Abstract
Compared with many well-studied enveloped viruses, herpesviruses use a more sophisticated molecular machinery to induce fusion of viral and cellular membranes during cell invasion. This essential function is carried out by glycoprotein B (gB), a class III viral fusion protein, together with the heterodimer of glycoproteins H and L (gH/gL). In pseudorabies virus (PrV), a porcine herpesvirus, it was shown that gH/gL can be substituted by a chimeric fusion protein gDgH, containing the receptor binding domain (RBD) of glycoprotein D fused to a truncated version of gH lacking its N-terminal domain. We report here the 2.1-Å resolution structure of the core fragment of gH present in this chimera, bound to the Fab fragment of a PrV gH-specific monoclonal antibody. The structure strongly complements the information derived from the recently reported structure of gH/gL from herpes simplex virus type 2 (HSV-2). Together with the structure of Epstein-Barr virus (EBV) gH/gL reported in parallel, it provides insight into potentially functional conserved structural features. One feature is the presence of a syntaxin motif, and the other is an extended “flap” masking a conserved hydrophobic patch in the C-terminal domain, which is closest to the viral membrane. The negative electrostatic surface potential of this domain suggests repulsive interactions with the lipid heads. The structure indicates the possible unmasking of an extended hydrophobic patch by movement of the flap during a receptor-triggered conformational change of gH, exposing a hydrophobic surface to interact with the viral membrane during the fusion process.
Keywords: herpesvirus entry, herpesvirus envelope proteins, membrane fusion, syntaxins and intracellular vesicle fusion, protein disulfide isomerase motif
The members of the Herpesviridae family of enveloped DNA viruses infect a broad range of organisms (1). Their classification into α-, β- and γ-subfamilies is based on evolutionary relatedness, tropism, and properties of the viral cycle. α-Herpesviruses have the widest host range and establish latency in the nervous system after a rapid lytic phase. β-Herpesviruses are characterized by a slower lytic cycle and cause latent infections of a variety of tissues. γ-Herpesviruses have oncogenic properties and cause latent infections of lymphoid cells.
Herpesviruses display about a dozen envelope proteins at their surface—the exact number depends on the virus. A subset of these glycoproteins is necessary for fusion of viral and host cell membranes during entry into target cells (2). This core subset is composed of glycoproteins B, H, and L (gB, gH, and gL) and is conserved across the three subfamilies. Crystallographic studies of the gB ectodomain from herpes simplex virus type 1 (HSV-1) (3) and Epstein-Barr virus (EBV) (4) revealed structural homology with the vesicular stomatitis virus envelope glycoprotein G (5), introducing a third structural class of viral membrane fusion proteins (6). Structural information on gH and gL has been lacking until the very recently reported structure of the gH/gL ectodomain complex of herpes simplex virus type 2 (HSV-2) (7). Despite the structural data now available on gB and gH/gL, the molecular mechanism of protein-induced membrane fusion during cell entry of herpesviruses remains to be understood.
gB and gH are both type I transmembrane (TM) proteins, with a large N-terminal ectodomain and a small cytosolic tail, whereas gL is not membrane anchored and associates noncovalently with the gH ectodomain. Despite its conservation in all herpesviruses, gH displays substantial variability in sequence and length, especially in its N-terminal half, even when considering viruses from the same subfamily (Tables S1 and S2). We have concentrated on structural studies of envelope proteins of pseudorabies virus (PrV), a porcine herpesvirus of veterinary concern. The ectodomain of PrV gH, with 622 amino acids, is shorter and likely more compact than its counterparts from other herpesviruses for which sequences are available (Table S1).
PrV belongs to the α-Herpesvirinae subfamily, together with notable human pathogens such as HSV-1, HSV-2, and varicella-zoster virus (VZV). PrV, which is a member of the same genus as VZV (Varicellovirus genus), is the causative agent of Aujeszky's disease in swine (8). The high morbidity and mortality rates associated with PrV infections cause substantial economic losses worldwide. Thus, PrV has been studied intensively and serves as a model to understand α-herpesvirus biology in general (9).
