Abstract
A role for microRNA (miRNA) has been recognized in nearly every biologic system examined thus far. A complete delineation of their role must be preceded by the identification of all miRNAs present in any system. We elucidated the complete small RNA transcriptome of normal and malignant B cells through deep sequencing of 31 normal and malignant human B-cell samples that comprise the spectrum of B-cell differentiation and common malignant phenotypes. We identified the expression of 333 known miRNAs, which is more than twice the number previously recognized in any tissue type. We further identified the expression of 286 candidate novel miRNAs in normal and malignant B cells. These miRNAs were validated at a high rate (92%) using quantitative polymerase chain reaction, and we demonstrated their application in the distinction of clinically relevant subgroups of lymphoma. We further demonstrated that a novel miRNA cluster, previously annotated as a hypothetical gene LOC100130622, contains 6 novel miRNAs that regulate the transforming growth factor-β pathway. Thus, our work suggests that more than a third of the miRNAs present in most cellular types are currently unknown and that these miRNAs may regulate important cellular functions.
Introduction
The role of microRNA (miRNA) is being increasingly recognized in several diverse cellular processes, including oncogenesis1,2 and immune cell function.3,4 Efforts toward a comprehensive characterization of miRNA expression in cellular systems are limited by the continuing discovery of new miRNAs. More than 100 new miRNAs have been identified in humans in the past 2 years.5 Thus, miRNA platforms used to measure miRNA expression in any biologic system are frequently obsolete by the time the studies are published.
Bioinformatic analyses of the genome suggest that there might be thousands of miRNAs encoded in the genome.6 However, thus far only approximately 700 unique miRNAs have been identified in humans.7 Traditional methods for discovering miRNAs have relied on cloning and Sanger sequencing.8,9 The vast dynamic range of miRNAs (5 log or higher) can result in the domination of the data by a few highly expressed miRNAs, limiting the identification of less abundant miRNAs. Massively parallel, high-throughput sequencing (deep sequencing) provides the means to identify the expression of millions of small RNA transcripts in a single tissue type, thereby providing a sensitivity of detection that is several orders of magnitude higher than conventional methods.
Mature B-cell differentiation is of interest, not only for its role in adaptive immunity, but also because malignancies derived from B cells are common. B-cell malignancies reflect defined stages of normal B-cell differentiation and constitute the majority of leukemias and lymphomas. Therefore, we carefully chose a total of 31 samples from normal human B-cell subsets and human B-cell malignancies to represent the spectrum of normal and malignant B cells (Table 1). We reasoned that deep sequencing of a large number of biologically related cases would provide the opportunity to discover new miRNAs and that shared expression of these miRNAs in separate cases would minimize the rate of false discovery.
Table 1.
Normal and malignant B cells used to identify small RNA transcriptome
| ID | Sample | B-cell stage/phenotype |
|---|---|---|
| S1 | Naive B cell (FACS-sorted primary human) | Naive |
| S2 | Naive B cell (FACS-sorted primary human) | Naive |
| S3 | Germinal center B cell (FACS-sorted primary human) | Germinal center |
| S4 | Germinal center B cell (FACS-sorted primary human) | Germinal center |
| S5 | Memory B cell (FACS-sorted primary human) | Memory |
| S6 | Memory B cell (FACS-sorted primary human) | Memory |
| S7 | Plasma cell (FACS-sorted primary human) | Plasma cell |
| S8 | Plasma cell (FACS-sorted primary human) | Plasma cell |
| S9 | Activated B cell (primary human) | Naive |
| S10 | Epstein-Barr virus–transformed B cell (primary human) | Naive |
| S11 | Lymphoblastic lymphoma (primary human tumor) | Precursor B cell |
| S12 | Transformed follicular lymphoma (primary human tumor) diffuse large B-cell lymphoma | Germinal center |
| S13 | Diffuse large B-cell lymphoma (Farage human cell line) | Germinal center |
| S14 | Diffuse large B-cell lymphoma (BJAB human cell line) | Germinal center |
| S15 | Diffuse Large B-cell lymphoma (LY3 human cell line) | Activated phenotype |
| S16 | Burkitt lymphoma (primary human tumor) | Germinal center |
| S17 | Burkitt lymphoma (Ramos human cell line) | Germinal center |
| S18 | Burkitt lymphoma (Daudi human cell line) | Germinal center |
| S19 | HIV lymphoma (primary human tumor) | Unknown |
| S20 | Mantle cell lymphoma (JVM2 human cell line) | Naive |
| S21 | Mantle cell lymphoma (JVM13 human cell line) | Naive |
| S22 | Mantle cell lymphoma (Mino human cell line) | Naive |
| S23 | Hodkgin lymphoma (L1236 human cell line) | Germinal center |
| S24 | Hodkgin lymphoma (L428 human cell line) | Germinal center |
| S25 | Multiple myeloma (U266 human cell line) | Plasma cell |
| S26 | Multiple myeloma (H929 human cell line) | Plasma cell |
| S27 | Multiple myeloma (KMS12 human cell line) | Plasma cell |
| S28 | Marginal zone lymphoma (primary human tumor) | MALT type |
| S29 | Marginal zone lymphoma (primary human tumor) | Splenic type |
| S30 | Chronic lymphocytic leukemia (primary human tumor) | IgVH mutated phenotype |
| S31 | Chronic lymphocytic leukemia (primary human tumor) | IgVH unmutated phenotype |
In this study, we addressed whether deep sequencing could be used to elucidate the small RNA transcriptome of normal human B cells and human B-cell tumors. We analyzed more than 328 934 149 separate sequences (6 billion bases) to identify the expression of small noncoding RNAs including miRNA, transfer RNA (tRNA), piwi-interacting RNA (piRNA), small nuclear RNA (snRNA), ribosomal RNA (rRNA), and small nucleolar RNA (snoRNA). In all, we identified 333 mature miRNAs, a number that is more than twice as high as previously reported in a single tissue type.8 We also identified the expression of 286 candidate novel miRNAs that satisfied the same structural and expression criteria10 that were used to identify known miRNA. Through deep sequencing of biologic replicates, as well as real-time polymerase chain reaction (PCR) of known and candidate novel miRNA, we established deep sequencing as a viable method for reproducibly measuring quantitative miRNA expression. It is anticipated that our work in delineating the expression of small RNAs, including the complete identification of their mature sequences, precursors, genome locations, and conservation patterns of miRNA, will enable a complete exploration of their functional role in normal and malignant B cells. We have thus developed a comprehensive framework that spans the spectrum of identifying new miRNAs using high-throughput sequencing and validating them, and then applying real-time PCR to provide a clear path to clinical utility.
