Abstract
Evidence from experimental studies suggests that fetal exposure to the endocrine-disrupting chemical bisphenol A (BPA) has adverse reproductive effects in both males and females. Studies from our laboratory suggest that exposure to the developing female fetus produces a unique, multigenerational effect. Specifically, maternal exposure affects the earliest stages of oogenesis in the developing fetal ovary, and the resulting subtle meiotic defects increase the likelihood that embryos produced by the exposed female in adulthood (i.e., the grandchildren) will be chromosomally abnormal. To understand the impact of BPA on the developing ovary, we conducted expression studies to characterize gene expression changes in the fetal ovary that result from BPA exposure. We first tested the validity of the approach, asking whether we could reliably detect temporal changes in expression levels of meiotic genes in controls. As anticipated, we were able to identify appropriate increases in expression in meiotic, but in few other, genes. Intriguingly, this analysis provided data on a small set of genes for which timing and expression changes suggest that they may have important and heretofore unrecognized meiotic roles. After verifying the utility of our approach, we focused our analysis on BPA-exposed animals. We found modest, but significant, changes in gene expression in the fetal ovaries from exposed fetuses. The first changes were evident within 24 h of exposure, and the most extensive changes correlated with the onset of meiosis. Furthermore, gene ontology analysis suggested that BPA acts to down-regulate mitotic cell-cycle genes, raising the possibility that fetal BPA exposure may act to limit expansion of the primordial germ cell population.
Keywords: bisphenol A, meiosis, meiotic gene expression, meiotic prophase, oocyte development
Significant changes in gene expression in the fetal ovary are evident in female mice whose mothers are exposed to low doses of bisphenol A.
INTRODUCTION
Studies conducted during the past decade provide compelling evidence that exposure to endocrine-disrupting chemicals (EDCs) during gestation has the potential to impair the developing reproductive tract of both males and females. Adverse reproductive effects have been ascribed to the actions of the phytoestrogen genistein (see, e.g., [1, 2]), synthetic estrogens used pharmacologically (e.g., diethylstilbestrol [DES] and ethinyl estradiol [3]), compounds such as bisphenol A (BPA) and phthalates that are found in plastics and a variety of other consumer products (for review, see [4, 5]), and chemicals used in fertilizers and pesticides (for review, see [6]).
The most complete data regarding the effects of an EDC on the developing reproductive tract and fertility have come from studies of the estrogenic chemical BPA. This does not mean that BPA is the only such chemical of concern, only that it has been the most widely studied. Because defects induced during fetal development can adversely impact adult fertility, a major challenge is understanding the molecular basis of the developmental changes induced by these chemicals and determining whether therapeutic approaches can be developed to diminish their impact on the reproductive potential of the adult.
Although the vast majority of studies have involved exposures to rodents, findings from DES-exposed human sons and daughters provide compelling evidence that the rodent data can, in many cases, be generalized to humans [7]. One of the most significant concerns is the suggestion that some of the effects resulting from prenatal and early postnatal exposure to EDCs are multigenerational; that is, exposure to one generation increases the likelihood of defects in subsequent generations. Multigenerational effects have now been described in mouse studies of DES [8], BPA [9], and vinclozolin and methoxychlor [10] (for review, see [11]). Although this mechanism currently is thought to be the result of epigenetic changes that are heritable, data remain limited. However, in addition to this type of heritable effect, we recently reported a different type of multigenerational effect that is not inherited [12]. Specifically, we found a grandmaternal effect whereby exposure to the pregnant female altered germ cell development in the fetal ovary, leading to an increased risk of chromosomally aberrant grandchildren.
To determine how BPA affects the earliest stages of oocyte development in the fetal ovary, we conducted detailed expression array studies to pinpoint the earliest changes. The results of these studies are summarized here. An initial analysis of the data from control females demonstrated the sensitivity of the technique for detecting meiotic gene expression and identifying candidate genes for which expression is consistent with a meiotic role. Subsequent analyses of gene expression following BPA exposure indicated significant BPA-induced changes in gene expression that correlated with the onset of meiosis in the fetal ovary. These results are consistent with the hypothesis that BPA exerts it effect on the fetal gonad by several distinct mechanisms.
MATERIALS AND METHODS
Mouse Information
The C57BL/6J inbred mice used for the present study were from a pathogen-free breeding colony derived from stock obtained from The Jackson Laboratory. They were housed in ventilated rack caging and provided water and chow (Purina 5010) ad libitum. Protocols for the care and use of the animals were approved by the Washington State University Animal Care and Use Committee and were in accordance with National Institutes of Health standards established by the Guidelines for the Care and Use of Experimental Animals. Washington State University is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Exposure Information
Timed matings were used to generate pregnant females. These females were given a daily dose of 20 ng of BPA (Sigma-Aldrich) or 0 ng of BPA (placebo) per gram of body weight beginning at 11 days postcoitus (dpc). The 20-ng/g dose was chosen because previous studies of in utero BPA exposure demonstrated marked meiotic defects in ovaries from fetal females [12]. Both placebo and BPA doses were delivered orally to pregnant females in corn oil carrier as described previously [13]. Briefly, BPA was dissolved in tocopherol-stripped corn oil (catalog no. 901415; MP Biomedicals, Inc.), females were weighed, and a small volume of BPA (20–30 μl) equivalent to 20 ng/g was delivered orally using an Eppendorf pipette with a disposable tip.
Ovary Collection and RNA Isolation
To obtain 12-, 12.5-, 13.5-, and 14.5-day fetal ovaries, pregnant females were killed after 1, 1.5, 2.5, and 3.5 days of treatment. Fetuses were removed from the uterine horns, and the ovaries were isolated from the female fetuses. To ensure that ovaries were collected from fetuses of the appropriate developmental stage, several criteria were used to stage individual fetuses, including snout, paw, and gonad development. Isolated ovaries were dissected free of the mesonephros in a drop of PBS, placed in 7.5 μl of Extraction Buffer from the PicoPure RNA Isolation Kit (Arcturus Bioscience, Inc.), and stored individually at −80°C until RNA isolation was performed.
Because fetal sex cannot be determined reliably until after 12.5 days of gestation, the sex chromosome constitution of all fetuses collected from 12- and 12.5-day litters was determined by PCR on collected fetal tissues. These tissues were placed in lysis buffer, digested at 95°C, and PCR amplified using primers for the Kdm5c and Kdm5d genes (also known as SMC X and SMC Y, respectively; 5′-CCGCTGCCAAATTCTTTGG-3′ and 5′-TGAAGCTTTTGGCTTTGAG-3′) as described previously [14].