As with other α-herpesviruses (2), an essential step during entry into target cells is binding of the PrV envelope glycoprotein D (gD) to a specific entry receptor, herpesvirus entry mediator C (HveC) (10). This interaction signals the activation of the viral fusogenic machinery, composed of gB and the gH/gL heterodimer. The ensuing fusion of viral and cellular membranes results in the release of the viral capsid and tegument into the cytoplasm of the target cell. In contrast to HSV-1 and -2, PrV does not require the presence of gD for spreading from an infected cell to a neighboring, contacting cell (11). Another difference is that PrV gH is detected at the surface of the cell expressing it—and gets incorporated into virions—in the absence of gL. In all other herpesviruses for which this process has been studied, gH does not fold properly in the absence of gL and remains in the endoplasmic reticulum to be degraded by the unfolded protein-sensing system of the cell. This feature of PrV was used to study the properties of recombinant virions lacking gL (ΔgL), as well as virions that lack gD (ΔgD). The ΔgL and ΔgD PrV deletion mutants, in spite of being noninfectious from the medium, retain a certain degree of cell-to-cell spreading ability (11, 12). This property was used to subject the recombinant ΔgD and ΔgL PrV deletion mutants to repeated passaging by coseeding infected and uninfected cells, to eventually isolate infectious PrV variants capable of invading cells from the medium in the absence of gD or gL (variants termed ΔgD-Pass and ΔgL-Pass, respectively) (11, 13). In the ΔgD-Pass strain, the absence of gD is compensated, at least partially, by a single mutation (Ala64 to Pro) in gH, which maps to the N-terminal segment not present in the truncated gH molecule described here (see below). In addition, three mutations in gB were observed (14): a short insertion in the N-terminal region, which is disordered in the available structures of HSV-1 (3) and EBV gB (4), and two point mutations (A380V and L416P) mapping to the plekstrin homology domain (domain II in the HSV-1 gB structure). In contrast to these local alterations to compensate for the absence of gD, the ΔgL-Pass variant showed a more dramatic genetic rearrangement, resulting in a chimeric gDgH fusion protein. The gDgH chimera contained the receptor binding domain (RBD) of PrV gD (comprised within the amino-terminal 271 residues) fused to a truncated gH lacking the “N-domain,” consisting of its ∼100 N-terminal residues (counting the signal peptide) (13). These results showed that under selective pressure, PrV gL and gD become dispensable for cell invasion from the external medium. gH and gB therefore have the inherent functional requirements to allow entry of PrV virions into cells and are both necessary and sufficient to cause fusion of the viral envelope with the target cell membrane.
The chimeric gDgH fusion protein found in the ΔgL-Pass variant was further shown to substitute for gD, gH, and gL in both virus-free fusion of transfected cells (15) and in complementation assays (13). The chimera thus carries out gL-independent PrV entry, indicating that neither the gH N-domain nor gL are required for membrane fusion. The gH Ala64 to Pro mutation in the ΔgD-Pass PrV variant suggests that the N domain, in association with gL, might be used for interactions with the cell, in line with the observation that it is substituted by the gD RBD in the ΔgL-Pass variant. In this work, we generated a core fragment of the PrV gH ectodomain (gHC), lacking the N domain, for structural studies. We report the 2.1-Å resolution structure of gHC in complex with the Fab fragment of mAb A13-d6.3 (Fab d6.3), which neutralizes the ΔgL-Pass PrV variant.
Results and Discusson
Structure of the PrV gHC:Fab d6.3 Complex.
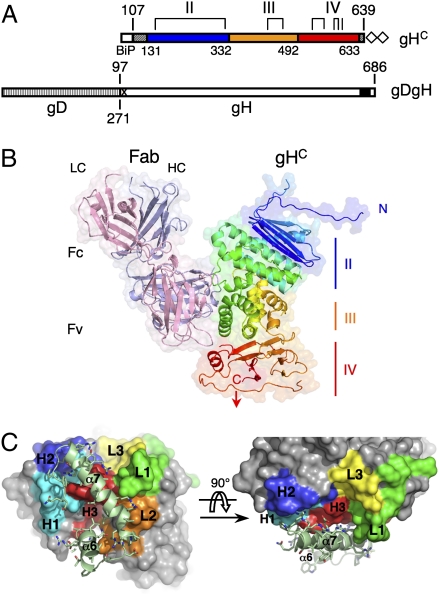
The constructs used for crystallization and the details of the structure determination are described in SI Materials and Methods, and the crystallographic statistics are reported in Table S3. The gHC protein includes amino acids 107–639 of PrV gH (Fig. 1A). The gHC:Fab d6.3 structure shows that gHC has an elongated, gently curved shape, roughly 70 Å in length and 50 Å in cross-section, folded as three sequential domains, with the N and C termini located at opposite ends. Fab d6.3 binds to the convex side, toward the C-terminal membrane proximal end (Fig. 1B). The crystals show clear electron density for 470 amino acids out of the 533 residues of the gHC construct. The disordered regions in the gHC structure are the N-terminal segment of 34 amino acids (107–130), five internal loops (198–210, 448–450, 469–482, 543–546, and 603–607) and the C-terminal five residues that connect to the TM region (634–639) (Fig. S1).
Fig. 1.