Methods
Patient sample processing
Patient samples were obtained using a protocol approved by the Institutional Review Board at Duke University Medical Center and the other collaborating institutions. Total RNA was extracted using the phenol-chloroform method to preserve miRNA, using Ambion reagents.
Selection of B-cell subsets
Tonsils from young patients undergoing routine tonsillectomy were disaggregated and separated by Ficoll. The mononuclear cell layer was harvested, washed in phosphate-buffered saline (PBS), and resuspended in ACK lysing buffer to remove small numbers of red blood cells. After a wash and resuspension with 10 mL of PBS with 10% bovine serum albumin, cells were counted, and 200 million were stained with fluorochrome-tagged monoclonal antibodies to CD19, immunoglobulin D (IgD), CD38, and CD27. The specific monoclonal antibodies used were anti–CD19-phycoerythrin-Cy5.5, anti–IgD-fluorescein isothiocyanate, anti–CD27-phycoerythrin, and anti–CD38-allophycocyanin, all from BD Biosciences and BD Pharmingen. Cells were sorted using the MoFlo Cell sorter (Dako Cytomation) into naive B cells (CD19+IgD+CD27−CD38+), germinal center B cells (CD19+IgD−CD38++), memory B cells (CD19+IgD−CD27+CD38dim), and plasma cells (CD19dimIgD−CD27++CD38+++). Two replicates of each B-cell subset were obtained from separate patients. The sample purity was verified by fluorescence-activated cell sorting and found to be more than 90% in all cases.
miRNA profiling using real-time PCR
miRNA expression profiling was conducted using the Applied Biosystems 384-well multiplexed real-time PCR assay using 400 ng of total RNA. Eight reactions, each containing 50 ng RNA and a multiplex looped primer pool with endogenous snoRNA controls, were used to reverse-transcribe the miRNAs in parallel fashion. The completed reactions were loaded onto the 384-well plate per manufacturer's instructions, and real-time PCR was run on the ABI 7900HT Prism instrument. For each 384-well plate, we used the automatically determined cycle-threshold (CT) using the SDS 2.2.1 software (Applied Biosystems). Consistent with manufacturer recommendations, a CT > 35 was treated as undetected. The probes deemed to be present were normalized to the average expression of a snoRNA control. The expression values were calculated as 2−ΔCT, then median centered to 500 and log2-transformed.
miRNA library preparation and sequencing
Total RNA (typically 5 μg) from each sample was run on denaturing polyacrylamide-urea gels. The approximately 17-25 nucleotide RNAs were excised from the gel, ligated to sequencing adaptors on both ends, and reverse-transcribed. The resulting cDNA library was PCR-amplified for 15 cycles and gel-purified on 6% acrylamide gel. The gel-purified amplicon quality and quantity were analyzed on a 6% acrylamide gel relative to oligonucleotides of known concentration and size. Library (120 μL 1-4pM) was loaded on to the Illumina cluster station, where DNA molecules were attached to high-density universal adaptors in the flow cells and amplified. The DNA clusters generated via this process were sequenced with sequencing-by-synthesis technology, where successive high-resolution images of the 4-color fluorescence excitation dependent on the base incorporated during each cycle were captured. Sequencing reads were generated for each of the 31 samples, and base calls were rendered using Illumina pipeline Version 1.3.2. All the primary sequencing data and gene expression data are publicly available through the Gene Expression Omnibus (GEO) archive through accession GSE22898
Activation of B cells
B cells were selected from peripheral blood of normal individuals using an antibody for CD19 and magnetic beads. These peripheral blood B cells were incubated with CpG and polyclonal antibodies to IgM and CD40 for 24 hours and then harvested.