All RNA samples were processed in a single day, resulting in three biological replicates (six sets of ovaries per sample) for each of four time points, or 72 sets of gonads in total. RNA was prepared using the PicoPure RNA Isolation Kit according to the manufacturer's instructions and included a DNase treatment step using the RNase-Free DNase Set (Qiagen). RNA quality was determined by agarose gel electrophoresis using a 2% gel and absorbance and concentration measurements using the NanoDrop 1000 spectrophotometer (NanoDrop Technologies).
Microarray Processing
For each sample, 100 ng of total RNA were processed using the Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay according to the manufacturer's directions. Double-stranded cDNA was synthesized using a random hexamer with a T7 promoter. Using in vitro transcription, cRNA was generated from the double-stranded cDNA template using the Whole Transcript cDNA Synthesis and Amplification Kit (Affymetrix). The cDNA was regenerated using a reverse transcription reaction randomly primed with a mix containing dUTP. After hydrolysis of the cRNA with RNase H, the sense strand of cDNA was purified using the Affymetrix sample cleanup module, fragmented by incubation with UDG (uracil DNA glycosylase) and APE1 (apurinic/apyrimidic endonuclease 1), and terminally biotin-labeled with terminal deoxynucleotidyl transferase using the Whole Transcript Terminal Labeling kit per the manufacturer's instructions (Affymetrix). Biotinylated sense-strand fragments were hybridized to Affymetrix Mouse Gene 1.0 ST GeneChips using the Hybridization Control and Hybridization Wash and Stain kits (Affymetrix). The stained array was then scanned using an Affymetrix GeneChip Scanner 3000 to generate the .CEL files for each experimental array.
Primary quality control was performed using the Affymetrix Expression Console. All samples were imported into the program and normalized using the default robust multiarray average (RMA) setting, and the relative log expression was examined to ensure that the data were properly corrected by normalization and that no outliers were present. MvA plots were also generated to examine the reproducibility of the replicates.
Data Analysis
Affymetrix .CEL files were imported into the Partek Genomics Suite using the default RMA normalization parameters. The probe set signal intensities were summarized using RMA, which includes RMA background corrections and quantile normalization across the arrays. The summarized signals were then subjected to a log base 2 transformation and median polish algorithm. To remove any batch effect that may have occurred during processing of the arrays, the data were additionally subjected to Partek's batch removal procedure. To evaluate and compare the global signal intensities from the normalized data, a principal component analysis was performed. The normalization and summarization procedures were performed in two sets: 1) control chips only and 2) BPA-treated versus placebo chips only.
Using the signal intensities from the control data, a one-way ANOVA was performed to assess significance and fold-change. The ANOVA was used to compare the signal intensities across the time points and between individual time points as follows: All days, 12 dpc versus 14.5 dpc, 12 dpc versus 12.5 dpc, 12.5 dpc versus 13.5 dpc, and 13.5 dpc versus 14.5 dpc. The control data were also subjected to multiple testing corrections using the false-discovery rate (FDR) step-down method to compensate for any testing errors.
To identify potential meiotic-like or novel meiotic players, filters were applied to the data for 12 dpc versus 14.5 dpc that required a P-value of less than 0.05 and a fold-change of greater than 2 at 14.5 dpc versus 12 dpc. Additionally, the resulting gene list was scrutinized visually to determine if these genes had the “meiotic-like” pattern of expression.
A two-way ANOVA was carried out on the BPA-treated and placebo data. This ANOVA was used to determine differential gene expression based on both fetal age and treatment. The program was asked to provide data based on treatment across the time points and between each time point as follows: BPA-treated 12 dpc versus placebo 12 dpc, BPA 12.5 dpc versus placebo 12.5 dpc, BPA 13.5 dpc versus placebo 13.5 dpc, and BPA 14.5 dpc versus placebo 14.5 dpc. In addition, the same one-way ANOVA used for the control chips was also carried out on the data from the treated mice. Filters were applied to each of the resulting two-way ANOVA lists that required a P-value of less than 0.05 and a fold-change of 1.2.
For the analysis of gene ontology (GO), significantly shared GO terms were used to describe common functions of a query gene set. To determine whether any GO terms were enriched in a query gene list at a frequency greater than what would be expected by chance, we calculated the probability from a hypergeometric distribution:
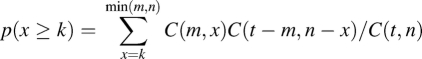 |
where C(j, k) is the general representation of the three “C( )” terms in the equation and represents the combinatorial factor j!/k!(j − k)!. In this equation, t is the total number of genes in the array, n is the number of genes in the array that are annotated by a GO term, m is the number of genes in a query list (for example, down-regulated genes in Fig. 3a), p stands for the hypergeometric P-value, x is a random variable, and k is the number of genes within the list that are annotated by the GO term.
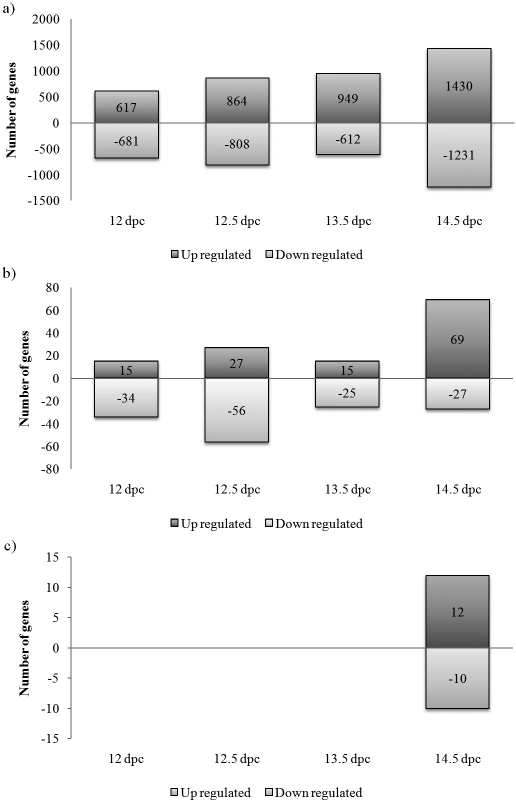
FIG. 3.
