Abstract
Background
The PI3K/AKT pathway plays a pivotal role in breast cancer development and maintenance. PIK3CA, encoding the PI3K catalytic subunit, is the oncogene exhibiting a high frequency of gain-of-function mutations leading to PI3K/AKT pathway activation in breast cancer. PIK3CA mutations have been observed in 30% to 40% of ERα-positive breast tumors. However the physiopathological role of PIK3CA mutations in breast tumorigenesis remains largely unclear.
Methodology/Principal Findings
To identify relevant downstream target genes and signaling activated by aberrant PI3K/AKT pathway in breast tumors, we first analyzed gene expression with a pangenomic oligonucleotide microarray in a series of 43 ERα-positive tumors with and without PIK3CA mutations. Genes of interest were then investigated in 249 ERα-positive breast tumors by real-time quantitative RT-PCR. A robust collection of 19 genes was found to be differently expressed in PIK3CA-mutated tumors. PIK3CA mutations were associated with over-expression of several genes involved in the Wnt signaling pathway (WNT5A, TCF7L2, MSX2, TNFRSF11B), regulation of gene transcription (SEC14L2, MSX2, TFAP2B, NRIP3) and metal ion binding (CYP4Z1, CYP4Z2P, SLC40A1, LTF, LIMCH1).
Conclusion/Significance
This new gene set should help to understand the behavior of PIK3CA-mutated cancers and detailed knowledge of Wnt signaling activation could lead to novel therapeutic strategies.
Introduction
Deregulation of the phosphatidylinositol 3-kinase (PI3K) signaling pathway is frequent in human cancers. Activation of PI3K, which catalyzes inositol lipid phosphorylation to produce phosphatidylinositol-3,4,5-trisphosphate, is one of the most important downstream molecular events following tyrosine kinase receptor activation. Phosphatidylinositol-3,4,5-trisphosphate activates the serine/threonine kinase AKT, which in turn regulates several signaling pathways controlling cell survival, apoptosis, proliferation, motility, and adhesion [1]. PI3K is a heterodimeric enzyme composed of a p110α catalytic subunit encoded by the PIK3CA gene, and a p85 regulatory subunit encoded by the PIK3R1 gene [2].
Gain-of-function mutations in PIK3CA have recently been found in several malignancies, including breast cancer [1], [3], [4]. PIK3CA is frequently mutated at “hotspots” in exons 9 and 20, corresponding to the helical (E542K and E545K) and kinase (H1047R) domains, respectively. P110α carrying a hotspot mutation has oncogenic activity, transforming primary fibroblasts in culture, inducing anchorage-independent cell growth, and causing tumors in animals [5], [6].
After the TP53 suppressor gene, the PIK3CA oncogene is the most frequently mutated gene in human breast cancers (up to 40% of breast tumors) [7], [8]. Activating somatic mutations of other oncogenes (EGFR, KRAS, HRAS, NRAF, BRAF and AKT1) involved in downstream molecular events following tyrosine kinase receptor activation are frequent in several malignancies but rare in breast cancer. Several studies suggest that PIK3CA mutations are more frequent in estrogen receptor alpha (ERα)-positive breast tumors (30–40%) than in ERα-negative breast tumors (10–20%) [7].
The pathological role of these gain-of-function PIK3CA mutations in breast tumors, and particularly in ERα-positive breast tumors, is largely unknown. Better knowledge of PIK3CA mutation impact requires the identification of downstream target genes and signaling pathways activated by aberrant PI3K/AKT signaling. Here, we compared gene expression in PIK3CA-mutated and PIK3CA wild-type ERα-positive breast tumors, using a genome-wide microarray and subsequently real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR).
Materials and Methods
Patients and Samples
We analyzed samples of 292 primary unilateral non metastatic ERα-positive postmenopausal breast tumors excised from women at René Huguenin Hospital (Saint-Cloud, France) from 1978 to 2008. Other characteristics of the patients are listed in Table S1. Each patient gave written informed consent and this study was approved by the Local Ethical Committee (Breast Group of René Huguenin Hospital). Immediately after surgery the tumor samples were stored in liquid nitrogen until RNA extraction. The samples analyzed contained more than 70% of tumor cells. ERα status was determined at the protein level by using biochemical methods (Dextran-coated charcoal method until 1988 and enzyme immunoassay thereafter) and was confirmed at mRNA level by real-time RT-PCR. Forty-three samples were used as a microarray and RT-PCR screening set to identify differentially expressed genes. These genes were then validated in the remaining 249 ERα-positive tumors by means of RT-PCR. Control samples consisted of eight specimens of normal breast tissue collected from women undergoing cosmetic breast surgery or adjacent normal breast tissue from breast cancer patients.
RNA extraction
Total RNA was extracted from breast tissue by using the acid-phenol guanidium method, and its quality was determined by agarose gel electrophoresis and ethidium bromide staining. The 18S and 28S RNA bands were visualized under ultraviolet light.
PIK3CA mutation screening
PIK3CA mutation screening was performed on cDNA fragments obtained by RT-PCR amplification of exons 9 and 20 and their flanking exons. Details of the primers and PCR conditions are available on request. The amplified products were sequenced with the BigDye Terminator kit on an ABI Prism 3130 automatic DNA sequencer (Applied Biosystems, Courtabœuf, France). Sequences thus obtained were compared with the corresponding cDNA reference sequence (NM_006218).
Microarray analysis
Microarray experiments used Human Genome U133 Plus 2.0 arrays from Affymetrix, containing 54675 probe sets. Gene chips were hybridized and scanned using standard Affymetrix protocols. Expression data were obtained as CEL files. BRB ArrayTools (version 3.6.0 available on http://linus.nci.nih.gov/BRB-ArrayTools.html) were used to import CEL files with Robust Method Average (RMA) normalization, and to analyze gene expression. A class comparison based on a univariate t test applied to log-normalized data was used to identify genes differentially expressed in breast tumors with and without PIK3CA mutations. Supervised class prediction analysis was implemented with the Prediction Analysis for Microarrays (PAM) algorithm to identify genes required for optimal prediction [9].
The Database for Annotation, Visualization and Integrated Discovery (DAVID, available on http://david.abcc.ncifcrf.gov/) was used to interpret the lists of differentially expressed probes and to identify statistically overrepresented biological function categories of Gene Ontology (GO) and biological pathways, as defined in the Kyoto Encyclopedia of Genes and Genomes (KEGG).
In compliance with the Minimun Information About a Microarray Experiment (MIAME) recommendations, raw data were deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under series accession number GSE22035.
Real-time quantitative RT-PCR
RT-PCR was applied to the selected genes, as well as ERα (NM_000125), MKI67 (NM_002417), and TBP (NM_003194; endogenous mRNA control). Primers and PCR conditions are available on request, and the RT-PCR protocol using the SYBR Green Master Mix kit on the ABI Prism 7900 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) is described in detail elsewhere [10]. The relative mRNA expression level of each gene, expressed as the N-fold difference in target gene expression relative to the TBP gene, and termed “Ntarget”, was calculated as Ntarget = 2ΔCt sample. The value of the cycle threshold (ΔCt) of a given sample was determined by subtracting the average Ct value of the target gene from the average Ct value of the TBP gene. The Ntarget values of the samples were subsequently normalized such that the median Ntarget value of the normal breast samples was 1. The relative expression of each gene was characterized by the median and range, and the differences in gene expression between tumors with and without PIK3CA mutations were analysed for significance with the non parametric Mann-Whitney U test.
