Abstract
Arabidopsis cyt1 mutants have a complex phenotype indicative of a severe defect in cell wall biogenesis. Mutant embryos arrest as wide, heart-shaped structures characterized by ectopic accumulation of callose and the occurrence of incomplete cell walls. Texture and thickness of the cell walls are irregular, and unesterified pectins show an abnormally diffuse distribution. To determine the molecular basis of these defects, we have cloned the CYT1 gene by a map-based approach and found that it encodes mannose-1-phosphate guanylyltransferase. A weak mutation in the same gene, called vtc1, has previously been identified on the basis of ozone sensitivity due to reduced levels of ascorbic acid. Mutant cyt1 embryos are deficient in N-glycosylation and have an altered composition of cell wall polysaccharides. Most notably, they show a 5-fold decrease in cellulose content. Characteristic aspects of the cyt1 phenotype, including radial swelling and accumulation of callose, can be mimicked with the inhibitor of N-glycosylation, tunicamycin. Our results suggest that N-glycosylation is required for cellulose biosynthesis and that a deficiency in this process can account for most phenotypic features of cyt1 embryos.
Cellulose, a high molecular weight polymer consisting of hydrogen-bonded chains of β-1,4-linked glucose, is the most abundant polysaccharide of plant cell walls. The mechanisms by which cellulose and other cell wall polysaccharides are synthesized and assembled are poorly understood (1–3). It is thought that cellulose synthase resides in the plasma membrane as a multisubunit complex called a rosette that extrudes β-1,4-glucan into the extracellular matrix. Close proximity of subunits in the rosette complex presumably facilitates the hydrogen bonding of individual glucan chains into tight bundles of characteristic diameter, the microfibrils.
Higher plants contain a large number of closely related genes, the CesA family, that encode a catalytic component of cellulose synthase. It is unknown whether other polypeptides are required to form an active cellulose synthase, and little is known about the regulation of its activity. However, because the deposition of cellulose microfibrils is typically oriented relative to the axis of cellular expansion, an interaction between cellulose synthase and the cortical cytoskeleton has been proposed. A temperature-sensitive mutation in one of the CesA genes, rsw1, leads to a mutant phenotype characterized by swollen cells that are thought to reflect the inability of the cells to resist or direct turgor-mediated expansion (4). Similar phenotypes have been generated with chemical inhibitors of cellulose biosynthesis (5) indicating that the presence of near-normal amounts of cellulose in the primary cell wall is required for normal growth.
A number of Arabidopsis mutations affecting cell wall biogenesis have been isolated (6). Some of these mutations change the overall composition of cell wall polysaccharides without interfering with viability of the plants (7, 8), others only affect specialized structures such as trichomes (9) or secondary walls (10). In contrast, mutations in the CYT1 gene are lethal and cause a complex phenotype indicating a severe defect in the formation of the primary cell wall (11). Mutant embryos develop normally until the early heart stage, at which point cell division becomes inhibited and the embryos assume a wider shape than their wild-type siblings. On a cellular level, cyt1 embryos are characterized by the absence of protein bodies, ectopic depositions of callose in a patchy pattern, and the occurrence of incomplete cell walls. An ultrastructural analysis revealed that the cell walls of mutants were variable in width and unesterified pectins, which normally are only found in the middle lamella, had a diffuse distribution (11). Some aspects of the cyt1 phenotype, including radial swelling, accumulation of callose, and the occurrence of incomplete cell walls, could be mimicked by treating wild-type embryos with the cellulose synthase inhibitor dichlobenil.
We here report that CYT1 encodes an enzyme catalyzing the production of GDP-mannose. Depletion of GDP-mannose leads to a number of secondary defects, such as changes in the cell wall composition, a deficiency in N-glycosylation, and a deficiency in ascorbic acid production. Indeed, a weak mutation in the same gene, termed vtc1, causes a reduction in ascorbic acid content, but does not interfere with viability under normal growth conditions (12). With regard to cell wall polysaccharides, the most notable change in cyt1 embryos was a dramatic decrease in cellulose content. Experiments with the inhibitor of N-linked glycosylation tunicamycin suggest that this reduction is due to a deficiency in N-glycosylation.
Materials and Methods
Plant Culture and Genetic Mapping.
Plants were grown at 22°C either with a 16-hour photoperiod (for ascorbic acid measurements; fluence rate about 80 μmol photons/m2/s) or in constant light (fluence rate about 60 μmol photons/m2/s). The cyt1-1 mutation was mapped in the F2 generation of a cross to the marker line W6 (stock number CS6, Arabidopsis stock center, Columbus, OH). Molecular markers used for mapping were m429 (13), nga168, and AthBIO2 (ATGC web site of the University of Pennsylvania; http://genome.salk.edu) and C005, C024, C029, C033, and C035 (see supplemental data on the PNAS web site, www.pnas.org, or TAIR database, www.arabidopsis.org).
Northern and Western Blotting.