BPA-induced transcription changes. a) Breakdown by day of the 7192 transcripts that were significantly (P < 0.05) up- or down-regulated in ovaries from BPA-exposed females. b) Increasing the selection criteria to transcripts with at least a 1.3-fold change in expression and P < 0.01 reduces the list of differentially expressed transcripts by approximately 95%, from 7192 to 268. c) The introduction of the FDR step-up correction to account for multiple tests further reduces the number of transcripts that were significantly up- or down-regulated from 268 to 22.
RESULTS
We recently reported that in the mouse, low-dose BPA exposure during fetal development alters the early stages of meiotic prophase in the fetal ovary [12]. Our studies focused on pachytene stage cells from 18.5-dpc fetuses exposed to BPA during the 7 days before analysis and revealed subtle, but significant, increases in recombination rates and synaptic defects in oocytes from exposed female fetuses. However, because the types of changes we observed (defects in synapsis and recombination between homologous chromosomes) have their genesis at the earliest stages of the meiotic prophase, we suspected that BPA was exerting its effect on the oocyte at the premeiotic or very early prophase stage. Thus, to understand the molecular mechanisms underlying these changes, we assessed the earliest changes in gene expression evident in the fetal ovary.
We initiated daily BPA exposure (20 ng/g) of pregnant females at 11 dpc, or approximately 1.5–2 days before the onset of meiosis in the fetal ovary, and collected fetal ovaries 24, 36, 60, and 84 h later (i.e., at 12, 12.5, 13.5, and 14.5 dpc). For expression analysis, RNA was prepared from six sets of ovaries for each time point, with three replicates for each. For each sample, 100 ng of total RNA were processed using the Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay and hybridized to Affymetrix Mouse Gene 1.0 ST GeneChips, and the stained array was scanned using an Affymetrix GeneChip Scanner 3000 to generate the .CEL files. These files were imported into the Partek Genomics Suite and corrected and normalized as detailed in the Materials and Methods.
Validation of the Approach: Meiosis-Specific Genes Exhibit Temporal Changes in Gene Expression
To test the sensitivity of Affymetrix Mouse Gene 1.0 ST GeneChips for the detection of temporal changes in the expression of genes involved with entry into, and progression through, meiotic prophase, we first examined expression levels in control animals (no treatment) of a set of genes for which meiotic functions are known. Specifically, we selected 16 representative genes with activities entirely, or almost entirely, restricted to meiosis and involving five broad functional categories: 1) a gene involved in meiotic entry (Stra8), 2) genes involved in formation of the synaptonemal complex (SC; Sycp1, Sycp2, Sycp3, Syce1, Syce2, and Tex12), 3) meiosis-specific cohesins (Rec8, Stag3, and Smc1b), 4) meiotic recombination pathway genes (Dmc1, Mei1, Msh4, and Msh5), and 5) a gene involved in chromatin remodeling (Prdm9). Strikingly, when the results of the individual genes were pooled within the groups, each of the five categories exhibited a dramatic increase in expression from 12 to 14.5 dpc (Fig. 1 and Table 1). In general, the increases observed mirrored the meiotic timeline. For example, increased expression was observed at 12.5 days for the meiotic entry gene Stra8, whereas increased expression of the later-acting recombination genes was not observed until 13.5 dpc. This suggests that the approach is, indeed, suitable for detecting temporal changes in the expression of genes involved in the meiotic prophase.
FIG. 1.
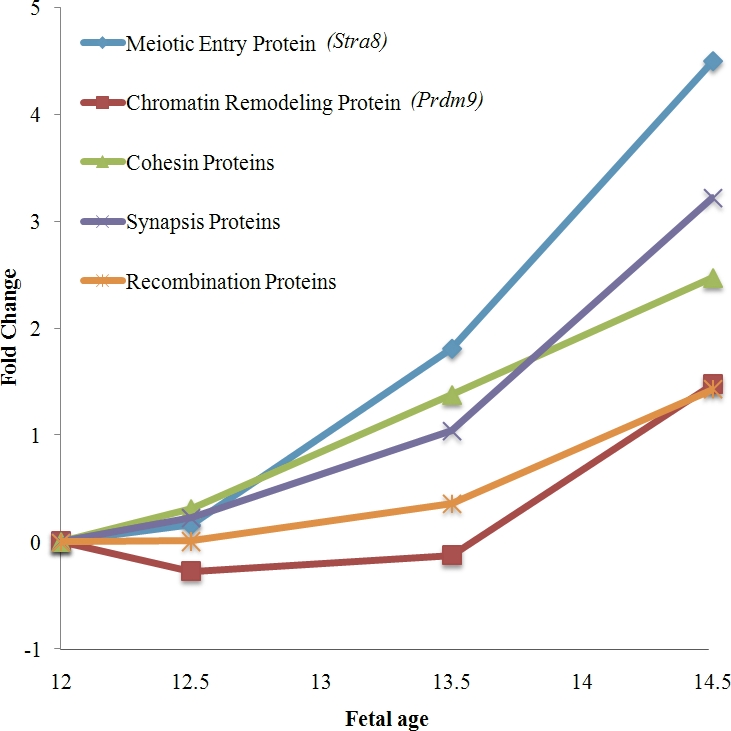
Expression changes in known meiotic genes in control ovaries between 12 and 14.5 dpc. The expression pattern obtained by Affymetrix Mouse Gene 1.0 ST GeneChips for genes with known meiotic functions was examined in ovaries from control fetuses at 12, 12.5, 13.5, and 14.5 dpc. The y-axis shows the fold-change in expression levels for each of the successive days of gestation. For gene categories that include multiple genes, expression values for individual genes were averaged. The meiotic entry protein gene, Stra8 (red line), shows a significant increase from 12.5 to 13.5 dpc and from 13.5 to 14.5 dpc. The expression of all other genes increases significantly during the 2.5-day interval (i.e., from 12 to 14.5 dpc).
TABLE 1.
Gene categories for initial analysis.