Structural organization of the PrV gHC:Fab d6.3 complex. (A) Schematic representation of the construct used for expression of PrV gHC (Upper) together with the gDgH chimera (Lower), for comparison. Domain boundaries are indicated, with the sequential domains colored blue, orange, and red. Residues 107–130 at the N terminus and residues 634–639 at the C terminus (stippled gray boxes) are disordered and are not resolved in the gHC structure. The four disulfide bonds are marked with brackets, and the last cysteine (which is free) with a vertical line. The double strep-tag used for purification is represented with empty diamonds. Boundaries of gD and gH within the gDgH chimera are marked Below and Above the schematic diagram, respectively. A black box represents the predicted transmembrane region of PrV gH (residues 647–667); the proline-rich region (PRR) excluded from recombinant gHC is marked by an X (SI Materials and Methods). (B) Structure of the PrV gHC:Fab d6.3 complex. The heavy and light chains (HC and LC) of Fab d6.3 are light blue and pink, respectively. PrV gHC is rainbow colored from N (blue) to C terminus (red). (C) PrV gHC: Fab d6.3 interactions. The epitope, composed of helices α6 and α7, is shown as light green ribbons. The Fab is shown as a solid molecular surface, with the CDR loops color coded: green (L1), orange (L2), yellow (L3), cyan (H1), blue (H2), and red (H3).
The d6.3 Fab contacts residues 332–373 of gH, in a region including 2 α-helices at the gHC surface (α6 and α7, Fig. 1C). Five of the six complementarity determinant regions (CDR) of the Fab interact with gH: all three CDRs from the heavy chain (loops H1, H2, and H3) and loops L1 and L2 of the light chain (Fig. 1C and Fig. S2), burying 950 Å2 of surface area per protomer. The interactions with the Fab are made mostly through side chain contacts. Tables S3 and S4 list all of the observed atomic antibody/antigen interactions, with details displayed in Fig. S2. The majority of the contacts involve gHC residues Glu361, Arg365, and Lys370 (Fig. S2D).
mAb A13-d6.3 is the only reported antibody binding to PrV gH, which is a poorly immunogenic glycoprotein (12, 13). This is in contrast to gH from HSV-1 and -2, for which numerous mAbs have been obtained, the majority of which maps to the N-terminal half of the protein (16–20). The lack of immunogenicity of the N domain of PrV gH is likely due to the absence of a rigid structure, given its high Pro/Gly content (29%) compared with HSV-2 and EBV gH (13 and 18%, respectively). In line with this observation, the gHE/gL heterodimer, containing the intact gH ectodomain (gHE), failed to crystallize in our hands (SI Materials and Methods), suggesting that domain I (which would be composed of gL and the N domain of gH (see below), is mobile and probably poorly structured.
Immunostaining experiments (Fig. S3A) showed binding of mAb A13-d6.3 to ΔgL-Pass virions and no binding to wild-type PrV, in line with previous results indicating that it neutralizes the ΔgL-Pass mutant but not wild type. However, our experiments demonstrate that the soluble recombinant gHE/gL heterodimer is efficiently recognized by Fab d6.3 in solution (Fig. S3B), indicating that the epitope is accessible in the presence of gL (the location of the mAb A13-d6.3 epitope in the context of the surface properties of gH is outlined in Fig. S4; it is also displayed as ribbons in Fig. S5). Immunostaining experiments with polyclonal anti-gH sera showed that the ΔgL-Pass variant appears to have a much higher density of the gHgD chimera at its surface, compared with the apparent density of gH/gL heterodimers in wild-type virions (Fig. S3A). The fact that the mutant would need a higher copy number of the gDgH chimera per virion is not surprising, given that the virus has not evolved to function that way, and the chimera is likely to be less efficient in entry than wild-type gH/gL. This feature would also make the gDgH chimera more easily neutralized by the mAb, explaining the previous observations. Additional studies on the relative copy number of the various herpesvirus entry glycoproteins are clearly necessary; they could provide further clues to understand the mechanism involved.
gHC Architecture.
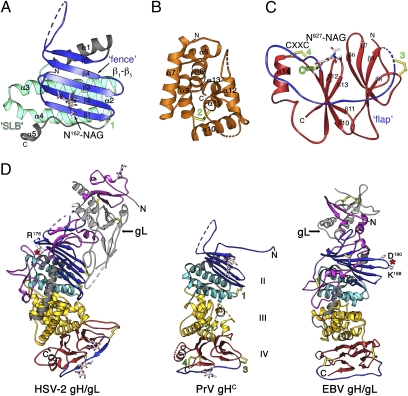
Despite poor sequence conservation (Fig. S1 and Table S1), PrV gHC displays the same domain organization as EBV gH, presented in the accompanying manuscript (21), and as HSV-2 gH (7) (Fig. 2; see also Table S2), although the domain boundaries reported for the latter are different. The N-terminal truncation in gHC (residues 25–106) results in the absence of domain I, with the structure displaying the remaining domains, II to IV. Comparison with the HSV-2 and EBV gH/gL structures indicates that domain I in PrV gH is formed by the N domain in association with gL and that it would be similar in size to domain I in EBV gH/gL, i.e., about half that of its counterpart in HSV-2 gH/gL.
Fig. 2.