RNA was extracted with Trizol reagent (Life Technologies, Rockville, MD). Native protein extract was prepared by grinding inflorescence tissue in PBS. To release N-linked oligosaccharides from glycoproteins, native extracts were digested with peptide:N-glycosidase F (Bio-Rad). For preparation of crude extracts, about 200 wild-type or 400 cyt1-1 embryos were dissected from the seeds before desiccation and homogenized in 50 μl of extraction buffer (18% sucrose, 2% SDS, 10 mM MgCl2, 40 mM 2-mercaptoethanol, 100 mM Tris⋅HCl, pH 8). Rabbit antiserum against Arabidopsis protein disulfide isomerase (PDI; Rose Biotechnology, www.rosebiotech.com) was used at a dilution of 1/2,500. Reacting material was visualized by incubation with goat anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (Bio-Rad) followed by a luminescence reaction (SuperSignal; Pierce).
Ascorbic Acid Levels and Cell Wall Composition.
Ascorbic acid levels were determined as described (14). For the analysis of cell wall polysaccharides, material was extracted with 70% ethanol and acetone and the dry weight determined with an AD2B autobalance (Perkin–Elmer). Neutral sugars of the noncellulosic cell wall fraction were analyzed by gas chromatography of alditol acetates (7). Myo-inositol was added as an internal standard. Cellulose was measured colorimetrically (15). A 14C-labeled crude cellulose preparation (American Radiolabeled Chemicals, St. Louis) was added to a subset of the samples as a tracer. Recovery of radioactive material in the acid insoluble fraction was similar for wild-type and cyt1-1 samples.
Root Growth and Callose Staining.
Seeds were germinated on MS media (Life Technologies, Rockville, MD), the seedlings transferred to MS media containing tunicamycin or mannose, and the roots imaged with a stereomicroscope. Measurements were taken from the images. The molarity of tunicamycin (Sigma, catalog no. T-7765) was calculated with respect to the sum of all isomers (A, B, C, and D, relative content: 3.3%, 42.3%, 30.5%, and 20.4%). Live root tips were stained for callose with sirofluor (Biosupplies, Parkville, Victoria, Australia; 25 μg/ml in water; excitation, 380 nm; emission, 480 nm) and viewed with a fluorescence microscope with a DAPI filter-set. All images were taken with a SPOT CCD camera (Diagnostic Instruments, Sterling Heights, MI).
Results
Map-Based Cloning of the CYT1 Gene.
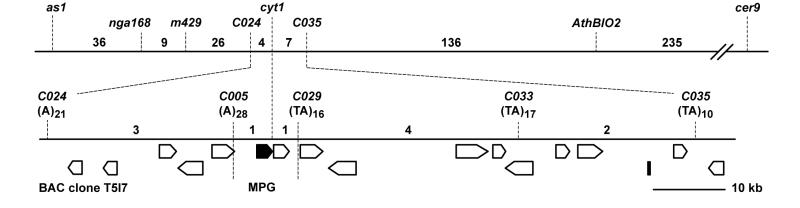
CYT1 had previously been mapped to chromosome 2 between the visible markers asymmetric leaf 1 (as1) and eceriferum 8 (cer8) (11). We made use of the flanking visible markers to select a large number of recombinants for fine mapping. Among the selfed progeny of an F1 hybrid with the genotype AS1 cyt1 CER8 (Wassilewskija ecotype)/as1 CYT1 cer8 (a hybrid between the Landsberg erecta and Columbia ecotypes), we identified 445 plants that were as1 but did not show a mutant cer phenotype. An analysis of these recombinants with PCR-based markers from the lower arm of chromosome 2 positioned CYT1 between m429 and AthBIO2 (Fig. 1). The use of additional markers from this region (16) made it possible to map the CYT1 gene to an interval of less than 10 kb within the BAC clone T5I7 (Fig. 1). This interval contained two ORFs—RNS2, an S-like RNase (17), and a putative mannose-1-phosphate guanylyltransferase (termed T5I7.8 in the sequence annotations; ref. 18).
Figure 1.
Map-based cloning of the CYT1 gene. (Upper) Genetic map of the CYT1 region of chromosome 2. (Lower) Map of the BAC clone T5I7 showing the location and properties of polymorphic repeats used for mapping. Predicted transcription units are shown as arrow bars. Numbers indicate recombination events between adjacent markers.
The predicted coding region of the T5I7.8 gene was PCR-amplified from homozygous cyt1-1 and cyt1-2 embryos and sequenced. The cyt1-1 allele had a point mutation in codon 89 (CCT→CTT; Fig. 2) resulting in a change from proline to leucine. In the cyt1-2 allele a single nucleotide was inserted in codon 306 (TCG→TTCG) causing a +1 frameshift and at the same time disrupting an XhoI site. No other difference to the published Columbia sequence was detected. We concluded that T5I7.8 corresponds to CYT1.
Figure 2.