Clustering analysis
Hierarchical clustering analyses of gene expression and samples were performed using BRB ArrayTools. Classification performance was calculated as overall accuracy, defined as the proportion of correctly classified tumors in each cluster, using Matthews' correlation coefficient (MCC) [11]. This parameter was used to discriminate identical accuracies. The chi-square test was used to determine the statistical significance of the clustering.
Results
Analysis of differentially expressed genes in 43 ERα-positive tumors
Overview of transcriptome changes in PIK3CA-mutated tumors
To identify PIK3CA mutation-related genes, microarray analysis (Affymetrix U133 Plus 2.0 arrays) was first applied to 43 ERα-positive breast tumors, of which 14 were PIK3CA-mutated and 29 were wild-type (Table S1). We found that 6124 probes were differentially expressed between breast tumors with and without PIK3CA mutations, with P values <0.05. Of these, 2538 probes (1630 unique genes) were up-regulated (Table S2) and 3586 (2672 unique genes) were down-regulated (Table S3). Only 216 up-regulated probes (153 unique genes) and 28 down-regulated probes (18 unique genes) showed at least a 2-fold change (FC).
Gene ontology analysis of differentially expressed genes
To identify families of genes that might have significant roles related to specific biological or molecular processes, we used the DAVID database to annotate the 6124 probes and categorize them by function. As shown in Table 1, these genes were mainly involved in the regulation of transcription, cell cycling, proliferation, death, adhesion and cytoskeleton organization, and also ion binding and transport, and ATP and RNA binding activity.
Table 1. Selected categories significantly over-represented in PIK3CA-mutated breast tumors.
| Up- and down-regulated genes | Up-regulated genes | Down-regulated genes | ||||
| Gene Category | Number of genes | P value | Number of genes | P value | Number of genes | P value |
| GENE ONTOLOGY | ||||||
| • Biological Process | ||||||
| Regulation of transcription | 581 (14%) | 0.0100 | 282 (17%) | <0.0001 | − | − |
| Regulation of cell cycle and proliferation | 203 (4.8%) | 0.0002 | 94 (5.8%) | 0.0004 | 115 (4.3%) | ns |
| Regulation of cell death | 198 (4.7%) | 0.0052 | 84 (5.2%) | 0.0360 | 120 (4.5%) | 0.0430 |
| Cell adhesion | 171 (4.1%) | 0.0073 | 81 (5.0%) | 0.0027 | 94 (3.5%) | ns |
| Ion transport | 169 (4.0%) | ns | − | − | 130 (4.9%) | 0.0003 |
| Cytoskeleton organization | 116 (2.8%) | 0.0014 | 58 (3.6%) | 0.0004 | 63 (2.4%) | ns |
| • Molecular Function | ||||||
| Ion binding | 936 (22%) | 0.0040 | 417 (26%) | 0.0007 | 543 (20%) | ns |
| Metal ion binding | 920 (15%) | 0.0019 | 411 (25%) | 0.0003 | 533 (20%) | ns |
| Zinc ion binding | 518 (22%) | 0.0140 | 268 (16%) | <0.0001 | 269 (10%) | ns |
| ATP binding | 339 (8.1%) | 0.0130 | 130 (8.0%) | ns | 218 (8.2%) | 0.0048 |
| RNA binding | 182 (4.4%) | 0.0009 | 87 (5.3%) | 0.0008 | 108 (4.0%) | 0.0260 |
| Acetylation | − | − | − | − | 378 (14%) | 0.0004 |
| KEGG PATHWAY | ||||||
| Pathways in cancer | 100 (2.4%) | <0.0001 | 55 (3.4%) | <0.0001 | 47 (1.1%) | ns |
| MAPK signaling pathway | 76 (1.8%) | 0.0011 | 32 (2.0%) | 0.0200 | 47 (1.1%) | 0.0190 |
| Calcium signaling pathway | 50 (1.2%) | 0.0093 | 10 (0.6%) | ns | 44 (1.0%) | <0.0001 |
| Jak-STAT signaling pathway | 43 (1.0%) | 0.0210 | 17 (1.0%) | ns | 28 (0.7%) | 0.0470 |
| Wnt signaling pathway | 41 (1.0%) | 0.0370 | 24 (1.5%) | 0.0015 | 17 (0.4%) | ns |
| Apoptosis | 27 (0.6%) | 0.0130 | 12 (0.7%) | ns | 15 (0.4%) | ns |
ns: not significant.
The biological processes, molecular functions and physiological pathways of genes were obtained from the DAVID database using GOTERM_BP_FAT, GOTERM_MF_FAT and KEGG PATHWAY, respectively. The two first tools (Gene Ontology) annotated 4202 genes (1630 up- and 2672 down-regulated genes) while KEGG annotated 960 genes (385 up- and 601 down-regulated genes). The gene enrichment of a given class was measured by determining the number of genes belonging to the class in the list of significantly altered genes, weighed against the total human genome, and was tested using Fisher exact probability test. Not all significant categories are included here in order to reduce redundancy. A given gene can belong to several processes.
The 2672 down-regulated genes were mainly associated with ATP binding, acetylation and ion transport (Table 1). Among the down-regulated genes with FC≥2, no significantly overrepresented GO categories appeared.
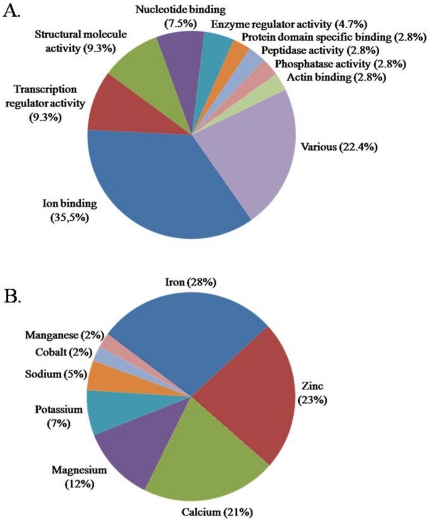
Most of the 1630 up-regulated genes were involved in transcriptional regulation (17.3%) (biological process) and ion binding (25.6%) (molecular function) (Table 1). The latter included the metal ion-binding and zinc ion-binding categories (Table 1). As shown in Figure 1A, the 216 probes most strongly up-regulated in PIK3CA-mutated tumors (153 unique genes) belonged mainly to the ion-binding category (35.5%) but also to categories of structural molecule activity (including structural cytoskeleton constituents) (9.3%), transcription regulatory activity (9.3%) and nucleotide binding (including ATP and GTP binding) (7.5%).
Figure 1. Molecular function classifications of genes up-regulated with a FC≥2 in the PIK3CA-mutated tumors.
Molecular functions were attributed to 107 of the 153 genes using GOTERM_MF_FAT from the DAVID database. Categories with at least three genes are represented in A. Subclassification of the 36 genes belonging to the metal ion-binding category is shown in B. All categories were represented and several genes were common to more than one category. Genes belonging to the metal ion-binding and transcription activity categories are listed in Table 2.
In the ion-binding category, the genes corresponded to genes encoding metal ion-binding proteins in 95% of cases: 28% encoding iron ion-binding and 23% with zinc ion-binding proteins (Figure 1B), pointing to a role of ion-binding proteins, and especially iron ion-binding proteins, in breast cancer with PIK3CA mutations. Interestingly, the genes belonging to the metal ion-binding category (Table 2) included two families of genes that were among the most strongly up-regulated in PIK3CA-mutated tumors. They comprised four genes of cytochrome P450 family 4 (CYP4Z1, CYP4X1, CYP4B1 and the pseudogene (CYP4Z2P) and two solute carrier genes (SLC4A4 and SLC40A1). All these genes, with exception of SLC4A4, are associated with iron ion binding. In addition to these genes, we found on the top of the list lactoferrin (LTF), also known to be involved in iron metabolism. Among the genes encoding zinc ion-binding proteins, three (ANPEP, LIMCH1 and NR2F2) are known to be cancer-related.