Transformation of B cells with Epstein-Barr virus
Peripheral blood B cells from normal individuals were incubated with high titers of the Epstein-Barr virus and incubated for 10 days. Those cells that were replicating autonomously were cultured separately for 7 days, and RNA was extracted.
Gene expression profiling
Gene expression profiling was performed on 3 replicates of each B-cell subpopulation using standard Affymetrix protocols, as described previously.11 Briefly, 1 μg of total RNA was reversed-transcribed using an oligodT primer, and cDNA was synthesized. In vitro transcription using a T7 primer was used to generate labeled cRNA, which was fragmented and hybridized to Affymetrix whole-genome U133 plus 2.0 microarrays. The arrays were scanned and data normalized as described previously. The data have been deposited in the publicly available GEO database (GSE12366).
Tumor samples from 101 patients with diffuse large B-cell lymphoma (DLBCL) were obtained at the time of diagnosis and freshly frozen. These cases were profiled using Affymetrix Gene 1.0 ST arrays. The molecular subgroups were distinguished using a Bayesian approach described previously.11
GO term enrichment analysis
TargetScan12 (release 5.0) was used to predict conserved target genes of known miRNAs that were differentially expressed during B-cell differentiation. For each pairwise comparison (naive vs germinal center, germinal center vs plasma cell, and germinal center vs memory), significantly overrepresented miRNA seed sequences were identified (χ2 test, P < .001). Genes that were differentially expressed in these binary distinctions for which ontology data for molecular function existed were analyzed using GOEAST program.13 Gene ontology terms were considered significantly enriched at P < .001.
Comparison of real-time PCR and sequencing data
Sequencing frequency for each miRNA was scaled to the total number of known miRNA sequences identified in the sample. These scaled frequencies were multiplied by 106 and log2-transformed for the purpose of comparison between samples to the normalized real-time PCR data.
Chromatin immunoprecipitation
Primary germinal center and memory B cells were crosslinked with 1% formaldehyde for 10 minutes at 37°C and terminated with 0.125M glycine for 5 minutes at room temperature. Cells were lysed in hypertonic lysis buffer (5mM Tris-HCl, pH 7.5 [Teknova], 85mM KCl, and 0.5% NP-40) on ice for 10 minutes. After centrifugation at 6000 rpm for 5 minutes at 4°C, the pellets of nuclei were lysed in chromatin immunoprecipitation (ChIP) lysis buffer (0.5% sodium dodecyl sulfate, 1% NP-40, 0.5% sodium deoxycholate in 1× PBS) and sonicated (Ultrasonic CV-18) to shear chromatin DNA to approximately 500-bp fragments. Each nuclear lysate (1/20) was kept for input, and the rest was incubated for 4 hours with Dynabeads (Invitrogen) that were coupled overnight with either the anti-H3K4me3 antibody (07-473; Millipore) or the anti-CD20 antibody (sc-15 361; Santa Cruz Biotechnology). Precipitated chromatin DNA was eluted with elution buffer (1% sodium dodecyl sulfate, 1% NaHCO3) and purified using the QIAGEN MinElute PCR purification kit. The library was subsequently sequenced, and semiquantitative PCR was carried out using primers to amplify the putative promoter region of the miRNA cluster (-226 to +49 relative to the predicted transcription start site, which is 4439 bp upstream of the first miRNA gene in the novel cluster).
Results
Deep sequencing identifies the small RNA transcriptome of normal and malignant B cells
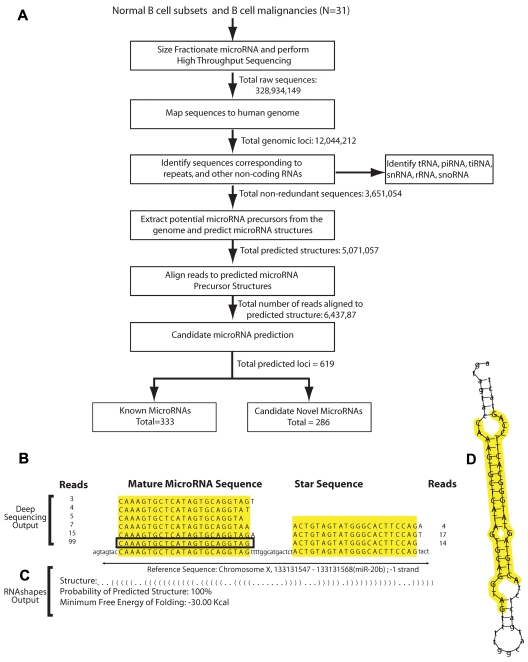
Small RNA libraries from these 31 cases were subjected to massively parallel, high-throughput sequencing using the Illumina platform to generate a total of 328 million separate reads (summarized in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Our approach to analyzing the sequences and discovering miRNAs broadly follows a previously described method14 and is summarized in Figure 1A. All bioinformatics analyses were performed using a cluster of 1024 Linux computer nodes. Preprocessing was carried out using locally written Shell and Perl scripts.
Figure 1.