Alignment of the Cyt1 and the yeast Vig9 amino acid sequences. Dots indicate identical residues, dashes indicate gaps, asterisks indicate conserved positions of the transferase hexapeptide repeat consensus motif: [LIV]-[GAED]-X-X-[STAV]-X-[LIV]-X-X-X-[LIVAC]-X-[LIV]-[GAED]-X-X-[STAVR]-X-[LIV]-[GAED]-X-X-[STAV]-X-[LIV]-X-X-X-[LIV]. Changes in the mutant alleles are noted above the sequences.
The CYT1 Gene Provides a Basic Metabolic Function.
Northern analysis of total RNA indicated that CYT1 transcripts were about 1.5 kb in size and expressed in all tissues examined (seedlings, roots, leaves, stems, inflorescences; data not shown). A putative full-length cDNA clone (96N23T7) was obtained from the Arabidopsis stock center and completely sequenced (GenBank accession no. AF108660). The intron/exon structure of CYT1 was as predicted (18). Stop codons in all frames were found in the sequence preceding the first ATG start codon suggesting that the cDNA clone contained the complete CYT1 coding region.
CYT1 encodes a polypeptide of 361 amino acids with strong similarity to the yeast VIG9 gene (60% identity; Fig. 2). The Vig9 protein has been demonstrated to have GTP:α-d-mannose-1-phosphate guanylyltransferase (MPG; EC 2.7.7.13) activity in vitro (19). CYT1 also shows similarity to bacterial MPGs and to the sequence of an N-terminal peptide from the small subunit of porcine MPG (data not shown). The C terminus of CYT1 contains a peptide signature found in a large group of enzymes with transferase activity (transferase hexapeptide-repeat, PROSITE motif PDOC00094; ref. 20). The corresponding sequence is predicted to be completely missing in the Cyt1-2 polypeptide (Fig. 2). Thus, we believe that cyt1-2 is a null-mutation. In contrast, the Cyt1-1 polypeptide is predicted to be of full-length and to differ from wild-type in a single residue. Because cyt1-1 mutant embryos show a slightly less severe phenotype (11), the Cyt1-1 polypeptide apparently retains some residual activity.
The high degree of sequence similarity to MPGs from different organisms suggests that the Cyt1 protein catalyzes the production of GDP-d-mannose from α-d-mannose-1-phosphate and GTP. In higher plants, GDP-d-mannose is required in a number of diverse processes. For example, it is a precursor of ascorbic acid (21) and GDP-l-fucose (22). GDP-d-mannose and GDP-l-fucose, in turn, are used to incorporate mannosyl and fucosyl residues into cell wall polymers. Furthermore, GDP-mannose is required for the synthesis of the core glycan chain attached to N-linked glycoproteins. We show below that all of these processes are affected in cyt1 embryos.
Cyt1 and vtc1 Are Alleles of the Same Genetic Locus.
As an unexpected corollary to our analysis it became evident that the previously described vtc1 mutation is caused by a single amino acid exchange in the same ORF disrupted by cyt1 mutations (ref. 12; Fig. 2). The vtc1 mutant was isolated by virtue of its sensitivity to oxidative stress (23) and found to be deficient in the production of ascorbic acid (14). In striking contrast to the lethality caused by cyt1 mutations, vtc1 plants are viable, fertile, and phenotypically similar to wild-type, indicating that the vtc1 mutation is weak. Consistent with this idea, MPG activity is reduced but not absent in protein extracts prepared from vtc1 mutant tissue (12).
To confirm allelism between vtc1 and cyt1, we generated vtc1/cyt1-1 and vtc1/cyt1-2 trans-heterozygous plants. Like vtc1 mutants, they are viable, fertile, and phenotypically similar to wild-type. Measurements of ascorbic acid levels in rosette leaves (Table 1) showed that vtc1/cyt1-2 trans-heterozygotes contained amounts similar to vtc1 mutants (29% of wild-type), indicating that cyt1-2 does not complement vtc1. In contrast, vtc1/cyt1-1 trans-heterozygotes contained amounts that were intermediate between vtc1 mutants and wild-type (45% of wild-type) suggesting partial interallelic complementation.
Table 1.
Allelism of vtc1 and cyt1 mutations
| Wild-type | vtc1/vtc1 | cyt1-1/vtc1 | cyt1-2/vtc1 | |
|---|---|---|---|---|
| AsA* | 3.1 | 0.9 | 1.4 (0.22) | 0.8 (0.15) |
| Man† | 9.5% (0.12) | 3.5% (0.33) | 4.5% (0.25) | 3.3% (0.13) |
Ascorbic acid content of rosette leaves (μmol per gram fresh weight). Average of three (cyt1-1/vtc1) and five (cyt1-2/vtc1) measurements, standard deviation in brackets. Values for wild-type (Col) and vtc1 were taken from reference (12).
Mannose content of the noncellulosic cell wall fraction of rosette leaves. Values indicate the relative weight with respect to all neutral sugars analyzed (rhamnose, fucose, arabinose, xylose, mannose, and galactose). Average of four measurements, standard deviation in brackets. Wild-type is the Col ecotype, similar values were also obtained for the Ws ecotype and cyt1-1/+, cyt1-1/+, cyt1-2/+, and vtc1/+ plants.