Domain organization of the PrV gHC fragment. (A–C) PrV gHC Individual domains II, III, and IV, respectively, with the N and C termini labeled. In domain II, the conserved structural elements, fence and SLB, are highlighted in blue and cyan, respectively, with the remainder in gray; domain III is colored orange and domain IV in red with the flap highlighted in blue. Green labels mark the four disulfides, displayed in yellow ball-and-stick, as is the free Cys573. The glycans attached to Asn162 and Asn627 (labeled) are displayed as sticks colored according to atom type. (D) Side-by-side comparison of the HSV-2 gH/gL (PDB code 3M1C), PrV gHC (this work), and EBV gH/gL (accompanying paper, ref. 21). The proteins were aligned structurally on the SLB (cyan) and are displayed in the same orientation, colored according to domains as in A–C. The assignment of domains II–IV of the three proteins was made in reference to the PrV domains represented in A–C. The N domain, missing in PrV gH, is shown in magenta and gL in pale gray which together constitute domain I in HSV-2 and PrV gH/gL. Regions that do not have a superposable counterpart in the different structures are colored in dark gray (in particular, the connection between domains II and III, which has different interfaces in the three structures). The location of the integrin binding sites RGD176–178 in HSV-2 gH and KGD188–190 in EBV gH are indicated with red stars.
Domain II.
Domain II comprises residues 131–332 (Fig. 2A and Fig. S6A) and has an α/β-fold. The segment 131–143, spanning the gHC N-terminal residues with visible electron density, is in an extended conformation and makes an antiparallel β-strand (termed β0) with its counterpart from a neighboring gHC molecule in the crystal (Fig. S5). Comparison with the structures of EBV and HSV-2 gH indicates that β0 is part of an upstream long loop that folds back against domain II in the full-length molecule. Immediately downstream is an antiparallel β-sheet (the “fence”) with up and down topology (strands β1 through β5), featuring a long β4β5 loop in which 13 residues are disordered (198–210). The fence is followed by helix α1, which connects to an elongated antiparallel 3-helix bundle, composed of helices α2–α4, which we term “syntaxin-like bundle” (SLB) because of the strong structural similarity to the N-terminal domain of syntaxins 1A and 6 (Dali scores of 6.2 and 7.5, respectively, Fig. S6B). The presence of the SLB went unnoticed in the description of the structure of HSV-2 gH (composed of helices α7–α9 in that structure), perhaps because it was grouped into a single domain with the α-helices of domain III (see below). The fence and the SLB are the conserved elements of domain II, with helices α3 and α4 packing tightly against the β1β2β3 portion of the β-sheet, burying aromatic and aliphatic side chains. This interface is relatively conserved in all three structures that we have compared (PrV, HSV-2, and EBV gH). In contrast, helix α1 is variable in length and orientation, and the packing of the SLB with domain III also differs in the various structures. This is illustrated in Fig. 2D, in which the three molecules, HSV-2 gH/gL, PrV gHC, and EBV gH/gL are structurally aligned on the SLB and displayed in the same orientation. In PrV and HSV-2 gH, the interactions with the SLB appear to force the β-sheet to be planar, like a fence, instead of presenting the standard right-handed twist typical of β-sheets, indicating that the SLB imposes an important structural constraint. The conserved, tight packing of the SLB against the fence argues against the distribution of these two tertiary structure elements into two separate domains (domains H1 and H2, as suggested for HSV-2 gH) (7). Furthermore, whereas the side of the fence packing against the SLB is very hydrophobic, its opposite side displays only polar residues, including a glycan attached to Asn162 on strand β2, which would not form the core of a separate domain. Disulfide bond 1, connecting the end of the last helix in the SLB with the tip of β1 in the fence (Fig. 2A and Fig. S6A) further clamps these two tertiary structure elements together.
Domain III.
Domain III (residues 333–492) is an α-helical bundle composed of eight consecutive helices (α6 to α13, Fig. 2B and Fig. S6C). The first four helices in the sequence are arranged as two imperfect HEAT repeats (22). Helices α8 and α11 are at the center of the bundle, and all of the others are at the periphery. The peripheral helices α6 and α7 contribute most of the residues that form the epitope recognized by mAb A13-d6.3, as described above.
The two central helices, α8 and α11, span patches of strong sequence conservation across herpesviruses, in particular helix α11, which breaks at the conserved sequence 437SerProCys439, giving rise to the 3/10 helix η4 at its C-terminal end (Fig. S1). Pro438 introduces a distortion so that Cys439 can make a strictly conserved disulfide bond with Cys404 (disulfide 2, Fig. S6C). A long, mobile loop connects the last two helices, α12 and α13, in which 14 residues are disordered. The same loop is shorter in gH from β-herpesviruses and is missing altogether in the γ-herpesviruses (Fig. S1).
Interface between domains II and III.
The contacts between these two domains in PrV gH are less extensive than between the SLB and the fence. The end of domain II features a stretch of extended polypeptide (between helices α4 and α5, Fig. S1 and Fig. 2A) wedged in between strand β1 in the fence and helix α4 in the SLB, with the short helix α5 at its very end. The other α-herpesviruses have a 20-aa-long insertion at the end of this segment (Fig. S1), as the chain enters domain III. In the structure of HSV-2 gH, this insertion contains a long α-helix (termed helix α11 in that structure (7), displayed in dark gray in Fig. 2D). Whereas domains II and III are individually similar and display high DALI scores (Table S2), the presence of helix α11 in HSV-2 gH alters considerably the domain interface, contributing to a different packing angle. Indeed, the domain II/III interface is at the “heel” of the boot-shaped HSV-2 gH/gL complex, whereas in PrV gH the three domains are along a rod, similar to EBV gH/gL.