Identifying miRNAs in normal and malignant B cells. (A) Analytic pipeline for the discovery and validation of known and novel miRNAs in B Cells. Small RNAs from B-cell subsets were subjected to massively parallel sequencing. Raw sequence reads (328 934 149) filtered, mapped to the genome, analyzed by miRDeep, and annotated. Identified miRNA loci were cross-referenced with miRBase13 to distinguish known from candidate novel miRNA. (B) Frequency of reads for miR-20b and their alignment to the genome. The sequence surrounded by a rectangle corresponds to the most abundant and longest mature miRNA sequence that perfectly matched the genome. (C) Outputs of the RNAshapes program that computed the abstract structure of the miRNA precursor, the probability of the predicted structure, and the minimum free energy of folding for the precursor structure for miR-20b. (D) Energetically favorable folding of the miR-20b precursor, with the miRNA mature sequence and the microRNA* sequence highlighted in yellow.
From the raw sequences generated by high-throughput sequencing, the 3′- and 5′- adaptor sequences were trimmed. Low-quality sequences were identified as those sequencing reads that contained stretches of consecutive identical bases or uncalled nucleotides (N) in the first 12 bases and sequencing reads shorter than 17 nucleotides. To minimize redundancy, reads were initially curtailed to the first 22 nucleotides, and identical sequences were represented with a single FASTA entry for analysis. Each unique sequence was mapped to the reference genome (Ensembl, build 50), and reads were filtered such that only perfect alignments (full-length, 100% identity) were retained. Reads that aligned to more than 5 positions in the genome and reads that overlapped with the University of California, Santa Cruz (UCSC) RNA genes were identified and excluded from miRNA analysis. Additional details regarding our methods are included in the supplemental materials.
Sequencing reads that mapped to known noncoding RNAs other than miRNAs15 were annotated separately. For each read, flanking sequences were retrieved from the genome, and potential secondary structures were predicted using RNAfold.16 Predictions of miRNA were made using miRDeep,14 which evaluated the sequences that indicated miRNA precursor processing and the thermodynamic energy of folding these precursors. The predicted probability and the thermodynamic energy of the miRNA precursor structures were consistent with previously described criteria for identifying miRNAs.17 An example of miRNA sequences that met the criteria for identifying a known miRNA (miR-20b) is shown in Figures 1B-D. Sequences that occurred 20 or more times in at least 1 sample were consolidated and annotated for the 31 samples. Genomic loci that overlapped with miRNAs described in miRBase (version 13) were identified as known miRNAs. The remaining genomic loci were identified as encoding candidate novel miRNAs.
To carefully calibrate the performance of our sequencing and bioinformatic approach, we performed multiplexed real-time PCR for 360 known miRNAs on biologic replicates of the same normal B-cell types. We found a high degree of overlap between the detection of miRNAs using both methods. For those 360 miRNAs that we measured using both multiplex real-time PCR and deep sequencing, we found that 64 of the 88 miRNAs were measured using both methods corresponding to a sensitivity of 72%, whereas 230/272 miRNAs were not detected by either method, corresponding to a specificity of 84%. Additional details regarding the calculation of sensitivity and specificity, as well as an alternative method of determining these statistics, which produced similar results, are described in supplemental Text 1 and supplemental Figure 1.
In all, we identified 619 miRNA precursor loci from normal and malignant B cells. The 333 miRNA precursor loci annotated in miRBase5 were identified as known miRNAs (supplemental Table 2), whereas the remaining 286 precursor loci were identified as candidate novel miRNAs (supplemental Table 3). Among these known miRNAs, there were 22 mature sequences (supplemental Table 2) that mapped to the ostensible microRNA* sequences and were expressed at levels that were comparable or, in some cases, higher than their complementary miRNAs. These previously unknown sequences likely reflect the influence of recently described pathways18–20 that allow both the miRNAs and the microRNA* strands to independently exert their regulatory functions.
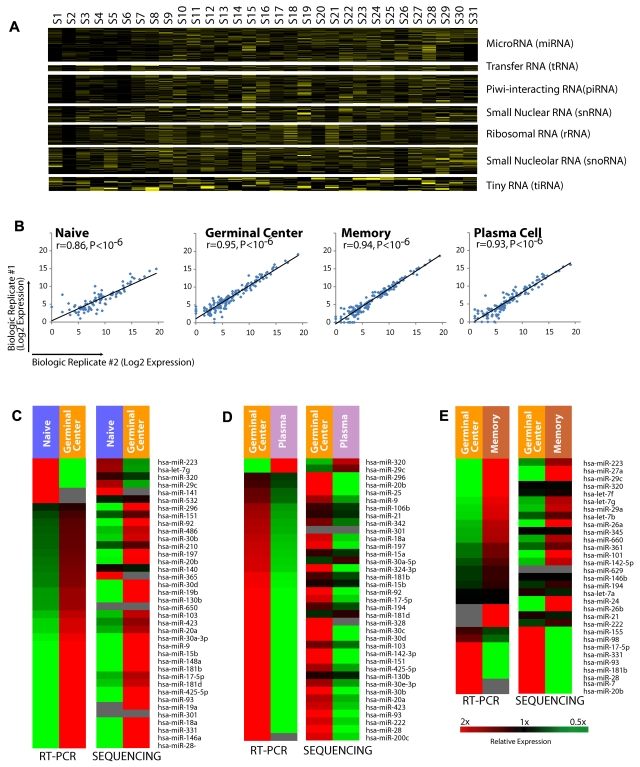
In addition to miRNAs, we identified the expression of other small RNAs, including tRNA, rRNA, snoRNA, snRNA, piRNA, and a recently described class of small RNAs, tiny RNA (tiRNA)21 in our data. The expression of these RNAs is shown in Figure 2A (primary data in supplemental Table 4).