Cyt1 Mutants Are Deficient in N-Glycosylation.
PDI is an abundant protein of the endoplasmic reticulum that is posttranslationally modified with N-linked glycans (24, 25). To examine the effect of cyt1 mutations on N-glycosylation, we analyzed the mobility of PDI on denaturing polyacrylamide gels (Fig. 3). Native PDI extracted from inflorescence tissue exhibited an apparent molecular mass of about 62 kDa. Enzymatic removal of the glycans resulted in a higher mobility of the protein that was detected as a band-shift. PDI extracted from wild-type embryos had a similar mobility as the PDI from inflorescence tissue. In contrast, PDI extracted from cyt1-1 mutant embryos showed a similar mobility to the enzymatically deglycosylated PDI. We conclude that cyt1 mutations cause a deficiency in N-glycosylation.
Figure 3.
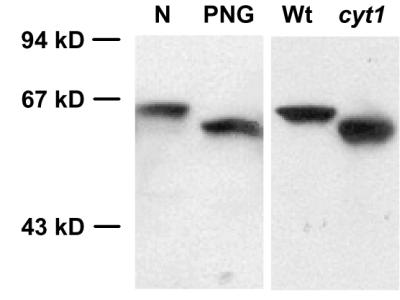
Effect of the cyt1-1 mutation on N-glycosylation. Western blot of an 8% denaturing polyacrylamide gel probed with a PDI antibody. N, native extract of inflorescence tissue; PNG, extract treated with N-glycosidase F; Wt, crude extract of wild-type embryos; cyt1, crude extract of cyt1-1 mutant embryos. Size standards are indicated to the left. The Wt and cyt1 lanes were not loaded with equal amounts of total protein.
The Cell Walls of cyt1 Mutant Embryos Contain Reduced Amounts of Fucose and Mannose.
Mannose and fucose residues are components of the noncellulosic cell wall polymers, and mannose residues have also been detected in the cellulose fraction (26). To examine the effects of cyt1 mutations on cell wall biogenesis, we compared the neutral sugar composition of the noncellulosic cell wall polysaccharides in wild-type and mutant embryos. Mutant embryos and their phenotypically wild-type siblings from the same siliques were dissected from the seeds before desiccation and the neutral cell wall sugars analyzed by gas chromatography.
The cell walls of cyt1-1 embryos contained reduced amounts of mannose (35% of wild-type) and fucose (45% of wild-type; Table 2). Other noticeable differences were elevated levels of rhamnose (152% of wild-type) and xylose (122% of wild-type). Because of their small size, only a single sample of cyt1-2 mutants was analyzed (1,000 embryos, dry weight 140 μg). Mannose and fucose levels were also reduced in the walls of cyt1-2 embryos, but apparently to a smaller extent (both 60% of wild-type). A decrease in the mannose and fucose content was expected from a depletion of GDP-mannose, although the observed effect might be considered small. The altered rhamnose and xylose levels, on the other hand, are more difficult to interpret. Because growth of cyt1 embryos is arrested, these changes might merely reflect differences related to embryonic development. Alternatively, they could result from an attempt to compensate for the lack of mannose and fucose.
Table 2.
Cell wall composition of wild-type and cyt1 embryos
| Wild-type | cyt1-1 | |
|---|---|---|
| Average dry weight | 2.6 μg/embryo | 1.0 μg/embryo |
| Neutral sugars | 61 μg/mg* | 64 μg/mg* |
| Rhamnose | 5.2% (0.30) | 7.9% (1.3) |
| Fucose | 2.0% (0.16) | 0.9% (0.07) |
| Arabinose | 54.9% (1.1) | 53.0% (1.4) |
| Xylose | 19.5% (1.1) | 23.5% (1.7) |
| Mannose | 4.8% (0.32) | 1.7% (0.21) |
| Galactose | 13.6% (1.7) | 13.0% (1.3) |
| Cellulose | 31 μg/mg (0.6) | 5.9 μg/mg (0.8) |
The total amount of neutral sugars of the noncellulosic wall and the amount of cellulose are expressed as a fraction of the dry weight. Values for individual neutral sugars indicate their relative weight with respect to all six sugars analyzed. Average of three measurements, about 500 cyt1-1 embryos or 200 wild-type embryos per sample, standard deviation in brackets.
Single measurement.
A similar analysis was carried out on rosette leaves of vtc1 mutants and vtc1/cyt1 trans-heterozygotes (Table 1). The only noticeable difference to wild-type was a decrease in the mannose content. The mannose content of vtc1 mutants, as well as vtc1/cyt1-2 trans-heterozygotes, was about 35% of wild-type, whereas vtc1/cyt1-1 trans-heterozygotes showed an intermediate mannose content (47% of wild-type). As the ascorbic acid measurements, this result indicates a partial interallelic complementation between vtc1 and cyt1-1.
