRV structure lacks D-I but exhibits slightly different packing at both the D-II/D-III and D-III/D-IV domain interfaces. However, much larger packing angle differences are observed for the HSV-2 D-I/D-II and D-II/D-III interfaces, of ∼90° and ∼45°, respectively. Thus, whereas the EBV structure seems linear and elongated, the HSV-2 structure adopts a boot-like (28) configuration (Fig. 2). The difference in D-I/D-II packing creates a prominent groove in the EBV gH/gL structure that is not present in HSV-2 gH/gL.
Fig. 2.
Structural comparison between EBV gH/gL, HSV gH/gL, and PRV gH. (A) EBV gH/gL was superimposed with the structures of HSV-2 gH/gL (28) and a three-domain fragment of PRV gH through D-II. The EBV structure is shown in dark blue, the HSV-2 structure is in yellow (33), and the PRV structure is in light red, with the EBV gH/gL domain boundaries indicated by labels. (B) A schematic representation of the EBV gH/gL structure is shown colored as in Fig. 1B and with the domains indicated. Interdomain packing changes between the structures are indicated, with approximated rotations given for the largest changes observed between the EBV and HSV-2 structures at the D-I/D-II and D-II/D-III interfaces. (C) Superposition of the EBV and HSV-2 structures through D-I residues. (D) Schematic representation of the EBV and HSV-2 D-I superposition showing the reorientation of the gH domains. (E) Superposition of the EBV and HSV-2 structures through D-III residues. (F) Schematic representation of the EBV and HSV-2 D-III superposition showing the reorientation of the N-terminal D-II and D-I domains. Although individual domains consist of similar core structures, the changes in interdomain packing create different overall shapes, especially for the EBV and HSV-2 structures.
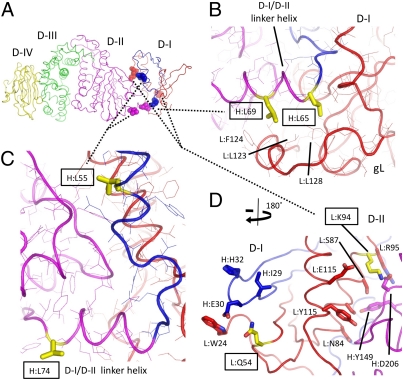
Mutations in D-I Influence Membrane Fusion Activity and gB Specificity.
Mutations that affect EBV gH/gL activity in membrane fusion map to D-I and the D-I/D-II interface (Fig. 3A) (32, 33). Mutants in leucine residues in gH were generated in the N-terminal region spanning residues 54–74, which are found in D-I or the helical junction to D-II (34). Mutations of L65A and L69A showed reduction in membrane fusion with both B cells and epithelial cells, whereas mutants L55A and L74A enhanced viral fusion. Residues 65 and 69 are both located in the N-terminal portion of the D-I/D-II linker helix and are buried in a hydrophobic interface with gL (Fig. 3 A and B). L55 is located at the interface of D-I/D-II and may destabilize the interdomain interface, whereas L74 is exposed on the surface of the D-I/D-II linker helix at the beginning of D-II (Fig. 3 A and C). The deleterious mutations of L65A and L69A may disrupt interactions with gL and the proper folding of D-I. The monoclonal antibody E1D1 recognizes an epitope generated by both gH and gL, and its binding to the L65A mutant is diminished, consistent with an alteration in the folding of D-I. The fusion-enhancing mutations of L55A and L74A are not buried in the D-I hydrophobic core and seem more likely to affect the D-I/D-II interdomain arrangement.
Fig. 3.