To confirm this, we compared the expression levels of these meiotic genes with those of genes known to function in both meiocytes and somatic cells and with genes having no known meiotic function (Table 1). We made no attempt to conduct an exhaustive survey of genes in these categories. Instead, we compared the 16 meiosis-specific genes described above with 17 genes having functions “shared” between meiotic and somatic cells and 17 genes commonly used as “housekeeping” genes [15]. We hypothesized that the expression of each of the meiosis-specific genes would increase significantly between 12 and 14.5 dpc; that some, but not all, “shared” genes might display a significant increase; and that expression levels would remain flat over time for the housekeeping genes. For this initial analysis, we scored genes as “increased” if they exhibited significant (P < 0.05, Bonferroni corrected for multiple comparisons) increases in expression levels and a 2-fold or greater difference between 12 and 14.5 dpc and as “nonincreased” if they did not fit either of these criteria. In general, our predictions were consistent with the observations. Specifically, for 14 of the 16 (87.5%) meiosis-specific genes, we observed significant increases of 2-fold or greater between 12 and 14.5 dpc (Fig. 2a). Those genes with the greatest fold-increases were associated with entry into meiosis (Stra8), formation of the SC (Sycp1, Sycp3, and Tex12), or establishment of sister chromatid cohesion (Smc1b). The only two genes on our list of meiosis-specific genes that did not display significant 2-fold differences were the SC protein-encoding gene Syce2 and the mismatch repair protein-encoding gene Msh5. However, even in these instances, expression levels increased over time, although they did not reach the 2-fold increase threshold (i.e., for Syce1, 1.9-fold, and for Msh5, 1.4-fold) and, in the case of Msh5, did not reach statistical significance.
FIG. 2.
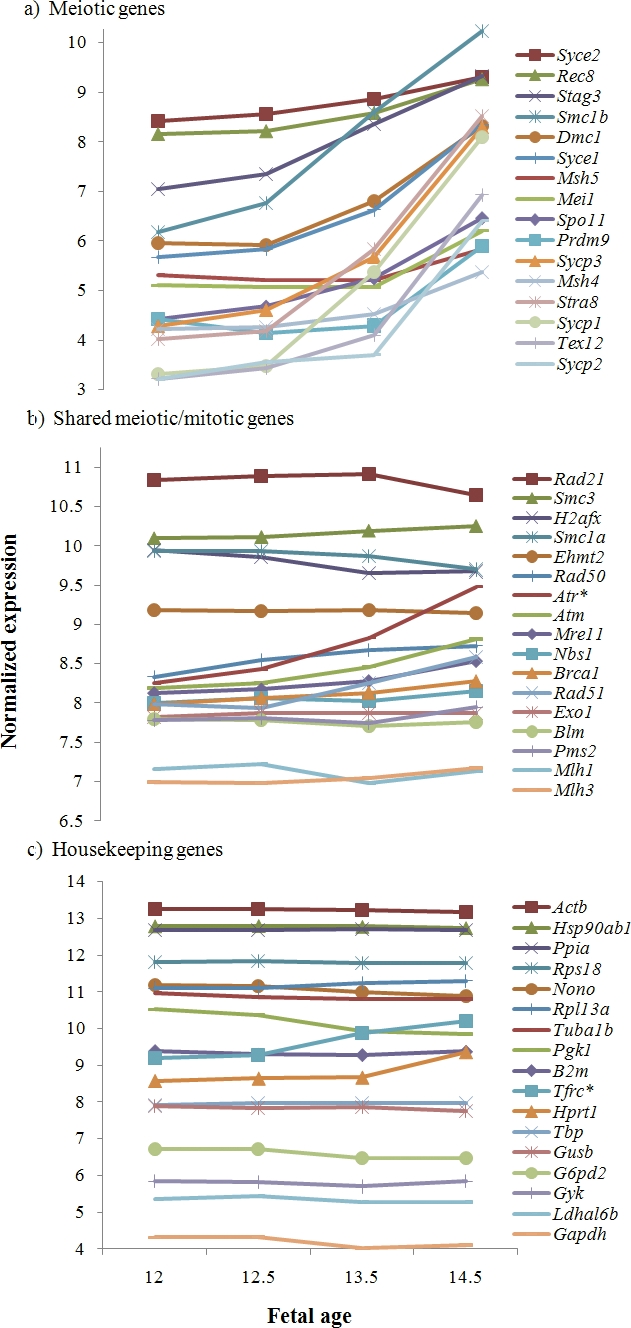
Comparison of expression profiles for meiotic genes, genes with both meiotic and mitotic functions, and genes with no known meiotic function. The lines in each panel show the normalized intensity of expression at the four different developmental time points for meiosis-specific genes (a), genes with both meiotic and mitotic functions (b), and a set of housekeeping genes (c). The genes within each panel are ordered from highest to lowest expression levels at 14.5 dpc; hence, the y-axis does not reflect relative levels of expression. a) The expression of 14 of 16 meiosis-specific genes exhibited a significant increase of 2-fold or greater during the 2.5-day developmental window (the expression of both Syce2 and Msh5 increased over time but did not reach significance). b and c) By comparison, of the 17 shared genes (b) and 17 housekeeping genes (c), only one in each category (Atr and Tfrc, respectively [indicated with an asterisk]) exhibited a significant 2-fold increase.
In contrast to the observations for the meiosis-specific genes, only 1 of the 17 “shared” genes (Atr) and 1 of the 17 housekeeping genes (Tfrc) showed significant increases of 2-fold or greater over this time period (Fig. 2, b and c, and Table 1); this yields a highly significant among-group difference (χ2 = 33.3, P < 0.001). Thus, our observations indicate that the vast majority of meiosis-specific genes are detected by comparing expression levels of genes at 12 and 14.5 dpc, but that this approach is less useful for identification of genes that function both in meiocytes and in somatic cells.
Identification of Candidate Meiosis-Specific Genes
Our initial observations indicated that it should be possible to identify novel meiosis-specific genes by examining genes with significant increases in expression between 12 and 14.5 dpc. Thus, we searched for all loci exhibiting significant 2-fold or greater increases in expression levels between 12 and 14.5 dpc. Because we were interested in identifying genes with steady increases over time, we excluded genes that did not have simple monotonic or curvilinear increases over time (e.g., genes with significant spikes or valleys at 12.5 or 13.5 dpc).
