Abstract
Immune cells take residence in metabolic tissues, providing a framework for direct regulation of nutrient metabolism. Despite conservation of this anatomic relationship through evolution, the signals and mechanisms by which the immune system regulates nutrient homeostasis and insulin action remain poorly understood. Here, we demonstrate that the IL-4/STAT6 immune axis, a key pathway in helminth immunity and allergies, controls peripheral nutrient metabolism and insulin sensitivity. Disruption of signal transducer and activator of transcription 6 (STAT6) decreases insulin action and enhances a peroxisome proliferator-activated receptor α (PPARα) driven program of oxidative metabolism. Conversely, activation of STAT6 by IL-4 improves insulin action by inhibiting the PPARα-regulated program of nutrient catabolism and attenuating adipose tissue inflammation. These findings have thus identified an unexpected molecular link between the immune system and macronutrient metabolism, suggesting perhaps the coevolution of these pathways occurred to ensure access to glucose during times of helminth infection.
Keywords: insulin resistance, obesity, cytokines, liver, Th2 immunity
Defense against pathogens, the primordial function of the immune system, is bioenergetically intensive and exquisitely sensitive to nutrient availability. For example, the sensing of bacteria by the Toll-like receptor 4 complex initiates signaling cascades that culminate in the activation of NF-κB, which, in turn, induces the expression of inflammatory and microbicidal molecules (1, 2). Interestingly, the ability of macrophages and neutrophils to support antimicrobial programs is completely dependent on the maintenance of high flux through the glycolytic pathways (3, 4). Because immune cells lack significant glycogen stores, the metabolic demands of these activated cells are primarily met by rapid uptake of extracellular glucose (5, 6). Thus, in the context of bacterial infection, the inhibition of insulin signaling by inflammatory molecules might be an adaptive response that redirects glucose from storage to host defense (7, 8). Although this nutrient reallocation strategy is protective during bacterial infection, it becomes maladaptive in the setting of obesity, resulting in insulin resistance and type 2 diabetes (7, 9–11).
In addition to bacteria, parasitic worms, the helminths, comprise another major group of vertebrate pathogens (12). Unlike bacterial infections that tend to be acute and limited, infections with extracellular helminths are chronic and widely prevalent. For example, although certain helminth species are known to reside in human hosts for decades (12), others exhibit near ubiquitous infestation of animals in the wild (13). Moreover, the immune response invoked by helminths is also distinct, one dominated by Th2 cytokines, such as IL-4 and IL-13 (12), whose functions in nutrient metabolism remain unexplored. Because infective helminths continually parasitize nutrients from the host, we hypothesized that signaling pathways mediating immunity against helminths might also regulate nutrient homeostasis via modulation of insulin action.
Th2 cytokines IL-4 and IL-13, which are locally produced in tissues, exert their biologically effects via activation of the transcription factor STAT6 (14). Confirming the importance of this pathway in antihelminth immunity and allergies, mice lacking STAT6 are severely compromised in their ability to mount allergic inflammatory responses (15, 16) or defend against various gastronintestinal helminths (12, 17). Thus, using STAT6-null mice and IL-4 as surrogates, we explored the role of Th2-type immune responses in regulation of nutrient metabolism and insulin action.
Results
IL-4 and STAT6 Regulate Hepatic Metabolism.
We first analyzed the expression and activity of STAT6 in major metabolic tissues: liver, fat, and skeletal muscle. Immunoblotting revealed that although STAT6 is expressed in liver and white adipose tissue (WAT) (Fig. 1A), treatment with IL-4 increased tyrosine phosphorylation of STAT6 in the liver with more variable effects in adipose tissue (Fig. 1A). These findings are consistent with presence of IL-4Rα, which mediates IL-4–dependent activation of STAT6, in hepatocytes but not adipocytes (Fig. S1) (18, 19). Because liver contains many hematopoietic cells, which express IL-4Rα and STAT6, we evaluated the patency of IL-4/STAT6 signaling in primary hepatocytes. Treatment of mouse hepatocytes with IL-4 increased tyrosine phosphorylation of STAT6 (Fig. 1B), suggesting its potential involvement in the regulation of hepatic metabolism.
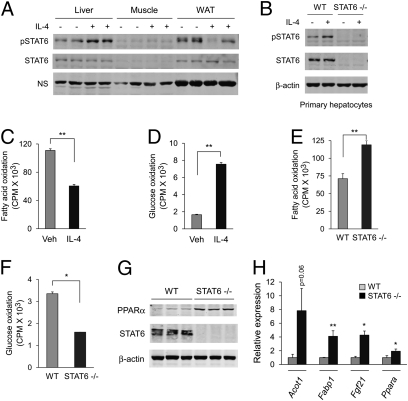
Fig. 1.