D-I residues implicated in membrane fusion activity of gH/gL. (A) Residues implicated in gH/gL function are shown as semitransparent CPK spheres colored by chain and domain as in Fig 1. The backbone trace is represented as coils, and the residues are labeled as described in Fig. 1. (B) Mutations of EBV gH L65A and L69A, referred to as H:L65 and H:L69, respectively, reduce gH/gL membrane fusion activity with both B cells and epithelial cells. H:L65 and H:L69 are shown in yellow sticks, with surrounding residues shown as lines colored by the gH/gL chain as in A and labeled. Both residues form hydrophobic contacts with gL at the edge of the D-I hydrophobic core. (C) gH mutants L55A and L74A enhance viral fusion activity. H:L55 is located at the D-I/D-II interface, whereas H:L75 is exposed on the surface of the D-I/D-II linker helix at the beginning of D-II. Residues are colored as in B. (D) EBV gL Q54 and K94 (yellow sticks and boxes) control the specificity of gB activation in membrane fusion. Mutation to Rhesus lymphocryptovirus (LCV) gL (82% identity with EBV gL) residues K54 and Q94 reduces membrane fusion with EBV gB. L:Q54 is exposed at the surface of D-I in the loop between the first and second β-strands (Lβ-1 and Lβ-2). Adjacent residues (L:W24, H:I29, H:E30, and H:H32) are also surface-exposed and form a prominent ridge adjacent to Q54. K94 is located at the D-I/D-II interface in an area rich in polar and charged residues (as described in the text). L:Q54 and L:K94 are show as yellow sticks, with identified neighboring residues shown as sticks colored by their associated chain and domain.
We have recently shown that two gL residues (54 and 94) determine the specificity of gB activation in membrane fusion (32), potentially through direct interactions with gB. Rhesus LCV gL (Rh-gL) and EBV gL are highly homologous (82% identity) (Fig. S4), but Rh-gL cannot mediate membrane fusion with EBV gB. The double mutation of residues K54Q/Q94K in Rh-gL incorporates the EBV amino acids and restores membrane fusion activity with EBV gB. The converse mutation of EBV gL to Rh-gL amino acids at these residues (Q54K/K94Q) significantly reduces membrane fusion activity with EBV gB.
Q54 is exposed at the surface of D-I in the loop between the first and second β-strands (Lβ-1 and Lβ-2) (Fig. 3D). The adjacent N53 seems to be glycosylated, but nearby residues gL-W24, gH-I29, gH-E30, and gH-H32 form a prominent protrusion from D-I. The prominent exposure of gL-W24 is particularly striking, and all of these residues are conserved in the Rh-gH and -gL sequences (Fig. S4).
The other key residue in determining gB specificity, K94, is located at the C-terminal end of the central α-helix (Lα-2) pointing to the D-I/D-II interface (Fig. 3D). This region of the D-I/D-II interface contains many charged and polar residues, including gL-N84, gL-S87, gL-S91, gL-K94, gL-R95, gL-E112, gL-Y115, gH-Y149, and gH-D206. Similar to the gH mutation L55A, which is on the neighboring gH helix of D-I, gL-K94 may affect D-I/D-II interactions and orientation, possibly leading to a enhanced fusogenic activity. Because K94 is located in a groove at the top of the D-I/D-II interface, it is also possible that this could form part of a binding surface that could engage gB (Fig. 3A).
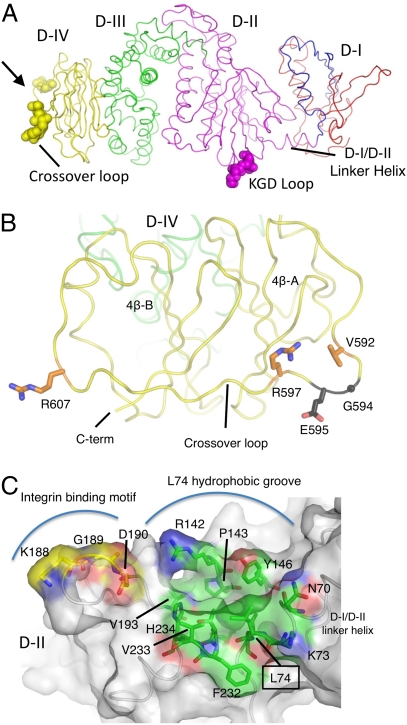
Residues in D-II and D-IV Are Implicated in Membrane Fusion.
Other regions of gH are also implicated in membrane fusion activity (Fig. 4A). The C-terminal end of gH has been identified as a critical functional region (35, 36). The anti-gH antibody CL59 blocks membrane fusion with epithelial cells but not B cells, and its binding epitope has been mapped to gH residues 501–628 (35, 36), forming the C-terminal end of D-III and the majority of D-IV. Insertion and point mutant studies have also implicated specific C-terminal residues in regulating differential gH fusion activity with B cells and epithelial cells (35, 36).