Cyt1 Mutants Are Deficient in Cellulose.
Some aspects of the cyt1 phenotype, including radial swelling, accumulation of callose, and the occurrence of incomplete cell walls, have been mimicked by treatment with the cellulose synthase inhibitor dichlobenil (11). This led us to compare the cellulose content of wild-type and cyt1 mutant embryos. Because of their small size, analysis of cyt1-2 embryos was not practical. Three samples of cyt1-1 and wild-type embryos were collected as described above and their cellulose content determined. Mutant cyt1-1 embryos contained about five times less cellulose than their wild-type siblings (Table 2). To the best of our knowledge, no other plant mutant causing an equally dramatic decrease in cellulose content has been described.
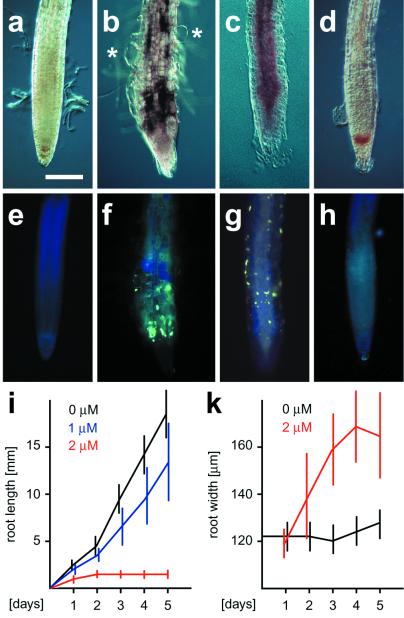
Inhibition of N-Glycosylation Causes Radial Swelling and Accumulation of Callose in Root Tips.
It was unclear how a depletion of GDP-mannose would affect the cellulose content of cyt1 embryos. To explore the possibility that cellulose synthesis was reduced because of a block in N-glycosylation, we examined the effect of tunicamycin on the growth of Arabidopsis roots (Fig. 4). Tunicamycin interferes with the synthesis of the core glycan chain attached to N-linked glycoproteins (27) and thus has a similar effect on N-glycosylation as cyt1 mutations. In the presence of 2 μM tunicamycin, Arabidopsis root tips showed significant radial swelling within 2 to 3 days (Fig. 4k). Bloated epidermal cells were frequently observed, and staining with the callose-specific dye sirofluor revealed ectopic deposition of callose in a patchy pattern (Fig. 4 b and f). Lower concentrations of tunicamycin inhibited the longitudinal growth of roots (Fig. 4i), but did not cause swelling or callose deposition (data not shown). In the presence of 5 μM tunicamycin, longitudinal growth ceased virtually instantaneously and no significant swelling was observed. However, the root epidermis had a slightly rough appearance, and patches of callose depositions were present (Fig. 4 c and g).
Figure 4.
Effects of tunicamycin and mannose on Arabidopsis roots. Bright field (a, b, c, and d) and epifluorescence images (e, f, g, and h) of Arabidopsis root tips stained with the callose-specific fluorescent dye sirofluor. Roots were cultured for 5 days on MS media with the following supplements: a and e, none; b and f, 2 μM tunicamycin; c and g, 5 μM tunicamycin; d and h, 0.1% mannose. Asterisks point out swollen epidermal cells. (Bar, 150 μm.) Time curve of longitudinal growth (i) and radial swelling (k) of Arabidopsis roots incubated with 0 μM (black), 1 μM (blue), and 2 μM tunicamycin (red). Vertical bars indicate standard deviations (sample size between 12 and 18). Tunicamycin concentrations of 0.5 μM and lower had no significant effect on longitudinal growth, whereas concentrations of 5 μM and higher completely arrested it. Concentrations of 1 μM and lower, as well as 5 μM and higher, did not cause significant radial swelling. For the sake of clarity these results are not included in the figure.
Tunicamycin is toxic to plant cells. To address the possibility that the observed effects were correlated to cell death, we also incubated roots on media containing levels of mannose that are cytotoxic and induce DNA laddering (28). Root growth was severely inhibited in the presence of 0.1% mannose. However, neither radial swelling nor accumulation of callose were observed (Fig. 4 d and h). Our results indicate that characteristic aspects of the cyt1 phenotype can be mimicked by treatment with tunicamycin, but not by treatment with cytotoxic doses of mannose.