IL-4 and STAT6 regulate liver nutrient metabolism. (A) IL-4 enhances phosphorylation of STAT6 in the liver, but not skeletal muscle or WAT. NS, nonspecific. (B) STAT6 is expressed in primary hepatocytes and becomes tyrosine-phosphorylated in response to IL-4. (C and D) IL-4 regulates macronutrient metabolism in isolated primary hepatocytes. Changes in rates of fatty acid (C) and glucose oxidation (D) in hepatocytes treated with IL-4 (10 ng/mL). (E and F) STAT6 regulates fuel oxidation in primary hepatocytes. Rates of fatty acid (E) and glucose oxidation (F) in wild-type and STAT6-null hepatocytes. (G and H) Induction of PPARα and its transcriptional program in livers of STAT6−/− mice. (G) Immunoblotting for PPARα and STAT6 in wild-type and STAT6-null livers. β-actin is used as a loading control. (H) Quantitative RT-PCR analyses for PPARα and its target genes, acyl-CoA thioesterase 1 (Acot1), fatty-acid binding protein 1 (Fabp1), and fibroblast growth factor 21 (Fgf21), in livers of wild-type and STAT6−/− mice. All results are displayed as means ± SEM n > 3. *P < 0.05; **P < 0.01.
To explore this possibility, we measured rates of nutrient utilization in primary hepatocytes treated with IL-4. In contrast to its effects in macrophages (20, 21), IL-4 suppressed β-oxidation of fatty acids in hepatocytes by ~50% (Fig. 1C) and increased hepatic glucose oxidation by ~450% (Fig. 1D). Genetic deficiency of STAT6 had the converse effect, shifting metabolic reliance of hepatocytes from glucose to fatty acids (Figs. 1 E and F). This shift toward fatty acid oxidation in STAT6-null hepatocytes was accompanied by increased activity of peroxisome proliferator-activated receptor α (PPARα), the pivotal regulator of hepatic β-oxidation pathways (22), as evidenced by induction of PPARα and its target genes Acot1, Fabp1, and Fgf21 (Figs. 1 G and H).
IL-4/STAT6 Immune Axis Inhibits PPARα Signaling.
The derepression of PPARα transcriptional activity in livers of STAT6-null mice led us to investigate the transcriptional crosstalk between these signaling pathways. Reporter gene assays demonstrated that, in an IL-4– and STAT6-dependent manner, STAT6 inhibited activation of the PPARα-responsive luciferase reporter (Fig. 2A). This transrepression of STAT6 required PPARα, because transfection of STAT6 alone had negligible effect (Fig. S2). Furthermore, coimmunoprecipitation experiments demonstrated that STAT6 readily associates with PPARα in the livers of mice treated with IL-4 and the PPARα ligand Wy14643 (Fig. 2B). Based on the physical interaction between these two proteins in vivo, we explored whether STAT6 represses PPARα transcriptional activity by preventing its recruitment to target genes. Indeed, ChIP assays demonstrated that recruitment of PPARα to Acot1 and Cyp4a10 promoters was abolished in hepatocytes simultaneously treated with the PPARα ligand Wy14643 and IL-4 (Fig. 2C).
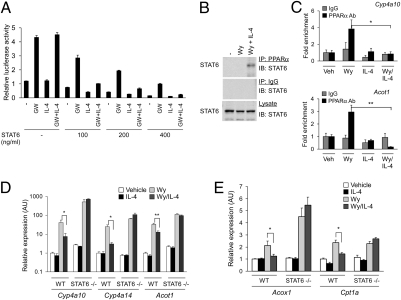
Fig. 2.
IL-4 and STAT6 inhibit PPARα transcriptional activity. (A) Suppression of PPARα transcriptional activity by STAT6 and IL-4. CV-1 cells were transfected with reporter plasmid (PPRE3-tk-Luc, 100 ng) and expression plasmids for PPARα (25 ng) and STAT6 (varying amounts). (B) Coimmunoprecipitation of STAT6 and PPARα. Liver lysates from treated animals were immunoprecipitated with anti-PPARα antibody and immunoblotted for STAT6. (C) ChIP analysis of PPARα target genes. Chromatin fragments were precipitated from hepatocytes treated with vehicle or Wy14643 (Wy) in the presence or absence of IL-4. Regions flanking the PPARα binding sites on Cyp4a10 and Acot1 promoters were amplified by qPCR and data were normalized to IgG control. (D and E) Activation of STAT6 by IL-4 represses PPARα transcriptional activity. Quantitative RT-PCR analyses of PPARα target genes in primary hepatocytes. Error bars are displayed as mean ± SEM (n = 3–4 for each mouse group). *P < 0.05; **P < 0.01.
To verify the importance of this transcriptional crosstalk on endogenous PPARα target genes, primary hepatocytes were stimulated with PPARα agonist Wy14643 in the presence or absence of IL-4, and target-gene expression was monitored. As expected, stimulation of wild-type and STAT6-null hepatocytes with Wy14643 led to induction of PPARα target genes, including those important in β- and ω-oxidation of fatty acids (Acot1, Acox1, Cpt1a, and Cyp4a10) (Figs. 2 D and E). Importantly, in a STAT6-dependent manner, costimulation with IL-4 inhibited their expression (Figs. 2 D and E), indicating that phosphorylated STAT6 transrepresses PPARα. Taken together, these data demonstrate that the IL-4/STAT6 immune axis controls nutrient metabolism by repressing the catabolic program of PPARα in hepatocytes.