Table 2. List of genes belonging to the metal ion-binding and transcription regulation categories.
| Probe set | FC | P value | Gene symbol | Probe set | FC | P value | Gene symbol |
| METAL ION BINDING | 221584_s_at | 2.11 | 0.0023 | KCNMA1 | |||
| Iron ion binding | 1564241_at | 2.07 | 0.0257 | ATP1A4 | |||
| 202018_s_at* | 10.52 | 0.0005 | LTF | 230364_at | 2.00 | 0.0217 | CHPT1 |
| 237395_at* | 7.76 | 0.0035 | CYP4Z1 | Sodium ion binding | |||
| 227702_at* | 5.57 | 0.0032 | CYP4X1 | 203908_at* | 4.81 | 0.0005 | SLC4A4 |
| 239723_at* | 4.42 | 0.0005 | SLC40A1 | 201242_s_at | 2.76 | 0.0001 | ATP1B1 |
| 210096_at* | 4.12 | 0.0011 | CYP4B1 | 201243_s_at | 2.71 | 0.0002 | ATP1B1 |
| 1553434_at* | 3.80 | 0.0009 | CYP4Z2P | 210738_s_at* | 2.13 | 0.0023 | SLC4A4 |
| 225871_at | 2.34 | 0.0188 | STEAP2 | 211494_s_at* | 2.13 | 0.0025 | SLC4A4 |
| 1555497_a_at* | 2.34 | 0.0061 | CYP4B1 | Potassium ion binding | |||
| 233123_at* | 2.29 | 0.0139 | SLC40A1 | 244623_at | 2.30 | 0.0152 | KCNQ5 |
| 223044_at* | 2.26 | 0.0066 | SLC40A1 | 221584_s_at | 2.11 | 0.0023 | KCNMA1 |
| 205542_at | 2.17 | 0.0266 | STEAP1 | 1564241_at | 2.07 | 0.0257 | ATP1A4 |
| 219232_s_at | 2.15 | 0.0006 | EGLN3 | Cobalt ion binding | |||
| 222453_at | 2.14 | 0.0119 | CYBRD1 | 205513_at | 2.87 | 0.0009 | TCN1 |
| 204446_s_at | 2.19 | 0.0003 | ALOX5 | Manganese ion binding | |||
| 224996_at | 2.10 | 0.0135 | ASPH | 230364_at | 2.00 | 0.0217 | CHPT1 |
| Zinc ion binding | |||||||
| 202888_s_at* | 3.52 | 0.0008 | ANPEP | TRANSCRIPTION REGULATION | |||
| 212774_at | 2.97 | 0.0320 | ZNF238 | 214451_at* | 6.68 | 0.0020 | TFAP2B |
| 212325_at* | 2.96 | 0.0002 | LIMCH1 | 1553394_a_at* | 4.34 | 0.0035 | TFAP2B |
| 225728-at | 2.72 | 0.0141 | SORBS2 | 223864_at | 4.25 | 0.0399 | ANKRD30A |
| 207981_s_at | 2.69 | 0.0213 | ESRRG | 230316_at* | 3.05 | 0.0006 | SEC14L2 |
| 212328_at* | 2.69 | 0.0001 | LIMCH1 | 204541_at* | 3.03 | 0.0004 | SEC14L2 |
| 204288_s_at | 2.69 | 0.0073 | SORBS2 | 209292_at* | 3.03 | 0.0002 | ID4 |
| 212327_at* | 2.49 | 0.0008 | LIMCH1 | 212774_at | 2.97 | 0.0320 | ZNF238 |
| 241459_at* | 2.35 | 0.0003 | LIMCH1 | 209291_at* | 2.96 | 0.0001 | ID4 |
| 227811_at | 2.20 | 0.0051 | FGD3 | 207981_s_at | 2.69 | 0.0213 | ESRRG |
| 211965_at | 2.18 | 0.0002 | ZFP36L1 | 226847_at | 2.61 | 0.0020 | FST |
| 215073_s_at* | 2.08 | 0.0063 | NR2F2 | 243030_at | 2.49 | 0.0006 | MAP3K1 |
| 231929_at | 2.07 | 0.0039 | IKZF2 | 226992_at | 2.23 | 0.0064 | NOSTRIN |
| 214761_at | 2.05 | 0.0016 | ZNF423 | 212762_s_at* | 2.18 | 0.0000 | TCF7L2 |
| Calcium ion binding | 210319_x_at* | 2.17 | 0.0011 | MSX2 | |||
| 219197_s_at | 3.08 | 0.0173 | SCUBE2 | 216511_s_at* | 2.16 | 0.0000 | TCF7L2 |
| 204455_at | 2.70 | 0.0065 | DST | 224975_at | 2.13 | 0.0003 | NFIA |
| 229030_at | 2.42 | 0.0370 | CAPN8 | 240024_at* | 2.12 | 0.0016 | SEC14L2 |
| 209369_at | 2.42 | 0.0174 | ANXA3 | 209706_at | 2.12 | 0.0292 | NKX361 |
| 203887_s_at | 2.20 | 0.0006 | THBD | 221666_s_at | 2.09 | 0.0050 | PYCARD |
| 204446_s_at | 2.19 | 0.0003 | ALOX5 | 215073_s_at* | 2.08 | 0.0063 | NR2F2 |
| 224996_at | 2.10 | 0.0135 | ASPH | 216035_x_at* | 2.08 | 0.0000 | TCF7L2 |
| 221584_s_at | 2.11 | 0.0023 | KCNMA1 | 231929_at | 2.07 | 0.0039 | IKZF2 |
| 1564241_at | 2.07 | 0.0257 | ATP1A4 | 214761_at | 2.05 | 0.0016 | ZNF423 |
| Magnesium ion binding | 220625_s_at | 2.02 | 0.0286 | ELF5 | |||
| 227556_at | 2.99 | 0.0007 | NME7 | 226806_s_at | 2.02 | 0.0006 | NFIA |
| 243030_at | 2.49 | 0.0006 | MAP3K1 | ||||
These genes are ranked according to the fold change (FC) in tumors with PIK3CA mutations relative to non mutated tumors. Several genes were common to more than one category. The genes marked with an asterisk were selected for RT-PCR validation.
Besides NR2F2, five other transcription factors, all known to be involved in tumorigenesis, were identified (Table 2): (a) TFAP2B, a tumor suppressor gene in breast cancer [12], (b) SEC14L2, a gene possibly involved in the antiproliferative effect of vitamin E in cancer [13], (c) ID4, which has been proposed to be involved in breast cancer, inhibiting mammary epithelial cell differentiation and stimulating mammary epithelial cell growth [14], (d) TCF7L2, also named TCF4, a cancer-promoting gene involved in the Wnt signaling pathway [15], and (e) MSX2, a gene implicated in mammary gland and breast cancer development [16], and which is also activated by Wnt signaling [17].
These five transcriptional factors (TFAP2B, SEC14L2, ID4, TCF7L2 and MSX2), as well as ten genes involved in metal ion binding (CYP4Z1, CYP4X1, CYP4B1, CYP4Z2P, SLC4A4, SLC40A1, LTF, ANPEP, LIMCH1 and NR2F2), were selected for validation by RT-PCR.