Figure 2.
Distribution of small RNAs in normal and malignant B cells. (A) Expression of small RNAs, including miRNAs, tRNA, piRNA, snRNA, rRNA, snoRNA, and tiRNA in each sample are shown more than a 2-fold range. Samples are listed in the same order as in Table 1. (B) Expression of miRNAs measured by sequencing across 2 biologic replicates for naive, germinal center, memory, and plasma B cells. The P values were computed using a correlation test against the null hypothesis that the data were not correlated; P < 10−6 in all 4 cases. (C) Relative expression of miRNAs differentially expressed in the naive to germinal center B-cell transition as measured by real-time PCR and sequencing. (D) Relative expression of miRNAs differentially expressed in the germinal center B-cell to plasma transition as measured by real-time PCR and sequencing. (E) Relative expression of miRNAs differentially expressed in the germinal center B to memory B-cell transition as measured by real-time PCR and sequencing.
Deep sequencing reproducibly measures miRNA expression in biologic replicates
We compared the expression of miRNAs identified by high-throughput sequencing and our analytic approach. In each of the biologic replicates (Figure 2B), we found that the measured miRNA expression in the biologic replicates were highly correlated (P < 10−6, correlation test for each comparison), confirming that our approach produces robust results. We further examined the expression of 107 miRNAs that we had previously identified as being differentially expressed among B-cell transitions.22 We compared the expression of these miRNAs as measured by real-time PCR and deep sequencing (Figures 2C-E) and found highly concordant results with more than 90% of the differentially expressed miRNAs, demonstrating identical patterns of expression regardless of the method of measurement (P < 10−6, χ2 test).
Therefore, we concluded that our sequencing and analytic approach reproducibly identified miRNA expression in biologic replicates, and the differential miRNA expression measured by deep sequencing and real-time PCR produced similar results.
Hundreds of known and novel miRNAs are expressed in normal and malignant B cells
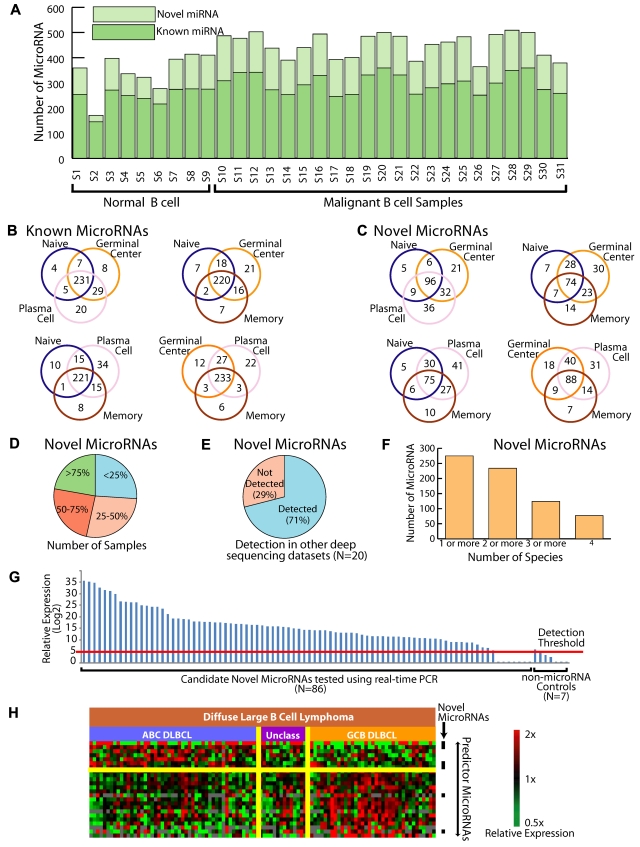
Roughly 2/3 miRNAs measured in each cell type were known miRNAs (Figure 3A), whereas the rest were candidate novel miRNAs. In general, the expression of candidate novel miRNAs was lower than that of the known miRNAs, although there was considerable overlap between their expression levels. We also found that the correlation of miRNA expression measured by real-time PCR and sequencing was similar for both known and candidate novel miRNAs (supplemental Figure 2). The number of known and novel miRNAs detected in each sample type did not vary significantly between individual sample types or between normal and malignant cases. We examined the known and novel miRNAs present in the different normal B-cell subsets and found that the majority of both known (Figure 3B) and novel miRNAs (Figure 3C) were shared across the B-cell lineage.
Figure 3.