STAT6-Null Mice Are Resistant to Diet-Induced Obesity.
Because dietary fats serve as endogenous ligands for PPARα (23), we next studied the role of STAT6 in high-fat diet (HFD)-induced obesity and insulin resistance. When challenged with a HFD, STAT6-null mice gained significantly less weight (Fig. 3A) and had smaller WAT depots (Fig. S3A). Dual-energy X-ray absorptiometry scans confirmed ~60% reduction in total body fat mass and ~43% reduction in adiposity (Fig. 3B). Moreover, histologic and morphometric analyses of epidydimal WAT showed that STAT6-null animals had much smaller adipocytes, which failed to undergo hypertrophic changes in response to a HFD (Fig. S3 B and C). This difference in weight gain and adiposity was not caused by impaired adipogenesis or decreased food intake (Figs. S3D and S4A), implying that resistance of STAT6-null mice to HFD-induced obesity stems from increased energy expenditure. Indirect calorimetry measurement of oxygen consumption was indeed 15% to 21% higher (Fig. 3C), whereas ambulatory activity was similar between the two genotypes (Fig. S4B). Similar to lean mice (Fig. 1G), expression of PPARα protein was increased in livers of STAT6-null mice maintained on the HFD (Fig. 3D). However, in this case, there was a global increase (~2- to 13-fold) in the expression of PPARα target genes, such as Acox1, Hmgcs2, Cpt1a, Pdk4, Cyp4a10, Fgf21, Fabp1, and Acot1 (Fig. 3E), findings that are consistent with activation of PPARα by diet-derived fatty acids (23).
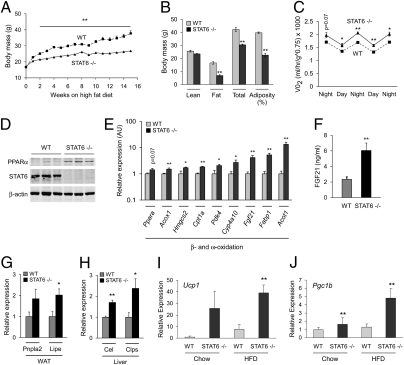
Fig. 3.
Resistance to dietary obesity and increased energy expenditure in STAT6-null mice. (A) Wild-type and STAT6−/− mice were placed on a HFD at 8 wk of age, and weight gain was monitored for 16 wk (n = 5 per genotype). (B) Dual-energy X-ray absorptiometry was used to determine body composition (n = 5 per genotype). (C) Indirect calorimetry was used to determine VO2 consumption in wild-type and STAT6−/− mice. (D) Immunoblotting for PPARα and STAT6 proteins in livers of wild-type and STAT6-null mice after 16 wk of HFD. (E) Quantitative RT-PCR analyses of total liver RNA from wild-type and STAT6−/− mice. Note induction of signature genes for β- and ω-oxidation in livers of STAT6−/− mice. (F) Serum Fgf21 levels were quantified by ELISA. (G and H) Quantitative RT-PCR analyses for adipose tissue and pancreatic lipases in WAT and liver of wild-type and STAT6−/− mice. Patatin-like phospholipase domain containing 2 (Pnpla2) and hormone sensitive lipase (Lipe) are expressed in WAT (G), whereas carboxyl ester lipase (Cel) and pancreatic colipase (Clps) are induced in liver (H). (I and J) Induction of uncoupled respiration in STAT6−/− WAT. Quantitative RT-PCR analyses for expression of Ucp1 (I) and Pgc1b (J) mRNAs in epididymal WAT of WT and STAT6−/− mice (n = 5 per genotype). Data presented as mean ± SEM. *P < 0.05; **P < 0.01.
PPARα plays an essential role in catabolic state of fasting by regulating the expression of genes important in mobilization and metabolism of fatty acids (24, 25). For example, in a PPARα-dependent manner, expression of fibroblast growth factor 21 (Fgf21) is induced in liver to promote breakdown of triglycerides in WAT (24, 26). Because transcriptional activity of PPARα is derepressed in livers of STAT6-null mice, we wondered whether there was also evidence for increased activity of Fgf21 in these animals. Indeed, expression of Fgf21 mRNA in liver (~fourfold) and its circulating levels (~threefold) were higher in STAT6-null mice (Fig. 3 E and F). The biological activity of Fgf21 was also increased, as evidenced by the induction of Fgf21 signature lipases in WAT (Pnpla2 and Lipe) (Fig. 3G) and liver (Cel and Clps) (Fig. 3H) (24). This increased breakdown of triglycerides in WAT of STAT6-null mice was also accompanied by induction of Ucp1 and the coactivator protein PGC-1β (Fig. 3 I and J), suggestive of “brownization” of WAT (27). In aggregate, these results suggest that the overall metabolic physiology of STAT6-null mice on a HFD is catabolic, resulting in increased breakdown and utilization of fatty acids.
Decreased Insulin Action in STAT6-Null Mice.