Fig. 4.
Residues in gH D-II to D-IV implicated in EBV entry. (A) Residues in EBV gH D-II and D-IV that are implicated in EBV entry are shown on the structure as CPK spheres, colored by domain as in Fig. 1. The D-IV residues are located at the extreme C-terminal end of the molecule in a cross-over loop between the two halves of the domain. The D-II residues, defining an integrin binding motif implicated in epithelial cell entry, are located in D-II, near the D-I/D-II linker helix. (B) gH residues G594 and E595 are located in the D-IV cross-over loop connecting the two β-sheets of the domain fold. The view shown is oriented ∼90° about the vertical axis from the view in A (bold arrow). The residues are shown as dark gray sticks with O and N atoms colored red and blue, respectively. gH residues 592, 597, and 607, which have moderate effects on gH/gL fusion activity, are also exposed on the D-IV cross-over loop. These residues are shown as orange sticks, with O and N atoms colored red and blue. (C) The gH/gL integrin binding site is adjacent to an extended hydrophobic groove including L74. The view shown is oriented ∼90° about the horizontal axis from the view in A. The gH/gL chain is shown as a coil beneath a transparent protein surface. The KGD motif is shown as sticks with carbon atoms colored yellow and mapped onto the gH/gL surface. The nearby residue L74, located in the D-I/D-II linker helix, is surrounded by gH residues that form a primarily hydrophobic groove lined with charged residues. These are also shown in stick representation, labeled, and colored with carbon atoms in green.
The mutation of G594 to alanine abolishes fusion with both B cells and epithelial cells, whereas the mutation of E595A has only moderate effects on epithelial cell fusion but dramatically increases fusion activity with B cells (35, 36). G594 and E595 are located on the cross-over loop in D-IV and exposed at the extreme C-terminal end of the gH structure (Fig. 4B). Additional mutations of V592A, R597A, and R607A have moderate effects on gH activity (35, 36), decreasing fusion with B cells ∼40%. These three residues are also located in the cross-over loop in D-IV (Fig. 4B), near the gH C terminus and the expected position of the transmembrane domain and viral membrane.
Recently, the interaction of EBV gH/gL with αvβ6 and αvβ8 integrins has been implicated in triggering viral fusion with epithelial cells (17). In the structure, the identified KGD motif is located in a prominent D-II loop between strands 2β-6 and 2β-7 (Fig. 4 A and C). The motif is near the D-I/D-II linker α-helix (2α-1) and the D-I/D-II interface, where mutations in both gH and gL affect the fusogenic activity of gH/gL. Nearby mutations in gH residues 65 and 69 disrupt E1D1 antibody binding. E1D1 inhibits membrane fusion with epithelial cells (but not B cells) (36) and blocks gH binding to integrin (37), consistent with the possibility that E1D1 and integrins may bind overlapping surfaces in this junctional region between D-I and D-II.
Final Remarks.
The structure of the EBV gH/gL complex adds to the previous structures of the EBV gB and gp42 proteins (6, 10, 18), which together define the essential membrane fusion machinery of this γ-herpesvirus. The EBV gH/gL protein has a four-domain architecture arranged linearly along the length of the polypeptide chain, with the N-terminal D-I being formed by both gH and gL subunits, similar to the structures of HSV-2 gH/gL (28) and a PRV gH fragment (33). Functional regions in EBV gH/gL have been mapped to at least four areas, including D-I, the interface between D-I and D-II, D-IV, and a prominently displayed KGD sequence in D-II implicated in integrin binding and epithelial cell entry (17). The integrin binding loop (residues 188–190) lies next to the functionally important L74 (34). L74 forms part of a ridge defining a hydrophobic groove adjacent to the integrin binding site and surrounded by a number of charged residues (Fig. 4C), potentially defining an extended surface region important to gH/gL function in EBV entry. gL residues that regulate the specificity of gB activation (32) have been mapped to the surface of D-I (Q54) and to the D-I/D-II interface (K94). Q54 is surface-exposed and surrounded by other prominently displayed D-I residues, which together could form a gB binding site. The binding region of gp42 for gH/gL has been mapped to a 33-aa peptide (14). We have not yet identified the corresponding binding site on gH/gL, but the prominent groove between D-I and D-II and the surface between the D-1/D-II linker helix and the integrin binding loop stand out as potential peptide binding sites. This putative site for gp42 binding is near to the integrin binding loop, and competitive binding between gp42 and integrin to gH/gL would be consistent with the observation that gp42 and gp42-derived peptides block fusion with epithelial cells (9, 13, 14, 16).