Identifying novel miRNAs through high-throughput sequencing. (A) Distribution of known and candidate novel miRNAs in normal and malignant B cells. Samples are listed in the same order as Table 1. (B) Distribution of known miRNA expression among the normal B-cell subsets. The majority of miRNAs are shared between the B-cell types. (C) Distribution of candidate novel miRNA expression among the normal B-cell subsets. The majority of miRNAs are shared between the B-cell types. (D) Proportion of samples in which candidate novel miRNAs were identified. The majority of miRNAs are expressed in more than one sample. (E) Proportion of candidate novel miRNAs identified in 20 additional sequencing datasets. (F) Conservation of novel human miRNAs in chimpanzee, rhesus monkey, mouse, and dog. Ninety percent of the 286 novel miRNAs were found to be conserved across humans and one or more additional species, and more than 80% were found to be conserved across humans and 2 or more additional species. (G) Validation of 86 novel miRNAs candidates by an alternative method of measurement, real-time PCR. Maximum measured values for miRNAs expressed in at least 10% of the samples are shown in log2-scale. Detection threshold is the expression level corresponding to CT ≤ 35. (H) miRNA profiling distinguishes the molecular subgroups of DLCBL that were defined based on gene expression profiles. ABC refers to activated B-cell–like DLBCL. GCB refers to germinal center B-cell–like DLCBL. Unclassified cases are those that did not meet criteria for either group. Black bars indicate candidate novel miRNAs that were differentially expressed between the 2 groups.
The majority (96%) of the candidate novel miRNAs were found in more than one sample, with only a small minority of miRNAs expressed exclusively in a specific B-cell subset or malignancy (Figure 3D). We further examined the expression of these miRNAs in all publicly available small RNA datasets from mammals. The combined number of publicly available samples (N = 20) from 4 separate studies14,23,24 (GEO datasets GSE16579, GSE15190, GSE19473, GPL6583, GSE10829) is considerably smaller than that in our study. Nevertheless, we confirmed the expression of 197 (71%) of the candidate novel miRNAs in those data (Figure 3E and supplemental Table 6). Many of the miRNAs that we identified in normal and malignant B cells were expressed at 10-fold or higher levels in these non B-cell cases. These findings suggest that the miRNAs we have identified are broadly expressed and may have roles in several diverse tissue types.
We also examined the conservation of the novel miRNAs across 4 mammals (chimpanzee, rhesus macaque, mouse, and dog). We found that more than 80% of the identified miRNAs were conserved with 2 or more species (Figure 3F). Our data suggest that there may be species-specific differences in miRNA expression, as has been noted previously.25 In our data, 75% of novel and 80% of known miRNA had more than 100 predicted target genes with a perfectly conserved match in the 3′untranslated region of the gene in the following species: human, mouse, rat, and dog. We found that 11% of novel and 9% of known miRNA had > 50 predicted target genes. The remaining miRNA had < 50 predicted target genes (supplemental Figure 4). Analysis of ontology26 of predicted target genes12 of both known and candidate novel miRNAs expressed during B-cell differentiation strongly implicated transcription factor activity as a regulated function, suggesting a potential role of miRNAs in the regulation of these processes (supplemental Table 5).
Real-time PCR independently confirms the expression of 92% of the novel miRNAs
For further validation, we selected 86 candidate novel miRNAs that were detectably measured in at the sequencing data from least one of 4 DLBCL cases. Using stem-loop reverse transcription27 for quantitative PCR, we tested the expression of the 86 miRNAs in 101 primary tumors from patients with DLBCL and found that 79 (92%) were detectably measured by real-time PCR (Figure 3G) in at least 10% of these cases, suggesting that real-time PCR reproducibly identifies miRNAs that are expressed in lymphomas. We also used real-time PCR to measure the expression of 90 known miRNAs in the same 101 samples and found that more than 90% of these were also detected in at least 10% of the cases using real-time PCR. We found that 6 of the 7 real-time PCR constructs that targeted RNA hairpins that had low probability of being a miRNA resulted in no detectable signal. These results suggest that our assays have high specificity for miRNAs and that the computational predictions based on our sequencing data correctly identified miRNAs.
miRNAs are efficacious in the distinction of clinically relevant groups of lymphoma
Gene expression profiling of patients with DLBCL has demonstrated that the tumors comprise at least 2 distinct diseases with different response rates to standard chemotherapy regimens.28 miRNAs have been shown to be robust biomarkers in malignancies.29 We hypothesized that miRNAs might be used to make this clinically important distinction for which gene expression profiling remains the gold standard. We performed gene expression profiling on 101 DLBCL cases and further subdivided these cases into the molecular subgroups. We found that 25 miRNAs with the highest t statistic were equally efficacious as the gene expression profiling in differentiating the 2 groups of DLBCL with more than 95% overlap between the classifications rendered by the 2 methods, using leaveout one cross-validation (Figure 3H). Interestingly, 6 of these 25 predictor miRNAs (listed in the supplemental materials) were candidate novel miRNAs, suggesting a biologic and clinical relevance for these candidate novel miRNAs in DLBCL tumors. The complete miRNA expression data are included in supplemental Table 8.