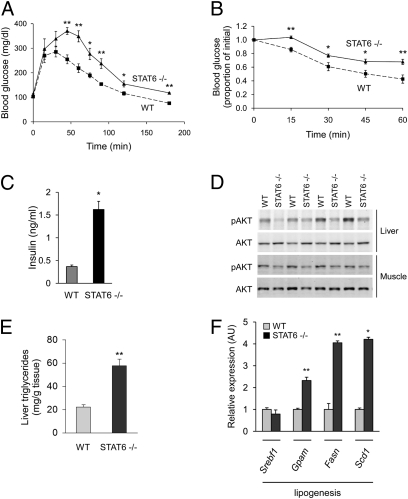
Although anabolic actions of insulin promote storage of glucose in adipose tissue, liver, and skeletal muscle, induction of PPARα enhances nutrient catabolism during fasting. In support of the polar opposite effects of insulin and PPARα signaling on nutrient metabolism, transgenic expression of PPARα in liver or skeletal muscle antagonizes anabolic actions of insulin (28, 29), whereas PPARα deficiency protects mice from diet-induced glucose intolerance and insulin resistance (29, 30). Because STAT6-null mice exhibit increased expression and activity of PPARα in the liver, we postulated that they might be susceptible to HFD-induced metabolic disease. Indeed, despite being lean, STAT6 null mice developed glucose intolerance on a HFD (Fig. 4A). This result was secondary to increased insulin resistance, as evidenced by decreased insulin sensitivity (Fig. 4B) and ~fourfold higher fasting levels of insulin in STAT6-null mice (Fig. 4C). In addition, STAT6-deficient animals exhibited other features of insulin resistance, such as increases in serum cholesterol (30%), serum triglycerides (24%), and serum levels of the adipokine resistin (~330%) (Table S1). To further explore the mechanisms behind this systemic insulin resistance, we measured the ability of insulin to activate AKT in obese animals. Insulin-stimulated phosphorylation of AKT was modestly reduced in the skeletal muscle of STAT6-null mice (Fig. 4D), whereas phospho-AKT levels were dramatically lower in STAT6-null livers (Fig. 4D), suggesting that genetic deficiency of STAT6 impairs insulin action in the liver. In agreement with this, hepatic steatosis, a cardinal feature of hepatic insulin resistance, was present in livers of STAT6-null mice, as evidenced by increased accumulation of triglycerides (Fig. 4E) and expression of lipogenic enzymes (Fig. 4F).
Fig. 4.
Insulin action is decreased in STAT6-null mice. (A and B) Impaired glucose homeostasis in STAT6-null mice fed a HFD. Wild-type (dashed line) and STAT6-null (solid line) mice (8–10 wk old) were fed HFD for 8 to 10 wk (n = 5 per genotype). Increased glucose intolerance (A) and insulin resistance (B), as assessed by intraperitoneal glucose tolerance test (2 g/kg) and insulin-tolerance test (0.75 U/kg). (C) Serum insulin levels were quantified by ELISA. (D) Decreased insulin signaling in STAT6−/− mice, as assessed by quantification of phospho-AKT. (E) Increased hepatic triglycerides in STAT6−/− mice (n = 5). (F) Induction of lipogenic gene expression in livers of STAT6−/− mice. All transcripts were measured by qRT-PCR (n = 5). Data presented as mean ± SEM. *P < 0.05; **P < 0.01.
IL-4 Administration and Th2 Polarization Improve Insulin Action.
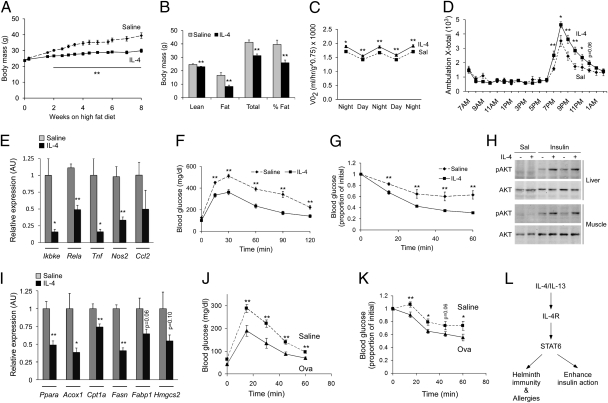
The repression of PPARα-regulated program of catabolic metabolism by the IL-4 and STAT6 raised the possibility that IL-4 signaling might exert salutary effects on metabolic disease. To test this idea, wild-type C57BL/6J mice were challenged with a HFD and concurrently treated with vehicle or IL-4. Unexpectedly, administration of IL-4 reduced weight gain and adiposity in C57BL/6J mice (Fig. 5 A and B). However, unlike STAT6-null mice, this reduction in adiposity resulted from energy expenditure associated with increased locomotor activity (Fig. 5 C and D).
Fig. 5.