Pathway analysis of differentially expressed genes
By applying KEGG pathway analysis to the 6124 probes differentially expressed in PIK3CA-mutated tumors, we identified physiological pathways directly or indirectly associated with PIK3CA mutations. The most significantly overrepresented pathways are shown in Table 1. In addition to signaling pathways in cancer cells, the following five signaling networks were thus identified: MAPK, Calcium, Jak-STAT, Wnt and apoptosis. The Calcium signaling pathway was specifically altered by the down-regulated genes, whereas the Wnt signaling pathway was specifically altered by the up-regulated genes. The same method applied to the 216 probes (153 unique genes) that were up-regulated with FC≥2 also revealed the Wnt signaling pathway (P = 0.015) (data not shown), highlighting the importance of this pathway in PIK3CA-mutated tumors. Five major genes of the Wnt signaling were thus recognized among the 216 probes (Table S2): MSX2 and TCF7L2 (already cited), and WNT5A, VANGL2 and TNFRSF11B/osteoprotegerin. These genes were also selected for RT-PCR validation.
Finally, among the genes up-regulated with FC≥2 (Table S2), we identified PIK3R1, the gene encoding the PI3K regulatory subunit, and two other genes of interest: HMGCS2, a nuclear gene encoding a mitochondrial matrix enzyme involved in ketogenesis and cholesterol synthesis, processes possibly implicated in the etiology or progression of breast cancer [18] and MAPT, a protein enhancing microtubule assembly and stability, that might be involved in taxane resistance [19]. These three genes were added to the RT-PCR validation set.
Two-class prediction analysis of differentially expressed genes
Two-class prediction analysis with the PAM algorithm was used to identify the group of genes that best characterized PIK3CA-mutated and wild-type tumors and that classified the tumors with the smallest number of predictive features. A threshold of 2.81, that minimized the error, identified 56 differentially expressed probes corresponding to 39 unique genes (Table S4). Thirty-eight of these 39 unique genes were over-expressed in ERα-positive breast tumors with PIK3CA mutations, 16 being up-regulated at least 3-fold, while only one gene (NKAIN1, encoding Na+/K+ ATPase interacting protein) was down-regulated, with a FC of 3.52. Interestingly, two major genes involved in the Wnt signaling pathway were also identified by PAM, namely WNT5A (the most predictive gene) and TCF7L2, further confirming the importance of this pathway in PIK3CA-mutated tumors. The previously selected up-regulated genes were almost all included in the list of the most predictive genes. PAM analysis identified five interesting new genes that were up-regulated with FC≥3, namely VTCN1, TMC5, NTN4, REEP1 and NRIP3, which were added to the RT-PCR validation set.
Among the down-regulated genes, NKAIN1 was selected for RT-PCR validation, along with two other genes known to be involved in cancer biology: TUSC3 and TPD52, that were among the 28 most strongly down-regulated probes (FC≥2) (Table S3) and that were also among the most predictive genes in PAM analysis with a lower FC threshold of 2.5 (data not shown).
Combined analysis of the GO, KEGG and PAM approaches identified 29 most promising genes (26 up-regulated and 3 down-regulated) for RT-PCR validation. The expression status of these genes was first confirmed in the same series of 43 breast tumors (Table 3). Strong positive correlations were observed between the microarray and RT-PCR expression levels of each gene (Spearman's correlation coefficients ranged from 0.69 to 0.97 and were all significant, at P<0.0001; data not shown).
Table 3. Microarray and RT-PCR analyses of the 29 genes in 43 ERα-positive breast tumors.
| Microarray analysis | RT-PCR analysis | ||||||
| Symbol Gene | GenBank | FC | P value | PIK3CA non mutated (n = 29) | PIK3CA mutated (n = 14) | FC | P value |
| UP-REGULATED GENES | |||||||
| ANPEP* | NM_001150 | 3.52 | 0.0008 | 0.17 (0.03–1.52) | 0.37 (0.10–23.7) | 2.16 | 0.0033 |
| CYP4B1* | NM_000779 | 4.12 | 0.0011 | 3.13 (0.11–71.5) | 10.6 (2.14–431) | 3.40 | 0.0033 |
| CYP4X1 | NM_178033 | 5.57 | 0.0032 | 1.04 (0.05–73.3) | 5.85 (0.63–97.7) | 5.62 | 0.0124 |
| CYP4Z1 | NM_171834 | 7.76 | 0.0035 | 0.36 (0.01–220) | 9.17 (0.10–311) | 25.15 | 0.0085 |
| CYP4Z2P* | NR_002788 | 3.80 | 0.0009 | 34.8 (0.12–1457) | 160 (22.9–2103) | 4.59 | 0.0007 |
| HMGCS2* | NM_005518 | 5.31 | 0.0003 | 0.10 (0.00–11.1) | 3.40 (0.07–16.3) | 32.56 | 0.0011 |
| ID4* | NM_001546 | 3.03 | 0.0002 | 0.07 (0.02–0.61) | 0.16 (0.05–1.03) | 2.13 | 0.0133 |
| LIMCH1* | NM_014988 | 2.96 | 0.0002 | 0.54 (0.10–3.83) | 1.66 (0.48–2.87) | 3.06 | 0.0014 |
| LTF* | NM_002343 | 10.52 | 0.0005 | 0.03 (0.00–11.3) | 0.86 (0.00–37.4) | 31.54 | 0.0012 |
| MAPT* | NM_016835 | 2.82 | 0.0004 | 1.09 (0.02–12.1) | 4.40 (0.04–10.2) | 4.02 | 0.0010 |
| MSX2 | NM_002449 | 2.17 | 0.0011 | 1.74 (0.09–4.56) | 3.32 (1.56–8.57) | 1.91 | 0.0025 |
| NR2F2 | NM_021005 | 2.08 | 0.0063 | 0.51 (0.14–2.02) | 1.06 (0.58–2.20) | 2.09 | 0.0009 |
| NRIP3* | NM_020645 | 3.28 | 0.0002 | 0.94 (0.05–18.9) | 3.64 (0.64–33.9) | 3.87 | 0.0025 |
| NTN4* | NM_021229 | 4.21 | 0.0008 | 0.48 (0.05–3.07) | 1.87 (0.68–3.19) | 3.91 | 0.0004 |
| PIK3R1* | NM_181523 | 2.45 | <0.0001 | 0.28 (0.07–0.89) | 0.49 (0.18–1.61) | 1.74 | 0.0053 |
| REEP1* | NM_022912 | 3.30 | 0.0005 | 1.15 (0.16–14.4) | 3.49 (1.36–8.99) | 3.04 | 0.0446 |
| SEC14L2* | NM_012429 | 3.03 | 0.0006 | 0.98 (0.13–16.1) | 5.54 (0.37–24.4) | 5.68 | 0.0049 |
| SLC4A4* | NM_003759 | 4.81 | 0.0005 | 0.28 (0.10–8.45) | 3.45 (0.00–116) | 12.15 | 0.0190 |
| SLC40A1* | NM_014585 | 4.42 | 0.0005 | 0.37 (0.11–7.79) | 1.14 (0.26–6.62) | 3.12 | 0.0068 |
| TCF7L2 | NM_030756 | 2.08 | <0.0001 | 0.24 (0.00–0.64) | 0.35 (0.23–0.91) | 1.45 | 0.0010 |
| TFAP2B* | NM_003221 | 6.68 | 0.0020 | 0.09 (0.00–26.0) | 1.32 (0.00–34.7) | 15.23 | 0.0164 |
| TMC5* | NM_024780 | 4.27 | 0.0022 | 2.53 (0.05–36.4) | 9.45 (1.26–37.8) | 3.74 | 0.0177 |
| TNFRSF11B | NM_002546 | 2.12 | 0.0023 | 0.67 (0.13–10.6) | 2.64 (0.44–31.3) | 3.91 | 0.0004 |
| VANGL2 | NM_020335 | 2.49 | 0.0009 | 0.64 (0.03–3.44) | 1.90 (0.13–5.37) | 2.99 | 0.0018 |
| VTCN1* | NM_024626 | 5.47 | 0.0007 | 0.19 (0.00–4.89) | 1.12 (0.22–23.3) | 5.97 | 0.0025 |
| WNT5A* | NM_003392 | 3.43 | <0.0001 | 0.56 (0.05–6.03) | 2.10 (0.37–6.17) | 3.75 | 0.0013 |
| DOWN-REGULATED GENES | |||||||
| NKAIN1* | NM_024522 | −3.52 | 0.0006 | 137.8 (0.94–560) | 12.13 (1.39–389) | −11.36 | 0.0124 |
| TPD52 | NM_005079 | −2.17 | 0.0014 | 6.29 (3.13–81.8) | 3.88 (1.20–11.68) | −1.62 | 0.0020 |
| TUSC3 | NM_006765 | −2.48 | 0.0026 | 0.58 (0.09–9.35) | 0.32 (0.12–0.63) | −1.83 | 0.0092 |
For each gene, we report the fold change (FC) between tumors with and without PIK3CA mutations. RT-PCR results are expressed as the median (range) mRNA level for each gene relative to normal breast tissues. Genes identified by PAM analysis are marked with an asterisk.