Discussion
Map-based cloning of the Arabidopsis CYT1 gene showed that it encodes an MPG required for the production of GDP-mannose. A weak mutation in the same gene, vtc1, was previously isolated because of its low content of ascorbic acid (14, 23). In contrast to the lethality caused by cyt1 mutations, vtc1 plants are viable, fertile, and phenotypically similar to wild-type, and MPG activity is reduced but still present in protein extracts prepared from vtc1 mutant tissue (12). Similarly, reduction of MPG activity by antisense techniques revealed a close correlation between enzymatic activity and ascorbic acid content, but had only subtle phenotypic consequences (29). The cyt1-2 mutation generates a frameshift that eliminates the C-terminal 20% of the wild-type amino acid sequence including a conserved peptide signature, termed a transferase hexapeptide-repeat (20). Thus, we believe cyt1-2 is a null mutation causing a complete lack of activity. Mutant embryos invariably die before or during desiccation and, so far, all attempts to rescue them in tissue culture have failed. Although the Arabidopsis genome contains a number of sequences with similarity to CYT1 (e.g., GenBank accession nos. AAD22341, AAD55285, CAB75917, and CAB79775), the mutants apparently can not generate enough GDP-mannose to sustain their basic metabolism. Two lines of evidence suggest, however, that during the first few days of development a pool of GDP-mannose is available to cyt1 embryos. First, cyt1 embryos are morphologically indistinguishable from wild-type before the transition stage, about 4 to 5 days after fertilization (11). Furthermore, their cell walls contain reduced but still substantial amounts of mannose and fucose. The origin of this pool is unclear. Possibly, MPG activity present in the megaspore mother cells persists long enough to sustain cyt1 embryos for a short time. Such cases of maternally inherited enzyme activities have been described in Drosophila. For example, the half-life of alcohol dehydrogenase (Adh) was estimated to be about 55 h in vivo (30), and maternally derived Adh activity can be detected in adh mutant larvae for 4 to 6 days after fertilization (31).
A depletion of GDP-mannose is predicted to cause a variety of secondary defects. GDP-mannose is a precursor of ascorbic acid (21) and GDP-fucose (22). GDP-mannose, as well as GDP-fucose, are used to incorporate mannose and fucose residues into cell wall polymers. Furthermore, GDP-mannose is required for two types of posttranslational protein modification, glycosylphosphatidylinositol (GPI) membrane-anchoring (32) and N-glycosylation (see below). With the exception of GPI-anchoring, which was not investigated, we have shown that cyt1 embryos are indeed defective in all these processes. What is their likely contribution to the cyt1 phenotype? Ascorbic acid is the most abundant antioxidant of higher plants (33). Among other roles, it has been implicated in the control of cell division. In the maize root meristem, for example, cells with a low ascorbic acid level also show low division rates (34). It seems possible that a lack of ascorbic acid might contribute to the slow growth of cyt1 mutants. However, supplementing cyt1 embryos with ascorbic acid in culture did not have any significant effect (T.C.N. and D.W.M., unpublished data), suggesting that a lack of ascorbic acid levels is not sufficient to explain their developmental arrest.
N-glycosylation is essential for proper folding, targeting, and function of many secreted proteins (35). The core glycan attached to N-linked glycoproteins is assembled on a dolichol phosphate and contains nine mannose residues, which are transferred to it from GDP-mannose. The N-linked glycans are thought to be recognized by ER-resident chaperones that aid folding of the nascent glycoproteins. Correct folding, in turn, allows further processing of the glycans and export of the proteins to other cellular compartments (36). In some instances, N-linked glycan chains have also been implicated in enzymatic activity. For example, activity of the xyloglucan endotransglycosylases TCH4 and Meri-5, enzymes involved in the modification of plant cell walls, can be substantially reduced by removing the N-linked glycans in vitro (37).
Our results demonstrate that cyt1 embryos are deficient in N-glycosylation, presumably because a lack of GDP-mannose prevents synthesis the core glycan. In budding yeast, mutations that block early steps in the assembly of core glycans (e.g., alg1, alg2, alg4, and the cyt1 ortholog vig9) are lethal (38). No Arabidopsis mutants specifically affecting these steps have been described so far. However, synthesis of the core glycan can be inhibited pharmacologically with tunicamycin, which blocks the first step in the process by preventing the transfer of N-acetylglucosamine to dolichol phosphate (27). On treatment with tunicamycin, cultured plant cells become deficient in protein processing and secretion into the medium (39, 40). Tunicamycin also triggers the unfolded protein response which eventually leads to a general shut-down of protein synthesis and an arrest of the cell cycle (41). Thus, it appears likely that the developmental arrest of cyt1 embryos is largely due to their deficiency in N-glycosylation.
The cell walls of cyt1 embryos show a number of severe defects (11). In part, these defects might be due to a failure of correctly modifying cell wall proteins, such as TCH4 and Meri-5 (see above) or GPI-anchored arabinogalactan-proteins (42). Unfortunately, there is only limited knowledge about the function of cell wall proteins, which makes it difficult to evaluate this possibility. An analysis of the cell wall polysaccharides of cyt1 embryos revealed only relatively minor changes in the composition of the noncellulosic fraction, namely an about 50% decrease in mannose and fucose content. Most likely these changes have little impact on the functionality of the walls. Arabidopsis mur1 mutants, for example, completely lack fucose in the walls of their stems and leaves, but are viable and fertile (7). On the other hand, cyt1 embryos showed a 5-fold decrease in cellulose content. By comparison, a conditional mutation in a cellulose synthase gene, rsw1, causes an approximately 50% reduction in cellulose concomitant with pronounced radial swelling of the root tips at the nonpermissive temperature (4). We infer that the cellulose deficiency of cyt1 embryos severely impairs the mechanical properties of their cell walls. Furthermore, it might also explain some of the observed structural changes, for example the ectopic deposition of callose. Callose is normally not found in the primary wall, but can be rapidly accumulated in response to wounding, pathogen attack, or mechanical stress (43). It is possible that cyt1 embryos accumulate callose as a response to mechanical stress.