IL-4 and Th2 polarization improve glucose homeostasis in mice maintained on a HFD. (A and B) IL-4 treatment protects from diet-induced obesity. Total weight gain (A) and DEXA assessment of adiposity (B). Eight-week-old C57BL/6J mice (n = 6–8 per group) were placed on a HFD for 8 wk, and concurrently treated with saline (dashed line) or IL-4 (solid line). (C and D) Indirect calorimetry measurement of VO2 consumption (C) and locomotor activity (D) (n = 6 per treatment group). (E) Quantification of inflammatory genes in WAT of saline- or IL-4–treated mice by qRT-PCR (n = 6–8 per group). (F–H) IL-4 treatment improves glucose homeostasis. Glucose tolerance (F), insulin tolerance (G), and insulin signaling (H) was assessed in saline and IL-4–treated C57BL/6J mice (n = 6–8 per treatment group). (I) IL-4 treatment suppresses expression of PPARα and its target genes in liver. (J and K) OVA-induced Th2 polarization improves glucose tolerance (J) and insulin sensitivity (K) in D011.10 mice. Eight- to 10-wk-old D011.10 mice maintained on HFD were treated with saline (dashed line) or OVA with aluminum hydroxide (solid line) for 8 wk (n = 5 per group). (L) Model of IL-4/STAT6 functions in immunity, nutrient metabolism and longevity. Data presented as mean ± SEM. *P < 0.05; **P < 0.01.
To explore this observation further, we examined the expression of NF-κB target genes in WAT because decrease in adipose tissue inflammation has been shown to increase energy expenditure and decrease weight gain on a HFD (31). Consistent with the anti-inflammatory effects of IL-4 in WAT macrophages (21), expression of IκB kinase ε (Ikbke), Rela and other NF-κB target genes, such as Tnf, Nos2, and Ccl2, was reduced by 50% to 80% in WAT of IL-4–treated mice (Fig. 5E). Thus, taken together, these data indicate that multiple different pathways contribute to the increased energy expenditure in IL-4–treated C57BL/6J mice.
Glucose and insulin tolerance tests revealed that treatment with IL-4 also improved glucose homeostasis in HFD-fed C57BL/6J mice (Fig. 5 F and G). This finding was further supported by the significantly lower levels of insulin (~44%), total cholesterol (~28%), and triglycerides (~40%) in sera of IL-4–treated animals (Table S2), indicating that therapy with IL-4 improves various metabolic indices of insulin resistance. Indeed, enhanced phosphorylation of AKT in response to exogenous insulin affirmed the improvement in insulin sensitivity in these animals (Fig. 5H). Finally, similar results on glucose clearance and insulin action were obtained when wild-type Balb/cJ mice were treated with IL-4 (Fig. S5 A and B). However, in this case, IL-4 treatment did not significantly affect total adiposity or weight gain, potentially reflecting the intrinsic Th1 and Th2 bias of C57BL/6J and Balb/cJ mice, respectively (32).
Because IL-4–mediated activation of STAT6 led to repression of PPARα and its transcriptional targets in livers of obese mice (Fig. 5I), we next examined the requirement of PPARα in the antidiabetic actions of IL-4. Although IL-4 treatment significantly improved glucose tolerance and insulin sensitivity in wild-type C57BL/6J mice, these metabolic improvements were largely absent in PPARα−/− mice (Fig. S6). Similarly, the ability of IL-4 to enhance insulin action showed complete dependence on STAT6 (Fig. S5). These data together demonstrate that the Th2 cytokine IL-4 signaling via STAT6 regulates peripheral nutrient metabolism by antagonizing the catabolic actions of PPARα in the liver and by reducing adipose tissue inflammation.
Increased production of Th2 cytokines, such as IL-4 and IL-13, normally occurs during chronic helminth infections or allergies. Therefore, to investigate whether polarization of endogenous immune response toward the Th2 axis is sufficient to protect against metabolic disease, we used a well-established model of allergic inflammation for inducing a Th2 bias in the immune response (15). DO11.10 transgenic mice, which express the rearranged T-cell receptor that recognizes chicken ovalbumin (OVA)-specific peptide sequence, were sensitized with aluminum hydroxide-adsorbed OVA. Two weeks later, mice were placed on HFD and challenged with OVA or saline for an additional 8 wk. This protocol of sensitization and challenge strongly biased the immune response toward the Th2 axis (33), as evidenced by ~14-fold higher serum levels of IgE and the robust recall responses of splenocytes to OVA and D011.10 peptide (Fig. S7). Strikingly, compared with control mice, OVA-challenged DO11.10 mice showed improvement in fasting glucose values (Saline 63.2 ± 3.7 mg/dL vs. OVA 43.4 ± 3.7 mg/dL; P = 0.003, t test), glucose tolerance, and insulin action (Fig. 5 J and K). This improvement in glucose homeostasis was independent of any change in body weight (saline 24.8 ± 0.7 g vs. Ova 25.6 ± 0.7 g, P = 0.22), suggesting that polarization of the immune response toward the Th2 axis primarily targets insulin sensitivity.