mRNA expression of the 29 genes of interest in 249 ERα-positive breast tumors
Overall expression of the 29 differentially expressed genes
The expression levels of the 29 genes selected by microarray analysis were then verified by RT-PCR in a large independent cohort of 249 ERα-positive breast tumors, of which 157 were PIK3CA wild-type and 92 were PIK3CA-mutated (Table S1). This PIK3CA mutation frequency of 37% was in keeping with the results of previous studies showing a mutation rate of up to 40% in ERα-positive breast tumors [7], [8]. Almost all the tumors had a single mutation, 44 (47.8%) in exon 9 (helical domain) and 46 (50%) in exon 20 (kinase domain) [7]. Two tumors (2.2%) carried two mutations, located in exons 9 and 20 in one case, and in exon 20 in the second case.
Nineteen (66%) of the 29 selected genes showed significantly different expression between mutated and wild-type tumors in the validation cohort, with a distribution similar to that observed in the screening cohort (Table 4). Among the three down-regulated genes of interest in the screening set, only NKAIN1 was significantly down-regulated in the validation set. Among the 26 up-regulated genes, 18 were also up-regulated in the validation set. With exception of VANGL2, up-regulation of the genes involved in Wnt signaling pathway, namely WNT5A, MSX2, TCF7L2 and TNFRSF11B, was confirmed in the validation set, further emphasizing the important role of the Wnt signaling pathway in PIK3CA-mutated breast cancer. Up-regulation was also confirmed for genes related to breast cancer (MAPT, HMGCS2, NR2F2 and TFAP2B), genes involved in metal ion binding (CYP4Z1, CYP4Z2P, SLC40A1, LTF and LIMCH1) and also NRIP3, NTN4, REEP1, SEC14L2 and TMC5. Deregulation of these genes was not related to ERα status or proliferation since similar expression levels of ERα and MKI67 were observed in PIK3CA-mutated and -non mutated tumors (Table 4). Only 2 of the 29 selected genes showed significantly different expression between PIK3CA exon 9- and exon 20-mutated tumors, namely TFAP2B and NRIP3 (Table S5). Interestingly, TFAP2B was over-expressed in exon 20-mutated tumors and NRIP3 in exon 9-mutated tumors.
Table 4. Relative mRNA expression levels of the 29 genes in 249 ERα-positive breast tumors.
| Symbol Gene | GenBank | PIK3CA non mutated (n = 157) | PIK3CA mutated (n = 92) | FC | P value | Rank in PAM |
| UP-REGULATED GENES | ||||||
| ANPEP | NM_001150 | 0.46 (0.00–154) | 0.39 (0.06–18.3) | −0.84 | ns | 15 |
| CYP4B1 | NM_000779 | 6.59 (0.00–222) | 5.72 (0.00–178) | −1.12 | ns | 20 |
| CYP4X1 | NM_178033 | 2.34 (0.02–59) | 3.78 (0.05–101) | 1.62 | ns | 11 |
| CYP4Z1 | NM_171834 | 1.15 (0.01–140) | 2.97 (0.01–254) | 2.58 | 0.0134 | 4 |
| CYP4Z2P | NR_002788 | 38.3 (0.00–1815) | 66.4 (0.00–1069) | 1.74 | 0.0060 | 8 |
| HMGCS2 | NM_005518 | 0.29 (0.00–24.8) | 0.60 (0.00–25.7) | 2.09 | 0.0487 | 10 |
| ID4 | NM_001546 | 0.13 (0.00–9.10) | 0.17 (0.02–9.57) | 1.30 | ns | 28 |
| LIMCH1 | NM_014988 | 0.73 (0.05–6.59) | 1.09 (0.08–8.58) | 1.49 | 0.0007 | 19 |
| LTF | NM_002343 | 0.08 (0.00–14.7) | 0.14 (0.00–41.8) | 1.74 | 0.0036 | 7 |
| MAPT | NM_016835 | 3.03 (0.07–71.1) | 4.52 (0.15–26.2) | 1.49 | 0.0039 | 14 |
| MSX2 | NM_002449 | 2.26 (0.00–13.9) | 3.69 (0.11–39.3) | 1.63 | 0.0003 | 5 |
| NR2F2 | NM_021005 | 0.79 (0.06–10.8) | 1.00 (0.11–7.27) | 1.25 | 0.0415 | 24 |
| NRIP3 | NM_020645 | 1.55 (0.00–168) | 2.69 (0.10–105) | 1.73 | 0.0250 | 16 |
| NTN4 | NM_021229 | 0.75 (0.03–5.47) | 1.17 (0.04–10.2) | 1.57 | 0.0002 | 12 |
| PIK3R1 | NM_181523 | 0.32 (0.06–1.38) | 0.37 (0.08–1.30) | 1.16 | ns | 27 |
| REEP1 | NM_022912 | 1.85 (0.00–12.1) | 2.59 (0.19–21.8) | 1.40 | 0.0053 | 6 |
| SEC14L2 | NM_012429 | 2.49 (0.00–24.0) | 4.51 (0.16–39.1) | 1.81 | <0.0001 | 2 |
| SLC4A4 | NM_003759 | 0.29 (0.00–178) | 0.42 (0.00–128) | 1.43 | ns | 17 |
| SLC40A1 | NM_014585 | 0.88 (0.03–7.81) | 1.22 (0.00–17.9) | 1.38 | 0.0311 | 13 |
| TCF7L2 | NM_030756 | 0.26 (0.03–1.05) | 0.32 (0.06–1.26) | 1.21 | 0.0373 | 26 |
| TFAP2B | NM_003221 | 1.28 (0.00–35.7) | 5.53 (0.00–179) | 4.31 | 0.0055 | 3 |
| TMC5 | NM_024780 | 4.79 (0.01–69.0) | 5.77 (0.11–46.2) | 1.20 | 0.0331 | 9 |
| TNFRSF11B | NM_002546 | 1.25 (0.00–50.3) | 1.90 (0.15–21.8) | 1.52 | 0.0068 | 21 |
| VANGL2 | NM_020335 | 0.73 (0.03–4.64) | 0.82 (0.07–9.09) | 1.12 | ns | 23 |
| VTCN1 | NM_024626 | 0.61 (0.00–10.3) | 0.64 (0.01–15.4) | 1.05 | ns | 22 |
| WNT5A | NM_003392 | 0.74 (0.03–12.4) | 1.17 (0.18–7.27) | 1.59 | <0.0001 | 18 |
| DOWN-REGULATED GENES | ||||||
| NKAIN1 | NM_024522 | 81.1 (0.54–1648) | 57.7 (0.71–560) | −1.41 | 0.0471 | 1 |
| TPD52 | NM_005079 | 6.01 (1.30–115) | 5.34 (1.75–80.9) | −1.12 | ns | 25 |
| TUSC3 | NM_006765 | 0.68 (0.08–3.72) | 0.63 (0.08–6.31) | −1.09 | ns | 29 |
| CONTROL GENES | ||||||
| ERα | NM_000125 | 8.77 (1.27–68.9) | 8.86 (1.59–39.8) | 1.01 | ns | − |
| MKI67 | NM_002417 | 12.1 (0.86–57.2) | 11.0 (1.79–313) | 0.91 | ns | − |
ns: not significant.