Differences in domain arrangements in the EBV, HSV-2, and partial PRV structures, along with the mapping of EBV mutants to the D-I/D-II interface, suggest that a D-I/D-II conformational change could be part of a mechanism for triggering membrane fusion. However, we do not observe large changes in the EBV structure in the 16 independent copies present in the crystallographic asymmetric unit. It will be important to determine whether the observed D-I/D-II conformations define a static structural difference across the α- and γ-herpesviruses or whether dynamic rearrangements of these domains are functionally significant.
Although it has been suggested that gH/gL could act as a direct participant in membrane fusion (for example, by triggering hemifusion intermediates) (38), more recent studies suggest that it may act as a regulator of gB activation (5, 28, 39) through direct interactions with homotypic gB protein from the same herpesvirus. The EBV gH/gL structure reveals that both ends of the protein are functionally important, with the N-terminal D-I region implicated in regulating gB activation and potentially defining a gB binding site. Mapping of EBV gH/gL functional mutants to D-IV provides evidence that this virus membrane-proximal domain is important in membrane fusion and would be consistent with direct interactions of gH/gL with the viral membrane during fusion. Finally, mapping of the integrin binding loop on gH/gL to D-II identifies a receptor binding region on gH/gL and places this adjacent to a functional region at the D-I/D-II interface. The clustering of EBV functional mutations in D-I and the integrin binding motif in D-II, both located toward the N-terminal end of gH/gL, implicates this end of the protein in interactions, which might orient the complex to a target cell membrane and promote gB contact with D-I and gB activation. PRV gH/gL can be substituted by a gD–gH fusion protein (40), consistent with a hypothetical requirement for orienting gH/gL toward a target cell during membrane fusion but seemingly inconsistent with the important role for D-I and the D-I/D-II interface highlighted by EBV mutations mapped here. Further studies are needed to clarify how gH/gL structures and functions vary across the herpesvirus family.
Materials and Methods
Production and Purification of gH/gL.
The expression and purification of soluble EBV gH/gL has been previously described (13). In the expression constructs, gH residues 18–679 and gL residues 24–137 were fused to the baculovirus gp64 signal sequence, resulting in the addition of three N-terminal residues (AMT) into gH and three (AMD) into gL. For protein production, Sf+ insect cells were grown to a density of 1.5 million cells/mL and infected with recombinant gH/gL baculovirus stock. Cell supernatants were harvested 72 h postinfection, and gH/gL was isolated using an anti-gH/gL mouse monoclonal antibody (E1D1) column. The E1D1 hybridoma was kindly provided by L. Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport, LA). Typically, 4 L of infected cell supernatant were passed through the E1D1 affinity column and washed with PBS. The protein was eluted with 0.1 M sodium citrate, pH 2.5, and immediately neutralized by the addition of 1 M sodium citrate Tribasic salt (pH ∼8) and 5 M NaCl, yielding a final pH of 5.5. The eluted protein solutions were combined, concentrated, and exchanged into 50 mM sodium citrate, pH 5.5, and 50 mM NaCl buffer. The protein was concentrated, filtered, and further purified using a S200 gel filtration column in 50 mM sodium citrate, pH 5.5, and 50 mM NaCl buffer.
gH/gL Crystallization.