Discussion
In all metazoans, defense against pathogens is tightly coupled to regulation of peripheral metabolism. For example, in mice and humans, infections with bacteria trigger Toll-like receptor signaling, which interferes with insulin action in liver and adipose tissue, leading to the release of glucose and fatty acids to fuel the activated immune system. However, infection with helminths poses distinct metabolic challenges because these pathogens chronically parasitize host nutrients for their own growth. Results presented here are unique in demonstrating that inflammation associated with helminth infection or allergies, notably the activation of IL-4/STAT6 signaling pathway, promotes glucose disposal by enhancing insulin action, in part, via antagonism of the catabolic program controlled by PPARα in the liver and by attenuation of adipose tissue inflammation (Fig. 5L).
Before our work, the functions of IL-4 and STAT6 have primarily been investigated in the immune system, where it exerts pleiotropic effects ranging from Th2 differentiation of helper T cells, activation of B cells to release IgE, and stimulation of alternative macrophage activation (34–36). Interestingly, as reported here, the transcription factor STAT6 is also expressed in metabolic tissues, such as liver and adipose tissue. However, the ability of IL-4 to stimulate STAT6 signaling is primarily restricted to hepatocytes, because adipocytes lack the IL-4Rα on their cell surface, findings that are consistent with previous published reports (18, 19). In hepatocytes, IL-4 signaling was found to regulate the switch between glucose and fatty acid oxidation via suppression of PPARα, a pivotal regulator of β-oxidation of fatty acids. Disruption of STAT6 relieved the brake on PPARα signaling, leading to the induction of β-oxidation pathways. Conversely, activation of STAT6 by IL-4 in wild-type hepatocytes repressed the transcriptional activity of PPARα. Mechanistically, this process was mediated by direct physical interaction between STAT6 and PPARα in the liver, which prevents PPARα from being recruited to its target genes. Congruent with these observations, stimulation with IL-4 failed to suppress PPARα transcriptional target genes in STAT6-deficient hepatocytes. Finally, a recent report demonstrated that infection of mice with the helminth Schistosoma mansoni, which induces a potent Th2 response in the liver, led to marked reduction in the expression of proteins involved in citric acid cycle, fatty acid oxidation, and oxidative phosphorylation (37), processes that are known to be regulated by PPARα (38).
As a consequence, the metabolic physiology of STAT6-null mice shares many features with that of transgenic mice overexpressing PPARα in the muscle (29). First, the metabolic program of β- and ω-oxidation of fatty acids are induced in the respective PPARα-expressing tissues (i.e., hepatocytes in STAT6-null and skeletal muscle in Mck-PPARα transgenic mice). Second, both strains of mice are markedly resistant to diet-induced obesity. Although the underlying mechanisms mediating resistance to diet-induced obesity were not investigated in Mck-PPARα transgenic mice, increased metabolic respiration associated with β-oxidation of fatty acids and uncoupled respiration likely accounts for the antiobesity phenotype of STAT6-null mice. Third, despite being lean, both strains of animals developed insulin resistance after HFD feeding, which was associated with ectopic deposition of lipids. In the case of Mck-PPARα transgenic mice, this was observed in skeletal muscle, whereas we detected marked increase in the hepatic triglyceride content. Because the accumulation of fatty acid intermediates, such as acyl-CoAs and diacylglycerol, has been implicated in the development of insulin resistance (39), it provides a potential mechanism for the observed decrease in insulin signaling in these animals.
Importantly, our work has revealed that the nature of the elicited immune response is a critical determinant of insulin action. Previous work by others has shown that increase in NF-κB activity in liver causes local and systemic insulin resistance, whereas decrease in NF-κB activity in liver or myeloid cells, as in liver- and myeloid-cell specific IKKβ knockout mice, protects mice from obesity-induced insulin resistance (10). In contrast, we found that skewing of the immune response toward the Th2 axis, as in the OVA-sensitized and challenged D011.10 mice or in IL-4–treated mice, confers protection against the metabolic sequelae of high-fat feeding. This improvement likely results from actions of IL-4 in multiple tissues, such as liver and adipose tissue, where it suppresses PPARα and attenuates inflammation, respectively. Finally, although these findings provide evidence for insulin-sensitizing effects of the Th2 cyotkine IL-4, the precise sites and mechanisms of its action will require generation of mice in which IL-4 signaling is disrupted in a cell- and tissue-specific manner.
Materials and Methods
Animals.
Wild-type and STAT6−/− mice were purchased from Jackson Laboratories and bred in our animal facility. Age-matched cohorts, 8 to 10 wk of age, were assembled and used for the in vivo studies. For experiments using IL-4, wild-type, STAT6−/−, or PPARα−/− mice on Balb/cJ or C57BL/6J background were fed a HFD and simultaneously given intraperitoneal injections of IL-4 (2 μg) complexed to anti-IL-4 antibody (10 μg, BVD4-1D11; BD Bioscience) or saline alone twice a week for a period of 8 wk (40). Delivery of biologically active IL-4 was verified by assaying for increase in serum IgE. The Bio-Serv diet F3282, which derives 60% of its calories from fat, was used to promote diet-induced obesity in all animal models. Cohorts of five animals per genotype per treatment were assembled, and all in vivo studies were repeated two to three independent times. All animal care was in accordance with Stanford University's Administrative Panel on Laboratory Animal Care committee guidelines.