In total, 265 genes fit these criteria (Supplemental Table S1, all Supplemental Data are available online at www.biolreprod.org). Importantly, these include a number of genes known to function in meiosis or to have putative roles in germ cell development that were not included in the original list of 16 meiosis-specific genes (e.g., Boll [16], Dazl [16], Hormad1 [17], Hormad2 [18], Tex11 [19], Tex13 [20], Tex14 [21], Tex101 [22], Sohlh2 [23], and Xlr [24]). Furthermore, genes thought to function in meiosis or in germ cell development were especially enriched among those exhibiting the highest fold-increases between 12 and 14.5 dpc. For example, if we consider the 66 genes with increases of greater than 4-fold, no fewer than 37 (56%) fit into one of these two categories (Supplemental Table S1). Thus, these results provide strong evidence that our approach identifies genes with activities that are largely, or entirely, restricted to early germ cell development and/or meiosis. Consequently, they provide confidence that careful examination of genes not known to function in meiosis but having a significant increase in expression between 12 and 14.5 dpc will uncover new meiotic genes. Indeed, among those with the highest fold-increases are several obvious candidates: several loci in the Xlr gene superfamily (e.g., Xlr4b, Xlr4c, Xlr5a, and Xlr5b) [24], three members of which are known to be imprinted in mice [25]; the germ cell-specific gene D6Mm5e [26]; the spermatogenesis-associated gene Spata22 [27]; and the putative germline-specific RNA helicase Mov10l1 [28]. Further characterization of these genes is presently underway.
BPA-Induced Transcription Changes
Previously, we have shown that low-level BPA exposure results in subtle, but significant, increases in abnormalities during meiotic prophase—specifically, an increased frequency of cells with incomplete synapsis, altered levels of recombination, and unusual end-to-end associations between the SCs of nonhomologous chromosomes [12]. To determine how BPA affects gene transcription in the fetal ovary, we first assessed the number of transcripts on the Affymetrix array that were significantly different in ovaries from BPA-exposed and placebo-treated females at comparable stages of development. Based on our previous study [12], we suspected that the differences would be subtle. Thus, in our initial analyses, we were simply interested in identifying general trends. Accordingly, we characterized all BPA-associated expression differences without correcting for multiple comparisons. If summed across all time points, a total of 7192 transcripts were significantly (P < 0.05) up- or down-regulated with BPA exposure (Fig. 3a). The number of genes affected by BPA increased markedly over the 4-day developmental window, with the largest number of significant changes (n = 2661) evident at 14.5 dpc. At the earliest time point (12 dpc), genes were more likely to be down- than up-regulated following BPA exposure, but this relationship was reversed at each of the following three time points examined.
Two aspects of the data are notable. First, the expression changes were all subtle, with fold-changes of less than 2 for almost all transcripts (Supplemental Table S2). Indeed, by modestly altering the stringency of the selection criteria (i.e., setting a minimal change in expression of 1.3-fold and reducing the significance level from P < 0.05 to P < 0.01), the number of transcripts with significantly increased or decreased expression in BPA-treated females was reduced by approximately 95%, from 7192 to 268 total transcripts (Fig. 3b). Furthermore, upon introducing a more stringent correction (FDR step-up, P < 0.1) to account for multiple comparisons, the number of significant up- or down-regulated transcripts was reduced from 268 to 22, with significant changes detected only at the 14.5-dpc time point (Fig. 3c). Second, despite the large number of gene changes (Fig. 3a), very little redundancy was observed between different developmental time points. Specifically, fewer than 5% of genes identified as being significantly up- or-down regulated at one time point were similarly up- or down-regulated at a successive time point. Nevertheless, 30 genes were significantly up- or down-regulated at three successive time points (i.e., 15 genes at 12, 12.5, and 13.5 dpc and 15 genes at 12.5, 13.5, and 14.5 dpc). Of this group of genes, 23 exhibited consistently elevated expression in ovaries from BPA-exposed females, and seven exhibited consistently decreased expression (Supplemental Table S3).
BPA-Induced Changes in Known and Candidate Meiotic Genes
In subsequent analyses, we were interested in further characterizing the genes that were up- or down-regulated by BPA. Specifically, we analyzed the list of genes exhibiting BPA-induced changes at P < 0.05 to determine the degree of overlap with the list of meiotic and putative meiotic genes obtained by the analysis of data from control animals (Supplemental Table S4).
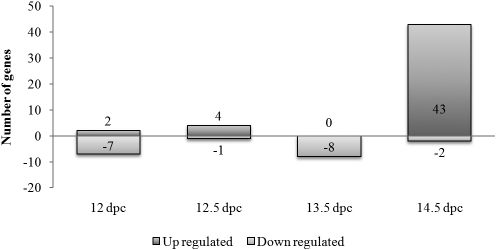
Of the 265 transcripts on our list of meiosis-specific and candidate meiotic genes, 67 were significantly up- or down-regulated by BPA at one or more of the four time points examined (Fig. 4). Furthermore, with increasing duration of exposure, more of these genes exhibited expression changes, with 45 of the 67 (67.2%) gene changes confined to the 14.5-dpc time point. Intriguingly, all 16 of the meiosis-specific genes (Table 1) were up-regulated at 14.5 dpc, although only three (Msh4, Dmc1, and Sycp2) reached statistical significance.
FIG. 4.
Meiosis-specific and candidate meiotic genes affected by BPA. Of the 265 transcripts identified as meiosis-specific or candidate meiotic genes (Supplemental Table S1), 67 were significantly up- or down-regulated by BPA. Strikingly, the majority of the changes in these transcripts involved up-regulation at 14.5 dpc.
Biological Processes Impacted by BPA
Analysis of GO was performed to identify biological processes associated with genes that were significantly up- or down-regulated by BPA. Separate analyses were conducted for genes identified using relaxed, intermediate, and stringent significance criteria, as outlined in Figure 3, a, b, and c, respectively. When genes significantly down-regulated by BPA at any of the four time points were considered (Fig. 3a), the most highly represented biological processes were cell cycle (P = 8.3E-08), mitosis (P = 1.6E-06), and DNA replication (P = 2.8E-05). Other categories found to be significant involved chromatin modification, remodeling, and chromosome condensation. Of the 20 biological processes that were associated with genes significantly down-regulated at the next most stringent level (P < 0.01) (Fig. 3b), half were related to mitosis/cell division. Finally, when the same analysis was applied to the 10 down-regulated genes passing the most stringent correction criteria (Fig. 3c), cell cycle (P = 0.0007) and mitosis (P = 0.03) remained the most represented processes. Cell-cycle activities were particularly notable, because 4 of the 10 down-regulated genes, including cyclin A2, cyclin F, aurora kinase A, and membrane protein palmitoylated, have known roles in the cell cycle.