Domain IV.
Domain IV (residues 493–633) is the most conserved domain of gH. It is a β-sandwich formed by two apposed four-stranded β-sheets, β6β7β9β8 and β13β12β10β11, which are sequential and are therefore termed N and C sheets, respectively (Fig. 2C and Fig. S6D). The β-sheets are oriented with the strands perpendicular to the long axis of gH, so that one edge of the sandwich packs against domain III and the other is at the distal end of the rod-like molecule. The topological arrangement of the β-strands is unlike any eight-stranded antiparallel β-barrel described previously. Helix α14, in the β11β12 loop, packs against the C sheet, which becomes sandwiched in between the N sheet and the helix. Domain IV has two disulfide bridges, 3 and 4, and a free cysteine (Cys573) at the beginning of strand β10.
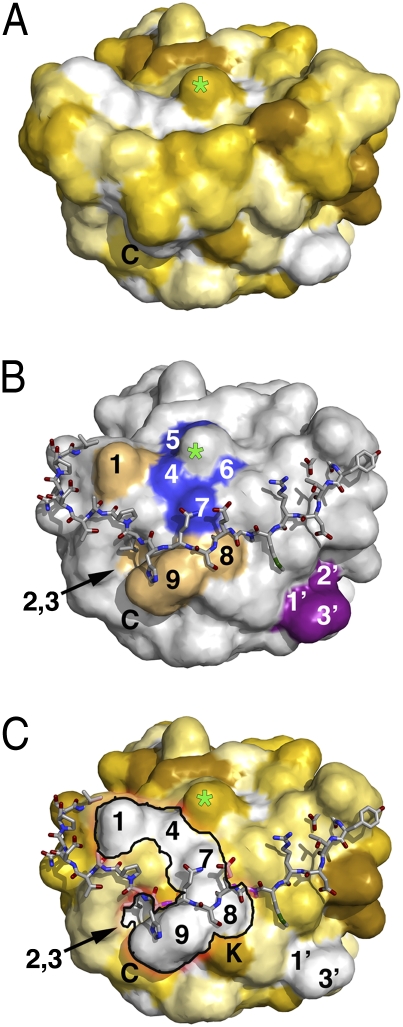
A glycan residue is attached to Asn627 in the loop connecting the last two strands, β12 and β13, near the C terminus of the ectodomain. This glycosylation site is conserved throughout the Herpesviridae family (Fig. S1), suggesting that it may have a functional role. Its removal was shown, however, not to affect the growth kinetics of a recombinant HSV-1 strain in tissue culture (18). The glycosylated Asn627 is part of a highly conserved segment of the gH sequence that encompasses amino acids 622–633 (Fig. S1 and Fig. 3B), which creates a hydrophobic surface patch, suggesting that a function for the glycan at this position could be to partially mask the hydrophobic patch from solvent (Fig. 3).
Fig. 3.
A flap capping the end of domain IV. (A) Solvent-accessible surface of gHC viewed down the C-terminal end (C terminus labeled “C”) and colored according to the Eisenberg hydrophobicity scale (39) from dark-yellow/brown (minimal) to white (maximal hydrophobicity). A green star marks the position of the conserved N-glycosylated Asn627 (the glycan was not included in the surface calculation) (B) The flap, shown as sticks, was removed from the surface calculation, displayed in gray. Strictly conserved residues in α-herpesvirus gH surrounding the glycosylated Asn627 are colored blue (5, Pro626; 4, Phe625; 7, Val630; and 6, Gly628). Additional hydrophobic residues (1, Val569; 8, Val631; 9, Leu633; 2, Phe575; 3, Leu599) forming a continuous hydrophobic surface, depicted in C), are colored light orange. The patch containing Val529, mentioned in the text, is colored purple (1′, Val529; 2′, Ile531; 3′, Pro534). (C) Same as B, but with the surface colored as in A. The hydrophobic patch (1′-2′-3′) containing residues Val529 and Ile531, makes a continuous hydrophobic surface with patch ‘1–4-7–8-9’, especially when counting the aliphatic part of the side chain of Lys620 (labeled “K”) lying underneath the flap.
Residue Val691 in HSV-1 gH, which corresponds to Val529 in PrV (Fig. S1) and is conserved in α-herpesviruses, has been identified as functionally important by insertional mutagenesis monitoring complementation of a gH-negative virus for fusion and infectivity (18). Val529 is located in the exposed loop between strands β8 and β9 in the N sheet (Fig. 3B). Together with Ile531 and Pro534, it forms a second hydrophobic patch in domain IV (Fig. 3C). The surface location of these residues strongly suggests that their perturbation by mutagenesis would be unlikely to interfere with the structural integrity of domain IV. Their conservation rather suggests a functional role, which could be providing binding sites for other viral envelope proteins, or participating in interactions with membranes, as proposed below.