Results are expressed as the median (range) mRNA level for each gene relative to normal breast tissues. For each gene, we report the fold change (FC) between tumors with and without PIK3CA mutations and the PAM rank.
Identification of the most discriminatory genes
PAM prediction analysis was then used to test the ability of each gene to classify the 249 ERα-positive breast tumors according to PIK3CA mutation status. NKAIN1 was the most predictive gene (PAM rank) (Table 4). NKAIN1 was also an essential classifier in supervised hierarchical clustering analysis. Indeed, the 19-gene set including NKAIN1 classified the 249 breast tumors significantly more accurately than the set of 18 up-regulated genes without NKAIN1 (accuracy 59% and 57%, Χ2 test P values of 0.0006 and 0.0141, respectively) (Table S6). Three different minimal sets of 4, 5 and 6 genes, all including both NKAIN1 and CYP4Z2P, showed the same overall clustering accuracy of 59.4% (Table S6). However, the 5-gene group (NKAIN1-CYP4Z2P-NRIP3-SEC14L2-TFAP2B) had the most significant discriminatory value (MCC = 0.2334, P = 0.0002) correctly clustering 66 of the 92 mutated tumors and 82 of the 157 non mutated tumors. Notably, this 5-gene set contained the two genes that were differently expressed between exon 9- and exon 20-mutated tumors, and thus had the best capacity to distinguish between these two tumor categories (data not shown). The other two gene sets, both comprising genes involved in Wnt signaling (NKAIN1-CYP4Z2P-WNT5A-TMC5 and NKAIN1-CYP4Z2P-WNT5A-MAPT-MSX2-TFAP2B), classified 65 mutated and 83 non mutated tumors correctly (MCC = 0.2286, P = 0.0003).
Discussion
We used a two-step strategy to identify downstream target genes and signaling pathways affected by PIK3CA mutations in breast tumors. We first applied a pangenomic oligonucleotide microarray approach to a series of 43 ERα-positive tumors with and without PIK3CA mutations, and then validated genes of interest by RT-PCR in an independent series of 249 ERα-positive tumors. A robust set of 19 genes differentially expressed in PIK3CA-mutated and wild-type tumors was thus identified.
Over-expression of several genes involved in Wnt signaling (WNT5A, TCF7L2, MSX2 and TNFRSF11B), regulation of gene transcription (SEC14L2, MSX2, TFAP2B and NRIP3) and metal ion binding (CYP4Z1, CYP4Z2P, SLC40A1, LTF and LIMCH1) was observed in PIK3CA-mutated tumors. Several of these genes have been linked to breast cancer (MAPT, HMGCS2, NR2F2, TFAP2B, NTN4, SEC14L2 and LTF).
The human Wnt signaling network is important for regulation of proliferation, differentiation, growth and survival from the embryo stage [20], [21]. Crosstalk of complex pathways belonging to Wnt signaling has been observed leading to, when altered, disparate effects in different tumor types [22]-[24]. We observed over-expression of four major genes involved in the Wnt pathway, namely WNT5A, TCF7L2, MSX2 and TNFRSF11B. WNT5A encodes a major Wnt ligand affecting tumor cell motility and metastasis, but its role in breast cancer is controversial [23]. The emerging view is that, in breast cancer, WNT5A has a suppressive effect, inhibiting migration and invasion of breast cancer cell lines [24]. Moreover, WNT5A over-expression observed in invasive breast tumors has been associated with a favorable outcome [24]. PIK3CA mutations have also been associated with favorable outcome of breast cancer patients [1], [3], [4], [25]. We can thus suggest a link between gain-of-function mutation in PIK3CA, up-regulation of WNT5A and favorable outcome in breast cancer. We also observed over-expression of TCF7L2, which encodes one of the four major transcription factors involved in the Wnt signaling pathway [20], [26], as well as two other genes (MSX2 and TNFRSF11B) known to be downstream targets of the Wnt signaling pathway [27]–[29]. Wnt signaling has a major role in cancer stem cell self-renewal and tumor maintenance [20], [30] and contributes to tumor invasion, metastasis and angiogenesis [31]. Recent studies have identified a role of Wnt pathway in epidermal-mesenchymal transition during breast cancer development [32], [33]. Thus, Wnt pathway activation appears to be an important consequence of PIK3CA mutations in breast tumors, in keeping with recently observed crosstalk between the PI3K/Akt and Wnt pathways in both physiological (myeloid progenitor cells) [21] and pathological conditions (medulloblastoma) [34].
Better understanding of the biological functions of the Wnt and PI3K/Akt pathways and their interplay could have therapeutic implications for breast cancer. Drugs targeting the PI3K/Akt pathway have given promising preliminary results in human malignancies [35], [36]. However, as the PI3K pathway is crucial for metabolic processes, PI3K inhibitors might also have side effects, especially affecting insulin signaling and cardiac functions [36], [37]. In contrast, targeting of downstream Wnt signaling events might have fewer adverse effects, considering their crucial importance in embryonic development [23], [38].
Genes encoding metal ion-binding proteins were also over-expressed in PIK3CA-mutated tumors. Such metal ion-binding proteins have regulatory roles in central cellular processes such as gene expression, proliferation, differentiation and survival. Increased expression of these proteins in ERα-positive breast tumors has also been reported by Abba et al. [39]. We observed over-expression of LIMCH1, a gene encoding zinc-binding protein, and also four genes encoding iron-binding proteins (LTF, SLC40A1, CYP4Z1 and CYP4Z2P) previously linked to breast cancer. LTF encodes lactoferrin, a protein involved in non specific immunity and that may inhibit carcinogenesis and tumor growth [40]. CYP4Z1 and its pseudogene CYP4Z2P are two members of cytochrome P450 family 4 which have been found to be over-expressed in about 50% of breast cancers relative to normal breast tissue from the same patients [41]. Here, we confirm that the pseudogene CYP4Z2P is expressed in both PIK3CA-mutated and -non mutated ERα-positive breast tumors, by using specific primers unambiguously distinguishing CYP4Z2P from CYP4Z1. Thus, CYP4Z2P is transcriptionally active, but its translation remains to be studied. CYP4Z2P is located in a head-to-head orientation close to CYP4Z1 in chromosome region 1p33 [41], raising the possibility that expression of these two genes is co-regulated in PIK3CA-mutated breast tumors.