The analysis of all up-regulated BPA genes (Fig. 3a) revealed only general pathways, such as translation and amino acid metabolism, to be most highly represented (P < 0.01). Similarly, analyses using more stringent selection criteria (Fig. 3, b and c) failed to reveal obvious related biological processes.
DISCUSSION
In the present study, we were interested in using an array-based approach to identify BPA-induced changes in gene expression at the time when germ cells are entering meiosis in the fetal ovary. However, before embarking on this analysis, we tested the utility of the approach—specifically, whether we could reliably detect temporal changes in expression levels of genes known to be involved in different aspects of early meiosis. The results of that analysis were compelling: Virtually all meiosis-specific genes, but relatively few others, that we tested exhibited significant increases in expression over the time period associated with the earliest events of meiosis. Furthermore, the earliest temporal change was observed for Stra8, a gene thought to be a key player regarding entry into the meiotic pathway.
These results are consistent with those of previous array-based analyses of the meiotic transcriptome in humans and mice from our own (see, e.g., [29]) and other groups (see, e.g., [30, 31]). Taken together, these studies demonstrate that array-based analyses can be used to monitor temporal changes in gene expression at successive stages of the meiotic prophase. Furthermore, they provide an important new approach for the identification of novel meiotic genes. For example, in our analysis, no fewer than 208 of the 265 genes exhibiting significant increases in expression between 12 and 14.5 dpc (Supplemental Table S1) are genes for which little or no information regarding function is available.
Importantly, our initial analysis of known meiotic genes also provided confidence that the array-based approach could identify BPA-induced changes in gene expression. Thus, in subsequent analyses, we compared gene expression profiles in the fetal ovaries in fetuses from pregnant females exposed to low levels of BPA and from placebo controls over the 4-day gestational interval that encompasses meiotic entry. Our results demonstrate that exposure to low levels of BPA—levels that are below the current U.S. Food and Drug Administration “safe” dose for daily exposure and that are thought to be analogous to current human exposure levels [32, 33]—induces gene expression changes in the developing fetal ovary within 24 h of the onset of exposure, and with time, increasing numbers of genes exhibit significant BPA-associated alterations in expression. Over the entire 3.5-day exposure window, no fewer than 7192 genes were significantly up- or down-regulated following BPA exposure.
It is notable that the BPA-induced changes in gene expression were subtle, with almost all differences observed being less than 1.6-fold. This is consistent with our previous cytological analyses in which we detected significant, but subtle, BPA-associated effects on meiotic prophase [12]. Specifically, at the 20-ng/g level, we observed a 3-fold increase in the incidence of subtle synaptic defects, a 10% increase in recombination rates, and a slight increase in length of the SC, the meiosis-specific scaffold on which recombination occurs. The nature of the expression changes is also consistent with our cytological observations, because expression of one quarter of the genes on our list of known and putative meiotic genes was significantly altered by BPA (Fig. 4), and all 16 of the meiosis-specific genes used in our analysis of the control data were subtly up-regulated in ovaries from BPA-exposed females. Because our most recent cytological studies suggest that the level of meiotic disturbance is directly correlated with the dose of BPA (Lawson and Hunt, unpublished data), it will be important in future expression studies to increase the BPA dose and ask whether changes of greater magnitude are also induced in this setting.
Frequently, reverse transcription PCR analysis of small sets of genes is used to validate the findings of expression array studies. In this case, our major finding—that the majority of early meiotic genes are up-regulated by BPA—is itself a validation of our previous cytological studies. As discussed above, those studies indicated increases in SC length and recombination levels following BPA exposure. Given those changes, the up-regulation of SC (i.e., Sycp1, Sycp2, Sycp3, Syce1, Syce2, and Tex12) and recombination machinery genes (i.e., Spo11, Dmc1, Mei1, Msh4, and Msh5) observed in the present analysis makes perfect biological sense: Increased availability of the necessary reagents would allow formation of a longer SC and more recombination.
In our cytological studies, we also found that the Esr2-knockout female mouse phenocopied most of these BPA-induced meiotic defects [12]. However, an additional abnormality—end-to-end associations between nonhomologous chromosomes—was observed in pachytene cells, but not in the Esr2 knockout mouse, following BPA exposure. Thus, we postulated that BPA was acting by two distinct mechanisms in the fetal ovary and that the end-to-end association defect resulted from a disruption of the cytoskeleton that impaired normal chromosome movement at the onset of meiosis. Intriguingly, our gene expression studies revealed four genes up-regulated by BPA at 14.5 dpc that are potentially involved in cytoskeletal regulation. These include Dock3, a guanine nucleotide exchange factor that may be involved in cytoskeletal reorganization; the coiled-coil domain-containing proteins (Ccdc79 and Ccdc36), which have roles in actin cytoskeletal processes; and Emerin, a nuclear membrane protein. Thus, our expression results also strongly support the conclusion from our meiotic studies that the effect of BPA on the fetal ovary is complex and likely involves several distinct mechanisms. However, simply confirming that BPA alters the expression of these specific players will not allow us to understand the potential effect of BPA on the oocyte cytoskeleton. This requires a detailed analysis of the way that chromosomes move on the nuclear envelope and undergo synapsis in the presence and absence of BPA, and these studies are currently ongoing in our laboratory.
The consistency of the findings between our meiotic and expression studies is notable, but our expression data do not provide a direct means of unraveling the role that estrogen signaling through ESR2 plays in the transition from germ cell to meiocyte in the fetal ovary. In retrospect, this is perhaps not surprising. By comparison with the male, the period of germ cell differentiation preceding meiotic entry remains poorly characterized in the female. Female germ cell differentiation is rapid, with the entire period of germ cell proliferation, meiotic commitment, and meiotic entry spanning a period of only several days. Thus, the time points used in our analysis (12, 12.5, 13.5, and 14.5 dpc) represent a developmental window that includes a multitude of important events. At the earliest stages of BPA exposure (11–12.5 dpc), germ cells are populating the genital ridges and undergoing mitotic proliferation (for review, see [34]). By 13.5 dpc, the first wave of oocytes is entering meiotic prophase [35]. The event commonly perceived as the “onset” of meiosis—the initiation of synapsis between homologous chromosomes—is preceded by a host of complex events in female germ cells, including X-reactivation, major chromatin remodeling events, premeiotic DNA replication, establishment of meiosis-specific cohesion complexes, and prealignment of homologous chromosome in preparation for synapsis and SC formation. Thus, it is not particularly surprising that a host of different genes are up- or down-regulated by BPA at different time points during this relatively short time frame.