The “Flap” and a Protein Disulfide Isomerase (PDI) Motif in gH.
An intriguing feature, conserved in the structures of gH from the various herpesviruses analyzed, is the connection between the N and C sheets of domain IV, via a long crossover segment of polypeptide chain, the flap (residues Asn542 to Cys573 connecting β9 and β10) (Fig. 2C and Fig. S6D). The flap runs across the bottom edge of the molecule, closing the β-barrel at the C-terminal membrane-proximal end, in a region displaying a strong negative electrostatic surface potential (Fig. S4). The flap is clamped at both distal ends by disulfide bonds 3 and 4, and its conformation appears as a flexible cap. The structure suggests that a modest conformational change moving the flap out of place would expose a single continuous hydrophobic surface, connecting the two conserved patches mentioned above (Fig. 3 A and C). Very similar hydrophobic patches covered by the flap, are also present in gH of HSV-2 and EBV (Fig. S7).
Disulfide 4, at the C-terminal side of the flap, is made by the two cysteines of a CXXC PDI motif, conserved across the Herpesviridae family with the notable exception of HSV-1 and -2 (Fig. S1). It is also noteworthy that in PrV, the PDI motif is extended to make a 568CXXCXC573 motif, with the last cysteine free (Fig. 2C). The cysteines are arranged such that a small distortion would allow disulfide reshuffling to connect C and C+5 instead of C and C+3. This possible reshuffling could be a way to stabilize a putative alternative conformation of the flap to expose the hydrophobic patch underneath. A PDI motif next to a free Cys has been observed in the envelope proteins of γ- and δ-retroviruses, where it is involved in releasing the disulfide bond across SU and TM subunits, allowing TM to undergo a fusogenic conformational change (23).
The fact that the free Cys573 of PrV gH is not conserved in the other herpesviruses indicates that stabilization of a putative “open” conformation of the flap by an alternative intramolecular disulfide bond is not an absolute requirement. On the other hand, the conservation of the PDI motif in so many distant herpesviruses (Fig. S1) is a strong indication of a possible functional relevance. The CXXC motif could be important only for proper folding in the endoplasmic reticulum (ER), a requirement that would not be needed for gH from HSV-1 and -2 in spite of sharing the same fold in domain IV. The location of the CXXC motif at the periphery of the molecule, at one end of the flap and not at the folding core of the domain, argues against a role during folding. Its absence in HSV-1 and -2 gH, which otherwise display the flap in the same location (Fig. S7), suggests that these viruses would have evolved a different way of activating exposure of the hydrophobic patch underneath the flap, if this is indeed the mechanism of interaction of gH with membranes. Such a role is supported by mutations of EBV gH residues that are part of the flap, which were shown to affect fusion (21, 24).
Functional Implications.
In contrast to gB, the structure of gH bears no structural homology to any of the characterized viral membrane fusion proteins. However, gB by itself is not sufficient to drive membrane fusion, requiring a concerted action with the gH/gL heterodimer. A role in membrane fusion for gH/gL is supported also by reports showing that it can cause hemifusion (25) as well as low levels of membrane fusion in the absence of gB (26, 27). The fact that herpesviruses need several proteins to induce membrane fusion has parallels with fusion of intracellular vesicles with their target membranes, for instance fusion with the plasma membrane to release neurotransmitters at synapses. Although the exact role of gH in the fusion process is not understood, the structural studies suggest that it could act like synaptotagmins, which work in conjunction with the SNARE proteins to drive fusion (28). Synaptotagmins have “C2” domains, which insert into the cytoplasmic leaflet of the vesicle membrane upon binding of Ca2+ ions, thereby inducing a strong, destabilizing local curvature in the lipid bilayer, lowering the energy required for the actual fusion step (29). In the case of herpesvirus fusion, the topology is opposite to that of intracellular vesicles, with the outer leaflet of the virus merging to the outer (or luminal) leaflet of the cellular membrane during hemifusion. Ca2+ ions are therefore not a likely signal, in contrast to the corresponding processes occurring at the cytoplasmic side. For α-herpesviruses, gD binding to its receptor signals the activation of the gH/gL and gB fusion machinery. In the case of HSV-1, there is evidence pointing to a physical interaction between gH/gL and gB upon the gD signal (30), similar to the interaction between synaptotagmins and the SNAREs. The trigger for fusion can be, in addition to binding to gB as discussed in (7), a change of conformation of the gH molecule itself, exposing a hydrophobic patch to interact with membranes and induce a strong, destabilizing local curvature, similar to the role of synaptotagmins.