We identified several genes previously implicated in breast cancer development or outcome. The proteins encoded by TFAP2B, NTN4 and SEC14L2 have been linked to tumors with less aggressive features and better outcome [12], [13], [42]. MAPT has been proposed as a predictive marker of taxane responsiveness in breast cancer [43]. NR2F2 has been also detected up-regulated in breast cancer, but its involvement in tumor development remains elusive because of its ability to affect both pro-oncogenic and anti-oncogenic proteins [44], [45]. HMGCS2 was recently shown to be regulated in response to hormonal stimulation [18].
NRIP3, TMC5, REEP1 and NKAIN1, whose expression had not previously been described in breast cancer, were also deregulated in the PIK3CA-mutated breast tumors. NRIP3, TMC5 and REEP1 are differentially expressed in various other tumor types [46]–[48]. Interestingly, NKAIN1 was the only gene under-expressed in PIK3CA-mutated tumors and was also the most discriminatory gene for these tumors. The role of these genes in breast cancer development remains to be evaluated in following studies.
Recently, Loi et al. identified a 278 gene-expression signature associated specifically with PIK3CA exon 20-mutated ER-positive/ERBB2-negative tumors [25]. These authors observed an unexpected significant down-expression of some Akt-regulated genes such as RPS6KB1 in their PIK3CA-mutated tumor series, but a normal level of AKT1 and mTOR transcripts. They also showed that phosphor-Akt expression was not significantly up-regulated at the protein level. In the present study, we did not identify RPS6KB1, AKT1 and mTOR in our final 19-gene set nor in the list of 6124 genes differentially expressed in PIK3CA-mutated tumors (Table S2 and S3). Interestingly, among the 168 significantly up-regulated genes detected by Loi et al., WNT5A and MSX2, as well as HMGCS2 and LTF, were identified in agreement with our results. The data of Loi et al. [25] confirm thus the positive association between PIK3CA mutation and Wnt signaling pathway activation reported in the present manuscript.
In conclusion, this gene expression profiling study suggests that over-expression of genes belonging to the Wnt signaling pathway may play a pivotal role in PIK3CA-mutated breast tumors, in particular WNT5A. Further studies of biological mechanisms affected by PIK3CA mutations may have therapeutic implications.
Supporting Information
Molecular, pathological and clinical characteristics of patients in relation to metastasis free survival (MFS) in the 43 ERα-positive and 249 ERα-positive patient series.
(PDF)
2538 probes up-regulated in tumors with PIK3CA mutations (Mutated) compared to tumors without PIK3CA mutation (Normal) with a P value <0.05 identified by a parametric t test using BRB ArrayTools. These genes were ranked according to fold change (FC) calculated between expression intensities of tumors with PIK3CA mutations and those of tumors without PIK3CA mutation. The 216 probes with a FC≥2 are put in bold. In this list, the probes belonging to Wnt signaling pathway are shaded in light grey, and PIK3R1, HMGCS2 and MAPT are shaded in dark grey.
(PDF)
3586 probes down-regulated in tumors with PIK3CA mutations (Mutated) compared to tumors without PIK3CA mutation (Normal) with a P value <0.05 identified by a parametric t test using BRB ArrayTools. These genes were ranked according to fold change (FC) calculated between expression intensities of tumors with PIK3CA mutations and those of tumors without PIK3CA mutation. The 28 probes with a FC≥2 are put in bold.
(PDF)
List of 56 probes (39 unique genes) deregulated in REα-positive breast tumors with PIK3CA mutations compared to those without PIK3CA mutation identified by PAM. These genes are presented according to the rank in PAM output. For each gene, we report the fold change (FC) calculated between expression intensities of tumors with and without PIK3CA mutations using BRB Arrays Tools. The genes with a FC≥3 are put in bold.
(PDF)
Relative mRNA expression levels of the 29 genes in the 44 ERα-positive breast tumors with exon 9 PIK3CA mutations compared to the 47 tumors with exon 20 PIK3CA mutations. The tumor with PIK3CA mutations in both exon 9 and exon 20 was excluded from the analysis. For each gene, we report the median (range) of the mRNA levels of each gene relative to normal breast tissue samples, the fold change (FC) between tumors with exon 9-mutated and exon 20-mutated PIK3CA and the P value associated to Mann-Whitney U test.
(PDF)
Supervised hierarchical clustering analysis of the 249 ERα-positive breast tumors. Classification performance of discriminating gene sets identified from the 19 significantly deregulated genes in tumors with PIK3CA mutations (18 up-regulated genes + NKAIN1). Each gene set separates the 157 tumors without PIK3CA mutation (N) and the 92 tumors with PIK3CA mutations (M) in two main clusters (cluster 1 and 2).
(PDF)
Acknowledgments
We thank Aurélien de Reyniès and Laetitia Marisa of the Ligue Nationale Contre le Cancer. We are extremely thankful for the contributions made by the CIT platforms (Affymetrix, IGBMC: Christelle Thibault, Philippe Kastner; BAC arrays, Institut Curie: Olivier Delattre; RNA analysis, Saint-Louis Hospital: Mira Ayadi).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by INCA (Institut National du CAncer), Programme Cancéropôles 2003-2007 Ile de France, and Comité Départemental des Hauts-de-Seine of the Ligue Nationale Contre le Cancer. This work is part of the Cartes d'Identité des Tumeurs (CIT) program (http://cit.ligue-cancer.net/index.php/en) launched and directed by the Ligue Nationale Contre le Cancer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 3.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 4.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saal LH, Holm K, Maurer M, Memeo L, Su T, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 8.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieche I, Onody P, Laurendeau I, Olivi M, Vidaud D, et al. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–1156. [PubMed] [Google Scholar]
- 11.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 12.Gee JM, Robertson JF, Ellis IO, Nicholson RI, Hurst HC. Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J Pathol. 1999;189:514–520. doi: 10.1002/(SICI)1096-9896(199912)189:4<514::AID-PATH463>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Johnykutty S, Tang P, Zhao H, Hicks DG, Yeh S, et al. Dual expression of alpha-tocopherol-associated protein and estrogen receptor in normal/benign human breast luminal cells and the downregulation of alpha-tocopherol-associated protein in estrogen-receptor-positive breast carcinomas. Mod Pathol. 2009;22:770–775. doi: 10.1038/modpathol.2009.24. [DOI] [PubMed] [Google Scholar]
- 14.Shan L, Yu M, Qiu C, Snyderwine EG. Id4 regulates mammary epithelial cell growth and differentiation and is overexpressed in rat mammary gland carcinomas. Am J Pathol. 2003;163:2495–2502. doi: 10.1016/S0002-9440(10)63604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, et al. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh K, Ginsburg E, Vonderhaar BK. Msx-1 and Msx-2 in mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9:195–205. doi: 10.1023/B:JOMG.0000037162.84758.b5. [DOI] [PubMed] [Google Scholar]