The gH/gL protein was concentrated to ∼10 mg/mL using Vivaspin 2 and Vivaspin 500 (Sartorius) concentrators, and the final concentration was confirmed using a Nanodrop spectrophotometer with an extinction coefficient of 87,515 (M−1·cm−1 at 280 nm). gH/gL crystals were grown in sitting drop format (Cryschem Plate HM3-158; Hampton Research), initially using a well solution consisting of 24% PEG1000 and 100 mM sodium citrate, pH 5.5, and drops containing 24% PEG1000, 100 mM sodium citrate, pH 5.5, 200–400 mM NDSB256-4T, and protein at room temperature. The crystallization was further optimized by seeding. For seeded crystal growth, the well solution contained 18–19% PEG1000, 100 mM sodium citrate, pH 5.5, and 200 mM NDSB201, and the drop consisted of a mixture of PEG1000, citrate buffer, NDSB256-4T, and protein. After >16 h of equilibration, 0.1 μL gH/gL crystal seed stock, generated using Seed Beads (Hampton Research), were added. In the optimized conditions, gH/gL crystals typically appeared after 2 d and grew up to a size of 50–100 × 100 × 200–300 μm, and occasionally larger, after 7 d. The gH/gL crystals were harvested in 2–3% higher PEG1000 concentrations and transferred in gradual steps of increasing PEG1000 concentration to a final concentration of 30% before freezing in liquid nitrogen.
Heavy-Atom Derivatization and Data Collection.
Initial phases were obtained from datasets with significant anomalous signals from crystals soaked in 2 mM Ta6Br12 for 4 h and crystals soaked in 1 mM K2IrCl6 for 4 d. The heavy atom-soaked crystals were transferred into freezing buffer conditions, maintaining constant concentrations of heavy atom before freezing. Selenomethionine-substituted protein was expressed in Hi5 cells by replacing the medium with methionine-free Ex-Cell 405 and by adding selenomethionine at the final concentration of 60.6 mg/L at the time of infection.
The native dataset was collected at the Advanced Light Source beamline 8.3.1 and processed to 3.5 Å using Mosflm and SCALA (41). The Ta6Br12 single-wavelength anomalous dispersion (SAD) data were collected at 24ID-C beamline operated by the Northeastern Collaborative Access Team at the Advanced Photon Source at the Ta peak wavelength of 1.2543 Å and processed with HKL2000 (42) to 4.6 Å. The K2IrCl6 dataset was collected at beamline BL9-2 at the Stanford Synchrotron Radiation Laboratory at 1.10519 Å and processed with HKL2000 to 5.5 Å. Additional heavy-atom datasets are collected in Table S1.
Structure Determination and Refinement.
The Native, Ta6Br12, and K2IrCl6 datasets were combined and scaled together (41), and SHARP (43) was used for heavy-atom refinement and phasing. Initial heavy atom-phased maps were generated at low resolution (∼6 Å) and used to identify noncrystallographic symmetry (NCS) relationships and helical features of the gHgL structure. Electron density maps were improved by a combination of density modification, NCS averaging, and heavy-atom refinement. A final total of 40 Ta6Br12 clusters, 34 K2IrCl6, 38 KReO4, 7 YbCl3, 14 (NH4)2WS4, 24 AuCN2, 27 SeMet sites, and 16 NCS-related gH/gL molecules were identified in the crystallographic asymmetric unit. The gH/gL model was built and refined using a combination of automated and manual approaches. Further details of the structure determination are included in SI Materials and Methods, and the final model statistics are presented in Table S1. The final model has an R-free of 31.3%, overall good geometry, Ramachandran statistics, and no unfavorable side-chain rotamers.
Supplementary Material
Acknowledgments
We thank Lindsey Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport, LA) for the E1D1 monoclonal antibody and members of the T.S.J. and R.L. laboratories. Portions of this research were conducted at the Advanced Photon Source, the Stanford Synchrotron Radiation Laboratory, and the Advanced Light Source (SI Materials and Methods). This work was supported by Public Health Service Grants AI076183 and CA117794 from the National Cancer Institute (to R.L. and T.S.J.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3PHF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011806108/-/DCSupplemental.