A further link to intracellular membrane fusion proteins is the presence of the SLB in domain II. Syntaxins are members of the SNARE family of proteins (31) and are anchored by their C-terminal end to the cytoplasmic side of target membranes (tSNAREs). Their N-terminal domain (called Habc domain in the case of syntaxin 1) is followed by a region that adopts an alpha-helical conformation to oligomerize with the other SNARE proteins to form the SNARE complex to drive membrane fusion. In gH, the SLB is followed directly by the α-helical bundle of domain III, which is in turn followed by the flap-containing domain IV. There is no evidence indicating that gH undergoes a major fusogenic conformational change in which the helices tightly folded within domain III would become available for oligomerization or for interaction with membranes. A more subtle conformational change, exposing the hydrophobic patch at the membrane proximal base of domain IV thus appears more plausible and is more in line with the existing mutational data.
Concluding Remarks.
The structure of the HSV-2 gH/gL complex was recently reported (7), providing the overall 3D organization of the molecule and revealing the intricate heterodimerization of its N-terminal segment with gL. In parallel, the accumulated biological data on PrV gH has allowed the dissection of regions of gH that are involved in membrane fusion and those that are likely to be used for interactions with the host cell, as evidenced by the fact that the N-domain together with gL can be replaced by the RBD of gD in a functional chimeric protein. The lack of sequence similarity in the N-terminal half precluded engineering similar chimeras for HSV-1 or -2, and the present structure now fills that void. In addition, the fact that neither gD nor gL is strictly necessary for PrV spread across cells, reinforces the notion that gD and domain I of gH/gL may be involved in interactions of α-herpesviruses with receptors that are necessary only for invasion from the medium. In line with this notion, the integrin αVβ3 binding motif in HSV-2 gH (32), composed of residues RGD176–178, maps to domain I (Fig. 2D).
Comparison with the structure of the more distant EBV gH/gL heterodimer highlights additional conserved structural features that were previously unnoticed. One is the SLB, with identical packing of the helices within the bundle. Although the structural similarity between the SLB and the syntaxin N-terminal domain is clear, there is a nonnegligible chance that it is fortuitous, given the simple nature of the three-helix motif. However, the parallel it brings with intracellular vesicular fusion is intriguing. Finally, the presence of the flap in all three structures, masking a conserved hydrophobic patch in the context of a strong negative electrostatic surface potential that would make repulsive interactions with the viral membrane, is another important feature emerging from this comparative structural analysis. Thus the various structures of gH, revealing intriguing structural similarities with other cellular and viral proteins involved in membrane fusion, open the way to structure-driven site-directed mutagenesis to unveil the mechanism of herpesvirus-induced membrane fusion.
Materials and Methods
Production of the Recombinant PrV gHC and Fab d6.3 Proteins.
The generation of the PrV gHC and gL expression constructs is described in SI Materials and Methods. Drosophila melanogaster Schneider 2 (S2) cells were cotransfected with 2 μg of each of the expression plasmids, and 10 ng of a plasmid carrying a hygromycin resistance gene. Stable S2 cell lines expressing gHC and gL were obtained by selection in medium containing 400 mg/L hygromycin using established protocols (33). Protein production was induced by addition of 0.5 M copper sulfate, and the supernatant was harvested 7 d after induction. gHC was purified from the supernatant using a Strep-Tactin Superflow high-capacity 1-mL column (IBA GmbH), followed by size exclusion chromatography on a HiPrep 16/60 Superdex S200 column (Pharmacia) in 10 mM Tris pH 8, 50 mM NaCl buffer. Typical yields of pure gHC were 8–12 mg/L of cell culture. The cloning, expression, and purification of Fab d6.3 were reported previously (34).
Crystallization of the PrV gHC:Fab d6.3 Complex.
The complex of PrV gHC with Fab d6.3, which was initially obtained by papain cleavage of the A13-d6.3 mAb (complexAb), was generated by mixing equimolar amounts of the two proteins. The in situ-generated complex was immediately used for setting up crystallization drops. Crystals appeared overnight in 0.1 M imidazole pH 8, 0.2 M sodium acetate, and 12–14% PEG 4,000.
The complex of PrV gHC with the recombinant Fab d6.3 (complexR) was obtained by mixing purified gHC and Fab in 1:1.3 M ratio. ComplexR was separated from the excess Fab by size exclusion chromatography. Crystals of complexR grew in 0.1 M imidazole pH 8, 0.7 M sodium formate and 10% PEG 4,000 and were soaked in the same solution containing 10% glycerol before being flash frozen in liquid nitrogen for data collection.
Structure Determination and Refinement of the PrV gHC:Fab d6.3 Complex.
Table S3 displays the data collection and processing statistics, and the details are provided as SI Materials and Methods. Briefly, initial phases were obtained by molecular replacement, using the Fab as a search model. This allowed building of a partial model of gH, using the data collected for the complexAb. The diffraction of complexR crystals improved to 2.1 Å by soaking in a potassium osmate (K2OsO4) heavy atom solution, allowing completion of the gH atomic model. All of the diffraction datasets were processed with autoPROC (35), using XDS (36) and SCALA (37). The atomic models were refined with REFMAC (37) and autoBUSTER (38).