Characterizing the molecular signals that control the germ cell-to-meiocyte transition in the female is also hampered by the fact that germ cell differentiation occurs during fetal development and the germ cell population is heterogeneous, with meiotic onset in the posterior gonad lagging a full 24–48 h behind that in the anterior region. Furthermore, unlike the male, where physical separation methods make it possible to obtain purified populations of germ cells and meiocytes, the size of the developing fetal ovary makes it difficult to isolate germ cells in the female. Thus, major changes in expression that are limited to germ cells may be masked by contamination from somatic cells, and vice versa. This will have the effect of dampening any temporal changes in expression levels involving one of the two cell types but, additionally, will make it less likely that we can detect germ cell or somatic cell-specific effects that carry over from one time point to the next. In addition, in our previous analyses of mouse oogenesis [12], we detected significant BPA-associated effects on meiosis. However, the methodology is highly sensitive, and at the dosage level used in both the previous and present studies (20 ng/g), the effects of BPA, while highly significant, were relatively subtle. Specifically, at the 20-ng/g level, we observed a 3-fold increase in the incidence of subtle synaptic defects and a 10% increase in recombination rates. Importantly, in contrast to the meiotic phenotypes induced by the knockout of individual meiotic genes (for review, see [36]), in which a block at a specific meiotic stage or a clear and consistent meiotic defect is induced, low-level BPA exposure induces subtle meiotic defects in a significant proportion of cells. Thus, expression level differences of the magnitude observed in the present study are consistent with the subtle, but significant, defects observed in our previous meiotic analyses. Clearly, in future expression studies, it will be important to increase the BPA dose and ask whether changes of greater magnitude are induced. Thus, although the findings reported here demonstrate that BPA induces changes in gene expression in the fetal ovary and provide data that can be mined by us and others, a careful molecular dissection of the germ cell-to-meiocyte transition is badly needed.
Potential Long-Term Consequences of BPA Exposure to the Fetal Ovary
In addition to providing candidate genes that will help unravel the effects of BPA on the fetal ovary, our results raise the possibility that BPA may affect female fertility by yet another mechanism. Specifically, our GO analysis revealed a striking down-regulation of mitotic/cell-cycle genes, raising the possibility that BPA exposure immediately before meiotic entry might act to shorten the reproductive life span of the female. That is, meiotic entry is preceded by successive rounds of mitotic proliferation that expand the germ cell population. Thus, if BPA exposure causes premature meiotic entry, this could act to reduce the total pool of fetal oocytes. In turn, this would have important reproductive ramifications, because females exposed to BPA in utero would be at an increased risk not only of producing aneuploid eggs and embryos [12] but also of premature reproductive senescence. Studies to test this hypothesis are currently underway.
Although to our knowledge this is the first report of an effect on primordial germ cells in the ovary, results of previous in vitro studies have suggested that high concentrations of estrogens stimulate the growth of primordial germ cells in culture [37], and several previous studies have reported effects of EDCs on male germ cell development. Exposure to phthalates in vivo has been reported to decrease the number of gonocytes in the fetal rat testis [38], and a similar decrease has been reported from in vitro exposures of fetal rat [39], mouse [40], and human [41, 42] testes. Furthermore, recent in vitro studies of human testicular seminoma cells revealed that very low concentrations of BPA stimulated cell proliferation by inducing rapid, nongenomic, membrane-initiated activation of the cAMP-dependent protein kinase and cGMP-dependent protein kinase signaling pathways. From this, it was postulated that the high affinity of BPA for G protein-coupled, nonclassical membrane-associated receptors might interfere with germ cell proliferation and differentiation in the developing fetus [43].
Summary and Future Directions
The present study demonstrates a large number of subtle changes in gene expression in the fetal ovary following low-dose (20-ng/g) BPA exposure. Because we have recently found that increasing doses of BPA are associated with increased levels of both recombination and synaptic defects (Lawson and Hunt, unpublished data), these relatively modest changes in gene expression likely reflect the low BPA dose used in the present study. Thus, in future studies, it will be important to re-examine those genes we have identified as being affected by BPA and ask how their expression levels change over a range of BPA exposures. This will provide a means to validate and refine a list of candidate genes responsible for the meiotic disturbances induced by prenatal BPA exposure and should aid in translating our studies of mice to humans. Importantly, in addition to supporting the conclusion from our previous meiotic studies that BPA acts by several different mechanisms to perturb the early stages of oogenesis, our expression studies provide evidence that BPA may also affect primordial germ cells, influencing mitotic proliferation and the timing of meiotic entry.
Supplementary Material
Footnotes
Supported by NIH grants ES013527 (to P.H.) and HD21341 (to T.H.).