Metabolic Phenotyping.
Metabolic phenotyping of animals was carried out after 8 to 12 wk of HFD feeding. Glucose and insulin tolerance tests were performed using 2 mg/kg of glucose and 0.75 units/kg of human insulin by intraperitoneal injection. Tissue lysates were probed for phospho-AKT and total AKT as described previously (41). Morphometric analyses of adipocyte cell size and body composition were performed as previously described (21). Oxygen consumption, food intake, and movement were monitored using an indirect calorimetry system (Oxymax series; Columbus Instruments) in mice on a HFD for 4 to 6 wk. Mice (n = 3–6 per genotype) were acclimated to metabolic cages for two dark cycles before collection of data, which was recorded at 15-min intervals over the next three dark cycles. Experiments were repeated two to three times with independent cohorts.
Th2 Biasing of Immune Response.
Chow fed male DO11.10 transgenic mice were sensitized by serial subcutaneous injections of 100 μg OVA in sterile PBS with 0.1 mL aluminum hydroxide adjuvant on day 1 and day 7; control mice were injected with aluminum hydroxide alone (33). On day 14, all mice were placed on the HFD and subsequently given weekly injections with OVA or saline for 8 wk.
Hepatocyte Gene Expression and Metabolism.
Primary hepatocytes were isolated, and fatty acid and glucose uptake and oxidation assays were performed as previously described (20). To examine the crosstalk between STAT6 and PPARα, cultured hepatocytes were treated with Wy14643 (50 μM) or GW7647 (100 nM) for 24 h in the presence or absence of IL-4 (10 ng/mL). Total mRNA was subsequently isolated and analyzed by qRT-PCR. All measurements were made in triplicate, and each experiment was repeated at least three independent times. Data from a representative experiment are shown in the figures.
Reporter Gene Assays.
CV-1 cells were transfected with plasmids using TransIT-293 (Mirus) reagent. Reporter gene activity was quantified 14 to 16 h after stimulation with combination of human IL-4 (10 ng/mL), Wy14643 (50 μM), or GW7647 (100 nM). All luciferase data were normalized to cell-associated β-galactosidase to control for transfection efficiency. Experiments were performed in triplicate and repeated at least three independent times.
Protein Analyses.
For immunoblot analyses, the following antibodies were used: PPARα (1:250, ABR), STAT6 (M-20, 1:1,000; Santa Cruz Biotechnology) pSTAT6 (1:1,000; Imgenex), total AKT (1:1,000; Cell Signaling), and S-473 phospho-AKT (Cell Signaling). Proteins were detected on Odyssey Li-Cor using infrared dye (IR Dye 800)-conjugated secondary antibodies (1:30,000; Rockland). For coimmunoprecipitation studies, liver proteins were extracted in lysis buffer and cleared by high-speed centrifugation. Protein extracts were precleared with Protein A agarose beads for 30 min at 4 °C. The resultant protein lysate (1.5 mg) was incubated with 4 mg of anti-PPARα monoclonal antibody (ABR) or 4 mg normal mouse IgG (Caltag Laboratories) for 2 h at 4 °C followed by incubation with protein A agrarose beads for 1 h. Immunoprecipitated proteins were analyzed by Western blotting. FACS analyses of IL-4Rα expression were performed using anti-mouse CD124 antibody antibody (#552509; BD). Serum levels of cholesterol, triglycerides, NEFFA, IgE, adiponectin, insulin, leptin, and resistin were quantified using commercially available colorimetric kits or ELISAs.
ChIP Assays.
Primary hepatocytes were seeded at a density of 8 × 106 cells per 15-cm tissue culture plate, and stimulated with Wy14643 (50 mM) for 2 h, followed by IL-4 (10 ng/mL) for 30 min. ChIP was performed using the EZ-ChIP kit (Upstate Biotechnology) except for the following modifications (24). Chromatin was sonicated to an average size between 200 and 1,000 bp, and immunoprecipitations were performed with 2.5 mg of rabbit polyclonal anti-PPARα (Affinity Bioreagents). Immunoprecipitated DNA was subjected to qRT-PCR analyses using the following primer pairs that amplify the region spanning the PPARα responsive site (DR-1) or a control region. Data are expressed as percentage of input normalized to untreated samples.
Gene Expression Analyses.
Total RNA was isolated from tissues using TRIzol reagent and reverse transcribed using a first strand cDNA synthesis kit (Marligen). Quantitative real time-PCR analyses were performed on cDNA templates using EvaGreen (Biotium), as previously described (21). Relative expression of mRNAs was calculated by the ΔΔCT method using L32 mRNA for normalization. All primer sequences are available upon request.
Statistical Analyses.
All data are presented as mean ± SEM. Statistical significance was determined using the Student's t test (two-tailed, two-sample unequal variance). P values < 0.05 were deemed to be statistically significant.