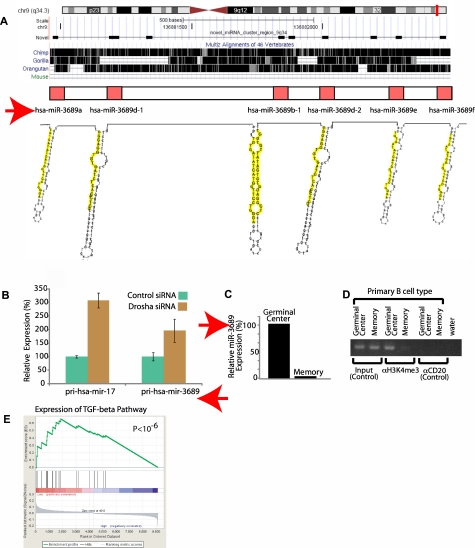
Deep sequencing reveals a new cluster of 6 novel miRNAs
Although miRNAs appear to be distributed throughout the genome, several miRNAs have been found in clusters such as miR-17-92 that are transcribed from a single primary transcript and cleaved into the individual miRNAs by the enzyme Drosha. The discovery of the miR-17-92 cluster greatly accelerated the identification of the function of those miRNAs in several biologic systems.3,4 We found 2 separate clusters of candidate novel miRNAs on chromosome 9 and chromosome 14 (within the IgH locus), respectively. The first cluster was previously annotated as a hypothetical gene LOC100130622 and subsequently discarded from Refseq when no associated protein was identified. Our data demonstrate that this cluster, conserved only in primates, encodes 6 separate miRNAs (Figure 4A). To evaluate whether the miRNAs encoded in these clusters originate from the same primary transcript, we took KMS12 multiple myeloma cells that express these miRNAs and used siRNA to knock down the expression of the miRNA processing enzyme Drosha. This enzyme acts at the first step of miRNA processing by cleaving miRNA precursors from the primary transcript. We found that decreased Drosha expression was associated with increased accumulation of primary transcripts of both the miR-17-92 cluster as well as the novel miR-3689 cluster. (Figure 4B). miRNAs from this cluster were found to be expressed more highly in normal germinal center (GC) B cells compared with memory cells (Figure 4C). We computationally predicted the core promoter region at 4 kb upstream from the primary transcript.30 We performed chromatin immunoprecipitation for H3K4Me3, the histone marker associated with gene expression. DNA bound to this histone marker was found to be selectively enriched in primary GC cells which express this miRNA cluster highly, but not in primary memory B cells (Figure 4D). The miRNAs of this cluster all share the same seed sequence, suggesting that they target the same genes. Among the computationally predicted targets of this miRNA cluster,12 we identified SMAD2 and SMAD3, which are well known mediators of the transforming growth factor-β (TGF-β) signaling pathway. We noted that gene expression both SMAD2 and SMAD3 in our set of 101 DLBCLs were inversely correlated with this cluster (P < .01, correlation test). Gene set enrichment analysis revealed that expression of the TGF-β pathway in DLBCL samples varied inversely with the expression of the miRNA cluster, with a higher expression of the miRNA association with a lower expression of the pathway31 (P < 10−6; Figure 4E and supplemental material), which has previously been noted to be important in the biology of these tumors.32
Figure 4.
Discovery and functional validation of a novel miRNA cluster. (A) Genomic locus of the mir-2355 cluster (chromosome 9q34.3), conservation across 3 primates and one additional mammal (mouse), and predicted secondary structure of each miRNA hairpin, with the mature sequences highlighted in yellow. The sequencess are conserved only in primates. (B) Knockout of the primary miRNA transcript processing enzyme, Drosha, by RNA interference results in accumulation of a known miRNA cluster primary transcript (pri-hsa-mir-17) and the novel miRNA cluster primary transcript (pri-hsa-mir-2355). (C) Expression of hsa-miR-2355 is higher in GCBs compared with memory B cells. (D) Chromatin immunoprecipitation with an antibody to H3K4me3 of GCBs and memory B cells shows enrichment of DNA from the predicted core promoter region of the miR-2355 cluster in GCBs compared with memory B cells. Three sets are shown: input (positive control for PCR using chromatin before immoprecipitation), immunoprecipitation with an antibody for H3K4me3, and immunoprecipitation with an antibody for CD20 (negative control). (E) Gene set enrichment analysis score and distribution of TGF-β pathway genes along the rank of transcripts differentially expressed in high versus low expressors of the miR-2355 cluster. Expression of the TGF-β pathway genes were inversely correlated with expression of the miRNA.
Discussion
The advent of widely accessible whole genome sequencing has created a new urgency to annotate the human genome to provide the context for understanding the genetic variation underlying health and disease. Noncoding RNAs have emerged as the “dark matter” of the genome33 that may play an important role in regulating the 1% of the total DNA that is expressed as protein. The ongoing identification of new miRNA genes in different tissue types is analogous to the discovery of protein-coding genes. Our work provides an exhaustive identification of the miRNAs in normal and malignant B cells that is a prerequisite to the delineation of their role. Furthermore, we have developed a comprehensive framework that spans the identification of miRNAs from deep sequencing data to measuring their expression using real-time PCR and validating their expression in primary human tumors.