Some aspects of the cyt1 phenotype, such as isodiametric swelling and accumulation of callose, can be mimicked by treatment with tunicamycin (this study) as well as the cellulose synthase inhibitor dichlobenil (11). In a previous study, tunicamycin has been demonstrated to inhibit the formation of cellulose microfibrils in the alga Oocystis solitaria (44). However, the aim of this study was to explore the role of dolichol-linked glucans as a possible substrate for cellulose synthase and, therefore, tunicamycin was applied at high concentrations where it prevents formation of these glucans. Because we have now found that cellulose content can also be affected by depletion of GDP-mannose, it appears more plausible that the observed effect on cellulose biosynthesis is due to a deficiency in N-glycosylation. How could a block in N-glycosylation affect cellulose biosynthesis? Perhaps the most obvious possibility is that N-linked glycans are required for proper folding or function of cellulose synthase or another unidentified component of the rosette complex. Consistent with this idea, the sequence of RSW1 contains four potential N-glycosylation sites that cluster around the presumptive catalytic domain (4). An alternative possibility is that an N-glycosylated polypeptide might act as a primer for polymer synthesis (45). Initiation of other glucan chains, such as starch and glycogen, requires polypeptide primers (46). This alternative might also explain why small amounts of mannosyl residues have been detected in cellulose preparations (26).
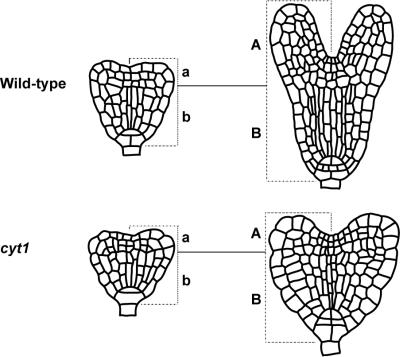
In a large-scale screen for embryos with altered morphology, we have identified nine mutations producing similar phenotypes to cyt1-1 and cyt1-2 (W.L., S. Gillmor, A. Roeder, and C.R.S., unpublished data). All of these mutations were in the CYT1 gene, indicating that cyt1 embryos have a distinct and easily recognizable shape. How is this shape determined? Between the early heart stage and the torpedo stage, wild-type embryos undergo morphogenetic changes realized by regional differences in cell expansion and cell division (Fig. 5). In contrast, growth of cyt1 embryos is essentially isodiametric. Their hypocotyls elongate to a similar extent as in wild-type (Fig. 5, B to b is about 1.6 in both cases), but there is no control over the direction of cell expansion and, as a consequence, they are radially swollen. In the cotyledons, fewer cell divisions occur than in wild-type such that they only grow with a similar rate as the hypocotyl (Fig 5, A to a is about 1.6 in cyt1, but greater than 3 in wild-type). Thus, two main defects appear to contribute to the shape of cyt1 embryos—a block of cell division and an inability to direct cell expansion. Although a number of factors might contribute to the block of cell division, the inability to direct cell expansion can in all likelihood be attributed to mechanically compromised cell walls. We expect that mutations in genes essential for cell wall biogenesis will cause comparable cell expansion defects in the embryo.
Figure 5.
Growth of wild-type and cyt1 mutant embryos. Drawings of whole mount cleared specimen at the early heart (Left) and the torpedo stage (Right).
Acknowledgments
We thank T. Richmond for deglycosylated protein extract, G.-J. De Boer for help with gas chromatography, and the members of the Somerville lab for comments on the manuscript. This work was funded in part by a grant from the U.S. Department of Energy (DE-FG02–97ER20133). W.L. was supported by the Human Frontiers Science Project Organization (Fellowship LT-594–96) and the Carnegie Institution of Washington (Barbara McClintock Fellowship). Support for P.L.C. was obtained from the U.S. Department of Agriculture, National Research Initiative. D.M. and T.C.N. were funded by the National Science Foundation Developmental Mechanisms Program.