Supplementary Material
Acknowledgments
We thank members of the A.C. laboratory and A. Loh for valuable comments, and Y. Ohmori for providing key reagents (Meikai University of School of Dentistry, Sakado, Saitama, Japan). This work was supported by National Institutes of Health Grants DK076760 and HL076746, the American Diabetes Association, a Larry L. Hillblom Foundation Network grant, and National Institutes of Health Director's Pioneer Award 1DP1OD6415-1 (to A.C.). Support was provided by National Research Service Award AI066402 (to R.R.R.-G.), the Stanford Medical Scientist Training Program (to J.I.O. and A.R.E.), the American Heart Association (J.I.O.), The Howard Hughes Medical Institute Gilliam fellowship (to A.R.E.), and the Larry L. Hillblom Foundation Postdoctoral fellowship (to H.J.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009152108/-/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Cramer T, Johnson RS. A novel role for the hypoxia inducible transcription factor HIF-1alpha: Critical regulation of inflammatory cell function. Cell Cycle. 2003;2:192–193. [PubMed] [Google Scholar]
- 4.Kellett DN. 2-Deoxyglucose and inflammation. J Pharm Pharmacol. 1966;18:199–200. doi: 10.1111/j.2042-7158.1966.tb07853.x. [DOI] [PubMed] [Google Scholar]
- 5.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: Energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 6.Newsholme P, Curi R, Gordon S, Newsholme EA. Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem J. 1986;239(1):121–125. doi: 10.1042/bj2390121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odegaard JI, Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab. 2008;4:619–626. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diangelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci USA. 2009;106:20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 10.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrante AW., Jr Obesity-induced inflammation: A metabolic dialogue in the language of inflammation. J Intern Med. 2007;262:408–414. doi: 10.1111/j.1365-2796.2007.01852.x. [DOI] [PubMed] [Google Scholar]
- 12.Maizels RM, Yazdanbakhsh M. Immune regulation by helminth parasites: Cellular and molecular mechanisms. Nat Rev Immunol. 2003;3:733–744. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]
- 13.Munene E, et al. Helminth and protozoan gastrointestinal tract parasites in captive and wild-trapped African non-human primates. Vet Parasitol. 1998;78:195–201. doi: 10.1016/s0304-4017(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 14.Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19:2577–2584. doi: 10.1038/sj.onc.1203485. [DOI] [PubMed] [Google Scholar]
- 15.Tomkinson A, Duez C, Lahn M, Gelfand EW. Adoptive transfer of T cells induces airway hyperresponsiveness independently of airway eosinophilia but in a signal transducer and activator of transcription 6-dependent manner. J Allergy Clin Immunol. 2002;109:810–816. doi: 10.1067/mai.2002.123531. [DOI] [PubMed] [Google Scholar]
- 16.Kuperman DA, Schleimer RP. Interleukin-4, interleukin-13, signal transducer and activator of transcription factor 6, and allergic asthma. Curr Mol Med. 2008;8:384–392. doi: 10.2174/156652408785161032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkelman FD, et al. Interleukin-4- and interleukin-13-mediated host protection against intestinal nematode parasites. Immunol Rev. 2004;201(1):139–155. doi: 10.1111/j.0105-2896.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 18.Deng J, et al. Interleukin-4 mediates STAT6 activation in 3T3-L1 preadipocytes but not adipocytes. Biochem Biophys Res Commun. 2000;267:516–520. doi: 10.1006/bbrc.1999.1993. [DOI] [PubMed] [Google Scholar]
- 19.Hua K, Deng J, Harp JB. Interleukin-4 inhibits platelet-derived growth factor-induced preadipocyte proliferation. Cytokine. 2004;25:61–67. doi: 10.1016/j.cyto.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4(1):13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarthy MV, et al. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Kersten S, et al. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badman MK, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal-Mizrachi C, et al. Dexamethasone induction of hypertension and diabetes is PPAR-alpha dependent in LDL receptor-null mice. Nat Med. 2003;9:1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- 29.Finck BN, et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 2005;1:133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Tordjman K, et al. PPARalpha deficiency reduces insulin resistance and atherosclerosis in apoE-null mice. J Clin Invest. 2001;107:1025–1034. doi: 10.1172/JCI11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang SH, et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell. 2009;138:961–975. doi: 10.1016/j.cell.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinzel FP, Sadick MD, Holaday BJ, Coffman RL, Locksley RM. Reciprocal expression of interferon gamma or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169(1):59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck L, Spiegelberg HL. The polyclonal and antigen-specific IgE and IgG subclass response of mice injected with ovalbumin in alum or complete Freund's adjuvant. Cell Immunol. 1989;123(1):1–8. doi: 10.1016/0008-8749(89)90263-3. [DOI] [PubMed] [Google Scholar]
- 34.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Kishimoto T, Akira S. STAT6: Its role in interleukin 4-mediated biological functions. J Mol Med. 1997;75:317–326. doi: 10.1007/s001090050117. [DOI] [PubMed] [Google Scholar]
- 36.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 37.Harvie M, Jordan TW, La Flamme AC. Differential liver protein expression during schistosomiasis. Infect Immun. 2007;75:736–744. doi: 10.1128/IAI.01048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: From genes to physiology. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 39.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finkelman FD, et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- 41.Odegaard JI, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.