Supplementary Material
Acknowledgments
We thank P. Weber, A. Haouz, and E. Stura for help with crystallization trials and Andrew Thompson (synchrotron SOLEIL), Clemens Schulze-Briese, and Takashi Tomizaki (Swiss Light Source, SLS) for assistance with data collection. We also thank colleagues at Global Phasing for help and advice with Global Phasing software and the members of the F.A.R. laboratory. Crystallization conditions were screened at the crystallogenesis core facility of Pasteur Institute (PF6) run by Ahmed Haouz. Diffraction data collection was performed at the SOLEIL synchrotron source (Proxima I beamline, dataset 1) and SLS (PXI beamline, datasets 2 and 3). This work was supported by the Ministère de l'Economie, de l'Industrie et de l'Emploi CristaLead Grant 06 2 90 6122 (to M.B. and F.A.R.), the Agence Nationale de Recherches (Grant Dentry to F.A.R.), and Merck-Serono.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2XQY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011507107/-/DCSupplemental.
References
- 1.Pellet PE, Roizman B. In: Fields Virology. Knipe DM, Howley PM, editors. New York: Lippincott Williams and Wilkins; 2007. pp. 2479–2499. [Google Scholar]
- 2.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 4.Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci USA. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 6.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdary TK, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mettenleiter TC. Immunobiology of pseudorabies (Aujeszky's disease) Vet Immunol Immunopathol. 1996;54:221–229. doi: 10.1016/s0165-2427(96)05695-4. [DOI] [PubMed] [Google Scholar]
- 9.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt J, Klupp BG, Karger A, Mettenleiter TC. Adaptability in herpesviruses: Glycoprotein D-independent infectivity of pseudorabies virus. J Virol. 1997;71:17–24. doi: 10.1128/jvi.71.1.17-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp BG, Fuchs W, Weiland E, Mettenleiter TC. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klupp BG, Mettenleiter TC. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt J, Gerdts V, Beyer J, Klupp BG, Mettenleiter TC. Glycoprotein D-independent infectivity of pseudorabies virus results in an alteration of in vivo host range and correlates with mutations in glycoproteins B and H. J Virol. 2001;75:10054–10064. doi: 10.1128/JVI.75.21.10054-10064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp BG, Nixdorf R, Mettenleiter TC. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng T, et al. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J Virol. 1998;72:6092–6103. doi: 10.1128/jvi.72.7.6092-6103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Showalter SD, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galdiero M, et al. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71:2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gompels UA, et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol. 1991;65:2393–2401. doi: 10.1128/jvi.65.5.2393-2401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cairns TM, et al. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J Virol. 2006;80:2596–2608. doi: 10.1128/JVI.80.6.2596-2608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. The crystal structure of the EBV gHgL complex. Proc Natl Acad Sci USA. 2010:22641–22646. doi: 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 23.Wallin M, Ekström M, Garoff H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004;23:54–65. doi: 10.1038/sj.emboj.7600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Hutt-Fletcher LM. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology. 2007;363:148–155. doi: 10.1016/j.virol.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian RP, Geraghty RJ. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duus KM, Hatfield C, Grose C. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: Analysis by laser scanning confocal microscopy. Virology. 1995;210:429–440. doi: 10.1006/viro.1995.1359. [DOI] [PubMed] [Google Scholar]
- 27.Pertel PE. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J Virol. 2002;76:4390–4400. doi: 10.1128/JVI.76.9.4390-4400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon TY, Shin YK. Progress in understanding the neuronal SNARE function and its regulation. Cell Mol Life Sci. 2009;66:460–469. doi: 10.1007/s00018-008-8372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon HT, Kozlov MM, Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Atanasiu D, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci USA. 2007;104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misura KM, Bock JB, Gonzalez LC, Jr., Scheller RH, Weis WI. Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog. Proc Natl Acad Sci USA. 2002;99:9184–9189. doi: 10.1073/pnas.132274599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parry C, Bell S, Minson T, Browne H. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol. 2005;86:7–10. doi: 10.1099/vir.0.80567-0. [DOI] [PubMed] [Google Scholar]
- 33.Schetz JA, Shankar EP. Protein expression in the Drosophila Schneider 2 cell system. Curr Protoc Neurosci. 2004 doi: 10.1002/0471142301.ns0416s27. Chapter 4:Unit 4. 16. [DOI] [PubMed] [Google Scholar]
- 34.Backovic M, et al. Efficient method for production of high yields of Fab fragments in Drosophila S2 cells. Protein Eng Des Sel. 2010;23:169–174. doi: 10.1093/protein/gzp088. [DOI] [PubMed] [Google Scholar]
- 35.Vonrhein C, et al. autoPROC version 0.5.3. Cambridge, UK: Global Phasing Ltd.; 2010. [Google Scholar]
- 36.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Bricogne G, et al. BUSTER version 2.8.0. Cambridge, UK: Global Phasing; 2009. [Google Scholar]
- 39.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.