References
- 1.Kieff E, Rickinson A. Epstein-Barr virus and its replication. In: Knipe D, et al., editors. Fields Virology. 5th Ed. Vol. 2. Philadelphia: Lippincott, Williams and Wilkins; 2007. pp. 2603–2654. [Google Scholar]
- 2.Pellett PE, Roizman B. The family: Herpesviridae, a brief introduction. In: Knipe D, et al., editors. Fields Virology. 5th Ed. Vol. 2. Philadelphia: Lippincott, Williams and Wilkins; 2007. pp. 2479–2500. [Google Scholar]
- 3.Rickinson A, Kieff E. Epstein-Barr virus. In: Knipe D, et al., editors. Fields Virology. 5th Ed. Vol. 2. Philadelphia: Lippincott, Williams and Wilkins; 2007. pp. 2655–2700. [Google Scholar]
- 4.Hutt-Fletcher LM. Epstein-Barr virus entry. J Virol. 2007;81:7825–7832. doi: 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longnecker R, Hutt-Fletcher L, Jardetzky T. Epstein-Barr virus entry. In: Blossom D, Pipas JM, editors. DNA Tumor Viruses. New York: Springer; 2009. pp. 355–378. [Google Scholar]
- 6.Mullen MM, Haan KM, Longnecker R, Jardetzky TS. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell. 2002;9:375–385. doi: 10.1016/s1097-2765(02)00465-3. [DOI] [PubMed] [Google Scholar]
- 7.McShane MP, Mullen MM, Haan KM, Jardetzky TS, Longnecker R. Mutational analysis of the HLA class II interaction with Epstein-Barr virus glycoprotein 42. J Virol. 2003;77:7655–7662. doi: 10.1128/JVI.77.13.7655-7662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva AL, Omerovic J, Jardetzky TS, Longnecker R. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J Virol. 2004;78:5946–5956. doi: 10.1128/JVI.78.11.5946-5956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol. 2007;81:9216–9229. doi: 10.1128/JVI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure. 2009;17:223–233. doi: 10.1016/j.str.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorem J, Jardetzky TS, Longnecker R. Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J Virol. 2009;83:6664–6672. doi: 10.1128/JVI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw PL, Kirschner AN, Jardetzky TS, Longnecker R. Characteristics of Epstein-Barr virus envelope protein gp42. Virus Genes. 2010;40:307–319. doi: 10.1007/s11262-010-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol. 2006;80:9444–9454. doi: 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, Marquardt G, Kirschner AN, Longnecker R, Jardetzky TS. Mapping the N-terminal residues of Epstein-Barr virus gp42 that bind gH/gL by using fluorescence polarization and cell-based fusion assays. J Virol. 2010;84:10375–10385. doi: 10.1128/JVI.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McShane MP, Longnecker R. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc Natl Acad Sci USA. 2004;101:17474–17479. doi: 10.1073/pnas.0404535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med. 2002;8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 17.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci USA. 2009;106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci USA. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 21.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 22.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 23.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 24.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian M, Rey FA. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suenaga T, et al. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc Natl Acad Sci USA. 2010;107:866–871. doi: 10.1073/pnas.0913351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browne H, Bruun B, Minson T. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J Gen Virol. 2001;82:1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- 28.Chowdary TK, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Turk SM, Hutt-Fletcher LM. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng T, et al. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J Virol. 1998;72:6092–6103. doi: 10.1128/jvi.72.7.6092-6103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Buranathai C, Grose C, Hutt-Fletcher LM. Chaperone functions common to nonhomologous Epstein-Barr virus gL and Varicella-Zoster virus gL proteins. J Virol. 1997;71:1667–1670. doi: 10.1128/jvi.71.2.1667-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plate AE, Smajlović J, Jardetzky TS, Longnecker R. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J Virol. 2009;83:7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backovic , et al. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc Natl Acad Sci USA. 2010;107:22635–22640. doi: 10.1073/pnas.1011507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omerović J, Lev L, Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol. 2005;79:12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Hutt-Fletcher LM. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology. 2007;363:148–155. doi: 10.1016/j.virol.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Borza CM, Hutt-Fletcher LM. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J Virol. 2005;79:10923–10930. doi: 10.1128/JVI.79.17.10923-10930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borza CM, Morgan AJ, Turk SM, Hutt-Fletcher LM. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J Virol. 2004;78:5007–5014. doi: 10.1128/JVI.78.10.5007-5014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian RP, Geraghty RJ. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson JO, Longnecker R. Reevaluating herpes simplex virus hemifusion. J Virol. 2010;84:11814–11821. doi: 10.1128/JVI.01615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klupp BG, Mettenleiter TC. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. In: Sweet RM, Carter CW, editors. Methods in Enzymology: Macromolecular Crystallography. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 43.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. In: Sweet RM, Carter CW, editors. Methods in Enzymology: Macromolecular Crystallography. Vol. 276. New York: Academic; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.