Abbreviations
- GPI-anchor
glycosylphosphatidylinositol membrane anchor
- MPG
mannose-1-phosphate guanylyltransferase
- PDI
protein disulfide isomerase
References
- 1.Brown R M, Saxena I M, Kudlicka K. Trends Plant Sci. 1996;1:149–156. [Google Scholar]
- 2.Delmer D P. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 3.Delmer D P, Amor Y. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arioli T, Peng L, Betzner A S, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 5.Heim D R, Skomp J R, Tschabold E E, Larrinua I M. Plant Physiol. 1990;93:695–700. doi: 10.1104/pp.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fagard M, Höfte H, Vernhettes S. Plant Physiol Biochem. 2000;38:15–25. [Google Scholar]
- 7.Reiter W D, Chapple C, Somerville C R. Plant J. 1997;12:335–345. doi: 10.1046/j.1365-313x.1997.12020335.x. [DOI] [PubMed] [Google Scholar]
- 8.Reiter W D, Chapple C, Somerville C R. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- 9.Potikha T, Delmer D P. Plant J. 1995;7:453–460. [Google Scholar]
- 10.Turner S, Somerville C. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickle T C, Meinke D W. Plant J. 1998;15:321–332. doi: 10.1046/j.1365-313x.1998.00212.x. [DOI] [PubMed] [Google Scholar]
- 12.Conklin P L, Norris S R, Wheeler G L, Williams E H, Smirnoff N, Last R L. Proc Natl Acad Sci USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 14.Conklin P L, Williams E H, Last R L. Proc Natl Acad Sci USA. 1996;93:9970–9974. doi: 10.1073/pnas.93.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Updegraff D M. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 16.Lukowitz W, Gillmor S, Scheible W. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor C B, Bariola P A, Del Cardayre S B, Raines R T, Green P J. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, Sakakibara A, Yamasaki M, Yoda K. J Biol Chem. 1997;272:16308–16314. doi: 10.1074/jbc.272.26.16308. [DOI] [PubMed] [Google Scholar]
- 20.Vuorio R, Harkonent T, Tolvanem M, Vaara M. FEBS Lett. 1994;337:289–292. doi: 10.1016/0014-5793(94)80211-4. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler G L, Jones M A, Smirnoff N. Nature (London) 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 22.Gibeaut D M. Plant Physiol Biochem. 2000;38:69–80. [Google Scholar]
- 23.Conklin P L, Pallanca J E, Last R L, Smirnoff N. Plant Physiol. 1997;115:1277–1285. doi: 10.1104/pp.115.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shorrosh B S, Subramaniam J, Schubert K R, Dixon R A. Plant Physiol. 1993;103:719–726. doi: 10.1104/pp.103.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimoni Y, Zhu X Z, Levanony H, Segal G, Galili G. Plant Physiol. 1995;108:327–335. doi: 10.1104/pp.108.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zablackis E, Huang J, Müller B, Darvill A G, Albersheimer P. Plant Physiol. 1995;107:1129–1138. doi: 10.1104/pp.107.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elbein A D. Plant Physiol. 1988;87:291–295. doi: 10.1104/pp.87.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein J C, Hansen G. Plant Physiol. 1999;121:71–79. doi: 10.1104/pp.121.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller R, Springer F, Renz A, Kossmann J. Plant J. 1999;19:131–141. doi: 10.1046/j.1365-313x.1999.00507.x. [DOI] [PubMed] [Google Scholar]
- 30.Anderson S M, McDonald J F. Can J Genet Cytol. 1981;23:305–313. doi: 10.1139/g81-032. [DOI] [PubMed] [Google Scholar]
- 31.O'Donnell J, Gerace L, Leister F, Sofer W. Genetics. 1975;79:73–83. doi: 10.1093/genetics/79.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson M A J. J Cell Sci. 1999;122:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 33.Smirnoff N. Curr Opin Plant Biol. 2000;3:229–235. [PubMed] [Google Scholar]
- 34.Klerk N M, Feldman L J. Development. 1995;121:2825–2833. [Google Scholar]
- 35.Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé A-C, Gomord V, Faye L. Plant Mol Biol. 1998;38:31–18. [PubMed] [Google Scholar]
- 36.Parodi A J. Biochem J. 2000;348:1–13. [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell P, Braam J. Plant J. 1998;18:371–382. doi: 10.1046/j.1365-313x.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- 38.Orlean P. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Pringle J R, Broach J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 229–362. [Google Scholar]
- 39.Faye L, Chrispeels M J. Plant Physiol. 1989;98:845–851. doi: 10.1104/pp.89.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okushima Y, Koizumi N, Sano H. J Plant Physiol. 1999;154:623–627. [Google Scholar]
- 41.Sidrauski C, Chapman R, Walter P. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 42.Schultz C, Gilson P, Oxley D, Youl J, Bacic A. Trends Plant Sci. 1998;3:426–431. [Google Scholar]
- 43.Aist J R. Annu Rev Phytopathol. 1976;14:145–163. [Google Scholar]
- 44.Quader H. Plant Physiol. 1984;75:534–538. doi: 10.1104/pp.75.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacLachlan G A. In: Cellulose and Other Natural Polymer Systems: Biogenesis, Structure and Degradation. Brown R M Jr, editor. New York: Plenum Press; 1982. pp. 327–339. [Google Scholar]
- 46.Smythe C, Caldwell F B, Ferguson M, Cohen P. EMBO J. 1988;7:2681–2687. doi: 10.1002/j.1460-2075.1988.tb03121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]