Abstract
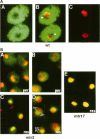
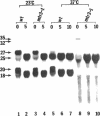
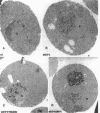
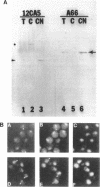
Synthesis of mRNA and rRNA occur in the chromatin-rich nucleoplasm and the nucleolus, respectively. Nevertheless, we here report that a Saccharomyces cerevisiae gene, MTR3, previously implicated in mRNA transport, codes for a novel essential 28-kDa nucleolar protein. Moreover, in mtr3-1 the accumulated polyA+ RNA actually colocalizes with nucleolar antigens, the nucleolus becomes somewhat disorganized, and rRNA synthesis and processing are inhibited. A strain with a ts conditional mutation in RNA polymerase I also shows nucleolar accumulation of polyA+ RNA, whereas strains with mutations in the nucleolar protein Nop1p do not. Thus, in several mutant backgrounds, when mRNA cannot be exported i concentrates in the nucleolus. mRNA may normally encounter nucleolar components before export and proteins such as Mtr3p may be critical for export of both mRNA and ribosomal subunits.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988 Jul;107(1):17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond V. C., Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol. 1993 Jun;13(6):3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R. A., Lehner C. F., Eppenberger H. M., Nigg E. A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989 Feb 10;56(3):379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Carter K. C., Taneja K. L., Lawrence J. B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991 Dec;115(5):1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deák I., Sidebottom E., Harris H. Further experiments on the role of the nucleolus in the expression of structural genes. J Cell Sci. 1972 Sep;11(2):379–391. doi: 10.1242/jcs.11.2.379. [DOI] [PubMed] [Google Scholar]
- Dietz H. C., Valle D., Francomano C. A., Kendzior R. J., Jr, Pyeritz R. E., Cutting G. R. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993 Jan 29;259(5095):680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991 May 31;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Ghetti A., Piñol-Roma S., Michael W. M., Morandi C., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992 Jul 25;20(14):3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessens G. Nucleolar structure. Int Rev Cytol. 1984;87:107–158. doi: 10.1016/s0074-7696(08)62441-9. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Traglia H. M., Dunst R. W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990 Aug;111(2):309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Deerinck T. J., Ellisman M. H., Spector D. L. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994 Aug;126(4):877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W. C., Izaurralde E., Mattaj I. W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994 Mar;124(5):627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Chen S., Hitomi M., Jacobs E., Kumagai C., Liang S., Schneiter R., Singleton D., Wisniewska J., Tartakoff A. M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994 Aug;126(3):649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Goldfarb D., Spitz L. M., Tartakoff A. M., Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993 Jul;12(7):2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi M., Chen S., Tartakoff A. M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol Biol Cell. 1994 Nov;5(11):1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalland K. H., Szilvay A. M., Brokstad K. A., Saetrevik W., Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994 Nov;14(11):7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M., Stevens A., McCammon M., Douglas M. G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993 Jan;13(1):341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T., Yamashita S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987 Nov 15;262(32):15428–15435. [PubMed] [Google Scholar]
- Lee W. C., Xue Z. X., Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991 Apr;113(1):1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. E., Malim M. H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994 Jul 1;8(13):1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- Mirzayan C., Copeland C. S., Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992 Mar;116(6):1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka T., Siomi H., Adachi Y., Ishibashi M., Kubota S., Maki M., Hatanaka M. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9798–9802. doi: 10.1073/pnas.86.24.9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M. W., Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol Cell Biol. 1993 Apr;13(4):2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Wente S. R. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994 Oct;4(10):357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Schneiter R., Kadowaki T., Tartakoff A. M. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995 Apr;6(4):357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Chiang A., Sadler I. Mutations that alter both localization and production of a yeast nuclear protein. Genes Dev. 1988 Jun;2(6):707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- Smitt W. W., Vlak J. M., Molenaar I., Rozijn T. H. Nucleolar function of the dense crescent in the yeast nucleus. A biochemical and ultrastructural study. Exp Cell Res. 1973 Aug;80(2):313–321. doi: 10.1016/0014-4827(73)90302-9. [DOI] [PubMed] [Google Scholar]
- Tartakoff A. M., Schneiter R. The nuclear GTPase cycle: promoting peripheralization? Trends Cell Biol. 1995 Jan;5(1):5–8. doi: 10.1016/s0962-8924(00)88925-4. [DOI] [PubMed] [Google Scholar]
- Tipper D. J. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol. 1973 Oct;116(1):245–256. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973 Feb 25;248(4):1412–1416. [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Harper F., Bachellerie J. P., Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993 Sep;208(1):19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Yung B. Y., Busch R. K., Busch H., Mauger A. B., Chan P. K. Effects of actinomycin D analogs on nucleolar phosphoprotein B23 (37,000 daltons/pI 5.1). Biochem Pharmacol. 1985 Nov 15;34(22):4059–4063. doi: 10.1016/0006-2952(85)90387-9. [DOI] [PubMed] [Google Scholar]