REFERENCES
- Jefferson WN, Padilla-Banks E, Newbold RR. Disruption of the female reproductive system by the phytoestrogen genistein. Reprod Toxicol 2007; 23: 308 316 [DOI] [PubMed] [Google Scholar]
- Jefferson WN, Padilla-Banks E, Goulding EH, Lao SP, Newbold RR, Williams CJ. Neonatal exposure to genistein disrupts ability of female mouse reproductive tract to support preimplantation embryo development and implantation. Biol Reprod 2009; 80: 425 431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howdeshell KL, Furr J, Lambright CR, Wilson VS, Ryan BC, Gray LE., Jr Gestational and lactational exposure to ethinyl estradiol, but not bisphenol A, decreases androgen-dependent reproductive organ weights and epididymal sperm abundance in the male Long-Evans hooded rat. Toxicol Sci 2008; 102: 371 382 [DOI] [PubMed] [Google Scholar]
- Richter CA, Birnbaum LS, Farabollini F, Newbold RR, Rubin BS, Talsness CE, Vandenbergh JG, Walser-Kuntz DR, Vom Saal FS. In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol 2007; 24: 199 224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howdeshell KL, Rider CV, Wilson VS, Gray LE., Jr Mechanisms of action of phthalate esters, individually and in combination, to induce abnormal reproductive development in male laboratory rats. Environ Res 2008; 108: 168 176 [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna CM, Skinner MK. Epigenetic transgenerational effects of endocrine disruptors on male reproduction. Semin Reprod Med 2009; 27: 403 408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RR. Prenatal exposure to diethylstilbestrol (DES). Fertil Steril 2008; 89: e55 e56 [DOI] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinology 2006; 147: S11 S17 [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A 2007; 104: 13056 13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005; 308: 1466 1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 2010; 21: 214 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Hassold TJ, Freeman E, Hunt PA. Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet 2007; 3: e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, Voigt RC, Thomas S, Thomas BF, Hassold TJ. Bisphenol A exposure causes meiotic aneuploidy in the female mouse. Curr Biol 2003; 13: 546 553 [DOI] [PubMed] [Google Scholar]
- Mroz K, Hassold TJ, Hunt PA. Meiotic aneuploidy in the XXY mouse: evidence that a compromised testicular environment increases the incidence of meiotic errors. Hum Reprod 1999; 14: 1151 1156 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet 2003; 19: 362 365 [DOI] [PubMed] [Google Scholar]
- Chen P, Ma M, Li L, Zhang S, Su D, Ma Y, Liu Y, Tao D, Yang Y. Phenotypic expression of partial AZFc deletions is independent of the variations in DAZL and BOULE in a Han population. J Androl 2009; 31: 63 68 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res 2010; 316: 158 171 [DOI] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 2009; 5: e1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 2008; 4: e1000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet 2005; 14: 2911 2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori N, Agno JE, Matzuk MM. Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod 2009; 80: 449 457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake H, Shirai Y, Mochizuki Y, Iwanari H, Tsubamoto H, Koyama K, Takamori K, Ogawa H, Hasegawa A, Kodama T, Hamakubo T, Araki Y. Molecular diversity of TEX101, a marker glycoprotein for germ cells monitored with monoclonal antibodies: variety of the molecular characteristics according to subcellular localization within the mouse testis. J Reprod Immunol 2008; 79: 1 11 [DOI] [PubMed] [Google Scholar]
- Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, Tashiro F, Yamato E, Miyazaki J. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol 2009; 325: 238 248 [DOI] [PubMed] [Google Scholar]
- Garchon HJ, Loh E, Ho WY, Amar L, Avner P, Davis MM. The XLR sequence family: dispersion on the X and Y chromosomes of a large set of closely related sequences, most of which are pseudogenes. Nucleic Acids Res 1989; 17: 9871 9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet 2005; 37: 620 624 [DOI] [PubMed] [Google Scholar]
- Arango NA, Huang TT, Fujino A, Pieretti-Vanmarcke R, Donahoe PK. Expression analysis and evolutionary conservation of the mouse germ cell-specific D6Mm5e gene. Dev Dyn 2006; 235: 2613 2619 [DOI] [PubMed] [Google Scholar]
- Huang X, Li J, Lu L, Xu M, Xiao J, Yin L, Zhu H, Zhou Z, Sha J. Novel development-related alternative splices in human testis identified by cDNA microarrays. J Androl 2005; 26: 189 196 [DOI] [PubMed] [Google Scholar]
- Lovasco LA, Seymour KA, Zafra K, O'Brien CW, Schorl C, Freiman RN. Accelerated ovarian aging in the absence of the transcription regulator TAF4B in mice. Biol Reprod 2010; 82: 23 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmard B, Small C, Yang L, Naluai-Cecchini T, Cheng E, Hassold T, Griswold M. Global gene expression in the human fetal testis and ovary. Biol Reprod 2009; 81: 438 443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod 2005; 72: 492 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen C, Nyeng P, Kalisz M, Jensen TH, Moller M, Tommerup N, Byskov AG. Global gene expression analysis in fetal mouse ovaries with and without meiosis and comparison of selected genes with meiosis in the testis. Cell Tissue Res 2007; 328: 207 221 [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA). Reprod Toxicol 2007; 24: 139 177 [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Chauhoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Urinary, circulating and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect 2010; 118: 1055 1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux K, Wylie C. Primordial germ cell migration. Int J Dev Biol 2004; 48: 537 544 [DOI] [PubMed] [Google Scholar]
- McLaren A. Somatic and germ-cell sex in mammals. Philos Trans R Soc Lond B Biol Sci 1988; 322: 3 9 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Sex matters in meiosis. Science 2002; 296: 2181 2183 [DOI] [PubMed] [Google Scholar]
- Moe-Behrens GH, Klinger FG, Eskild W, Grotmol T, Haugen TB, De Felici M. Akt/PTEN signaling mediates estrogen-dependent proliferation of primordial germ cells in vitro. Mol Endocrinol 2003; 17: 2630 2638 [DOI] [PubMed] [Google Scholar]
- Ferrara D, Hallmark N, Scott H, Brown R, McKinnell C, Mahood IK, Sharpe RM. Acute and long-term effects of in utero exposure of rats to di(n-butyl) phthalate on testicular germ cell development and proliferation. Endocrinology 2006; 147: 5352 5362 [DOI] [PubMed] [Google Scholar]
- Chauvigne F, Menuet A, Lesne L, Chagnon MC, Chevrier C, Regnier JF, Angerer J, Jegou B. Time- and dose-related effects of di-(2-ethylhexyl) phthalate and its main metabolites on the function of the rat fetal testis in vitro. Environ Health Perspect 2009; 117: 515 521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehraiki A, Racine C, Krust A, Habert R, Levacher C. Phthalates impair germ cell number in the mouse fetal testis by an androgen- and estrogen-independent mechanism. Toxicol Sci 2009; 111: 372 382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrot R, Muczynski V, Lecureuil C, Angenard G, Coffigny H, Pairault C, Moison D, Frydman R, Habert R, Rouiller-Fabre V. Phthalates impair germ cell development in the human fetal testis in vitro without change in testosterone production. Environ Health Perspect 2009; 117: 32 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habert R, Muczynski V, Lehraiki A, Lambrot R, Lecureuil C, Levacher C, Coffigny H, Pairault C, Moison D, Frydman R, Rouiller-Fabre V. Adverse effects of endocrine disruptors on the fetal testis development: focus on the phthalates. Folia Histochem Cytobiol 2009; 47: S67 S74 [DOI] [PubMed] [Google Scholar]
- Bouskine A, Nebout M, Brucker-Davis F, Benahmed M, Fenichel P. Low doses of bisphenol A promote human seminoma cell proliferation by activating PKA and PKG via a membrane G-protein-coupled estrogen receptor. Environ Health Perspect 2009; 117: 1053 1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.