Several miRNAs have previously been suggested as being tissue-specific. For instance, miR-223 has been suggested as a myeloid-specific miRNA,34 whereas miR-9, miR-124, and miR-128 have been suggested as brain-specific miRNAs.8 However, our study found expression of all of these miRNAs in the B-cell lineage. Our data indicate that the highly increased sensitivity of deep sequencing will challenge currently held notions regarding tissue-specificity of miRNAs and suggest that the same miRNAs could play different context and lineage-specific roles in different cell types.
miRNA expression has a wide dynamic range from a few transcripts to hundreds of thousands of transcripts per cell. Deep sequencing is especially efficacious for the identification of miRNAs expressed at low abundance in a cell. The number of miRNA transcripts needed to exert their effects in a cell is not known, and it is likely to be context-dependent. Further, it has been demonstrated that some miRNAs containing the hexanucleotide motif AGUGUU can relocalize to the nucleus to potentially regulate the transcription of target genes.35 These findings suggest that a few copies of a miRNA in a cell may be adequate to exert profound down-stream effects. We found the same hexanucleotide motif in 5 miRNAs expressed in B cells. It is also conceivable that some of the low-abundance miRNAs that we have identified in our study may be expressed at higher levels in other development stages or in other cell types. This notion is confirmed by our examination of the novel miRNAs in non–B-cell data. For instance, several the miRNAs that we discovered were also present at 10-fold or higher levels in cell lines derived from breast cancer and cervical cancer, suggesting that the miRNAs that we have discovered in B cells have broad biologic significance.
miRNAs have shown promise as biomarkers in several malignancies.22,29,36,37 DLCBL is the most common form of lymphoma and is known to comprise at least 2 molecularly distinct subgroups with different responses to standard therapy. There are 2 important clinical applications for the molecular subgrouping of DLBCL patients. First, the prognostic information can inform the choices and expectations of patients and their physicians. Second, the important molecular differences in these subgroups might dictate the use of different therapies in these patients. For instance, the benefit of receiving a proteosome inhibitor, bortezomib, appears to be predominantly limited to those patients who have activated B-cell (ABC) DLBCL.38 However, the current methods can only distinguish germinal center B cells (GCBs) from non-GCB DLBCL in a limited fashion,39 and have yielded sometimes conflicting results.40,41 Gene expression profiling remains the gold standard for the distinguishing the 2 molecular subgroups; however, this is not routinely performed in clinical laboratories. Our data suggest that miRNAs can render this distinction with equal efficacy. Since miRNAs are stable in tissue processed for routine paraffin sections using standard methods,42 miRNA-based assays would be much easier to implement in the clinical laboratory.
Emerging data have demonstrated a regulatory role for small RNAs in nearly every cellular system examined thus far. A comprehensive delineation of their role in any cellular system must be preceded by a complete identification of the small RNA transcriptome in that system. The miRNAs that we have identified will likely be included in future commercial microarray platforms that would be widely available. The expression and role of individual miRNAs can also be queried using existing methods as we have demonstrated. Thus, we anticipate that our data, which represents the most in-depth examination of small RNA sequences using second-generation methods, will serve as a foundation for the exploration of the role of small RNAs in several cellular systems.
Supplementary Material
Acknowledgments
We thank Drs Joseph Nevins and Jen-Tsan Chi for a critical review of the manuscript. We thank Susan Sunay for assistance with and the Georgia Cancer Coalition for support for sample collection.
S.S.D. was supported by the Doris Duke Charitable Foundation and the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.D.J., J.Z., C.J., K.L.R., C.H.D., W.W.L.C., W.Y.A., G.S., M.B.C., D.A.R., A.S.L., P.L.L., K.P.M., C.R.F., L.B.-M., K.N.N., A.M.E., L.I.G., M.L., D.R.F., J.B.W., M.A.T., J.I.G., Q.L., T.H., V.G., Y.G., A.P., H.W., J.Z., G.C.B., P.E.L., A.C., and S.S.D. performed research; D.J., J.Z., C.J., and S.S.D. conducted data analysis; and S.S.D., J.Z., and D.J. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sandeep S. Dave, 101 Science Dr, Duke University, Box 3382, Durham, NC 27710; e-mail: sandeep.dave@duke.edu.
References
- 1.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 2.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 3.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basso K, Sumazin P, Morozov P, et al. Identification of the human mature B-cell miRNome. Immunity. 2009;30(5):744–752. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354(23):2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36(Web Server issue):W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedlander MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.I. L., Hofacker WF, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie. 1993;125(2):167–188. [Google Scholar]
- 17.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38(Suppl):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 18.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36(3):431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czech B, Zhou R, Erlich Y, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36(3):445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2009;16(1):43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taft RJ, Glazov EA, Cloonan N, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113(19):4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyman SK, Parkin RK, Mitchell PS, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4(4):e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berezikov E, Thuemmler F, van Laake LW, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38(12):1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, et al. microRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26(1):51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani KM, Lefebvre C, Wang K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294(5543):797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 34.Chen CZ, Li L, Lodish HF, Bartel DP. microRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 35.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 36.Malumbres R, Sarosiek KA, Cubedo E, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113(16):3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 38.Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113(24):6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 40.Nyman H, Adde M, Karjalainen-Lindsberg ML, et al. Prognostic impact of immunohistochemically defined germinal center phenotype in diffuse large B-cell lymphoma patients treated with immunochemotherapy. Blood. 2007;109(11):4930–4935. doi: 10.1182/blood-2006-09-047068. [DOI] [PubMed] [Google Scholar]
- 41.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.