- 17.Cheng SL, Shao JS, Cai J, Sierra OL, Towler DA. Msx2 exerts bone anabolism via canonical Wnt signaling. J Biol Chem. 2008;283:20505–20522. doi: 10.1074/jbc.M800851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riggins RB, Lan JP-J, Klimach U, Zwart A, Cavalli LR, et al. ERR Mediates Tamoxifen Resistance in Novel Models of Invasive Lobular Breast Cancer. Cancer Res. 2008;68:8908–8917. doi: 10.1158/0008-5472.CAN-08-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andre F, Mazouni C, Liedtke C, Kau SW, Frye D, et al. HER2 expression and efficacy of preoperative paclitaxel/FAC chemotherapy in breast cancer. Breast Cancer Res Treat. 2008;108:183–190. doi: 10.1007/s10549-007-9594-8. [DOI] [PubMed] [Google Scholar]
- 20.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 21.Nteliopoulos G, Marley SB, Gordon MY. Influence of PI-3K/Akt pathway on Wnt signalling in regulating myeloid progenitor cell proliferation. Evidence for a role of autocrine/paracrine Wnt regulation. Br J Haematol. 2009;146:637–651. doi: 10.1111/j.1365-2141.2009.07823.x. [DOI] [PubMed] [Google Scholar]
- 22.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt pathway. Dev Cell. 2003;5:367–77. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 23.Leris AC, Roberts TR, Jiang WG, Newbold RF, Mokbel K. WNT5A expression in human breast cancer. Anticancer Res. 2005;25:731–734. [PubMed] [Google Scholar]
- 24.Ford CE, Ekström EJ, Howlin J, Andersson T. The WNT-5a derived peptide, Foxy-5, possesses dual properties that impair progression of ERalpha negative breast cancer. Cell Cycle. 2009;8:1838–42. doi: 10.4161/cc.8863. [DOI] [PubMed] [Google Scholar]
- 25.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, et al. Proc Natl Acad Sci U S A. 2010;107:10208–13. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh M. WNT/PCP signaling pathway and human cancer. Oncol Rep. 2005;14:1583–1588. [PubMed] [Google Scholar]
- 27.Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bu G, Lu W, Liu CC, Selander K, Yoneda T, et al. Breast cancer-derived Dickkopf1 inhibits osteoblast differentiation and osteoprotegerin expression: implication for breast cancer osteolytic bone metastases. Int J Cancer. 2008;123:1034–1042. doi: 10.1002/ijc.23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malanchi I, Huelsken J. Cancer stem cells: never Wnt away from the niche. Curr Opin Oncol. 2009;21:41–46. doi: 10.1097/CCO.0b013e32831d1faf. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y. Wnt/Planar cell polarity signaling: A new paradigm for cancer therapy. Mol Cancer Ther. 2009;8:2103–2109. doi: 10.1158/1535-7163.MCT-09-0282. [DOI] [PubMed] [Google Scholar]
- 32.Yook JI, Li XY, Ota I, Hu C, Kim HS, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 33.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baryawno N, Sveinbjörnsson B, Eksborg S, Chen CS, Kogner P, et al. Small-molecule inhibitors of phosphatidylinositol 3-kinase/Akt signaling inhibit Wnt/beta-catenin pathway cross-talk and suppress medulloblastoma growth. Cancer Res. 2010;70:266–276. doi: 10.1158/0008-5472.CAN-09-0578. [DOI] [PubMed] [Google Scholar]
- 35.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Carnero A. Novel inhibitors of the PI3K family. Expert Opin Investig Drugs. 2009;18:1265–1277. doi: 10.1517/13543780903066798. [DOI] [PubMed] [Google Scholar]
- 37.Weng LP, Smith WM, Brown JL, Eng C. PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet. 2001;10:605–616. doi: 10.1093/hmg/10.6.605. [DOI] [PubMed] [Google Scholar]
- 38.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 39.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benaïssa M, Peyrat JP, Hornez L, Mariller C, Mazurier J, et al. Expression and prognostic value of lactoferrin mRNA isoforms in human breast cancer. Int J Cancer. 2005;114:299–306. doi: 10.1002/ijc.20728. [DOI] [PubMed] [Google Scholar]
- 41.Rieger MA, Ebner R, Bell DR, Kiessling A, Rohayem J, et al. Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Res. 2004;64:2357–2364. doi: 10.1158/0008-5472.can-03-0849. [DOI] [PubMed] [Google Scholar]
- 42.Esseghir S, Kennedy A, Seedhar P, Nerurkar A, Poulsom R, et al. Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin Cancer Res. 2007;13:3164–3173. doi: 10.1158/1078-0432.CCR-07-0224. [DOI] [PubMed] [Google Scholar]
- 43.Rouzier R, Rajan R, Wagner P, Hess KR, Gold DL, et al. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc Natl Acad Sci U S A. 2005;102:8315–8320. doi: 10.1073/pnas.0408974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakshatri H, Mendonca MS, Bhat-Nakshatri P, Patel NM, Goulet RJ, Jr, et al. The orphan receptor COUP-TFII regulates G2/M progression of breast cancer cells by modulating the expression/activity of p21(WAF1/CIP1), cyclin D1, and cdk2. Biochem Biophys Res Commun. 2000;270:1144–1153. doi: 10.1006/bbrc.2000.2562. [DOI] [PubMed] [Google Scholar]
- 46.Maxwell GL, Chandramouli GV, Dainty L, Litzi TJ, Berchuck A, et al. Microarray analysis of endometrial carcinomas and mixed mullerian tumors reveals distinct gene expression profiles associated with different histologic types of uterine cancer. Clin Cancer Res. 2005;11:4056–4066. doi: 10.1158/1078-0432.CCR-04-2001. [DOI] [PubMed] [Google Scholar]
- 47.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 48.Wu F, Ivanov I, Xu R, Safe S. Role of SP transcription factors in hormone-dependent modulation of genes in MCF-7 breast cancer cells: microarray and RNA interference studies. J Mol Endocrinol. 2009;42:19–33. doi: 10.1677/JME-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular, pathological and clinical characteristics of patients in relation to metastasis free survival (MFS) in the 43 ERα-positive and 249 ERα-positive patient series.
(PDF)
2538 probes up-regulated in tumors with PIK3CA mutations (Mutated) compared to tumors without PIK3CA mutation (Normal) with a P value <0.05 identified by a parametric t test using BRB ArrayTools. These genes were ranked according to fold change (FC) calculated between expression intensities of tumors with PIK3CA mutations and those of tumors without PIK3CA mutation. The 216 probes with a FC≥2 are put in bold. In this list, the probes belonging to Wnt signaling pathway are shaded in light grey, and PIK3R1, HMGCS2 and MAPT are shaded in dark grey.
(PDF)
3586 probes down-regulated in tumors with PIK3CA mutations (Mutated) compared to tumors without PIK3CA mutation (Normal) with a P value <0.05 identified by a parametric t test using BRB ArrayTools. These genes were ranked according to fold change (FC) calculated between expression intensities of tumors with PIK3CA mutations and those of tumors without PIK3CA mutation. The 28 probes with a FC≥2 are put in bold.
(PDF)
List of 56 probes (39 unique genes) deregulated in REα-positive breast tumors with PIK3CA mutations compared to those without PIK3CA mutation identified by PAM. These genes are presented according to the rank in PAM output. For each gene, we report the fold change (FC) calculated between expression intensities of tumors with and without PIK3CA mutations using BRB Arrays Tools. The genes with a FC≥3 are put in bold.
(PDF)
Relative mRNA expression levels of the 29 genes in the 44 ERα-positive breast tumors with exon 9 PIK3CA mutations compared to the 47 tumors with exon 20 PIK3CA mutations. The tumor with PIK3CA mutations in both exon 9 and exon 20 was excluded from the analysis. For each gene, we report the median (range) of the mRNA levels of each gene relative to normal breast tissue samples, the fold change (FC) between tumors with exon 9-mutated and exon 20-mutated PIK3CA and the P value associated to Mann-Whitney U test.
(PDF)
Supervised hierarchical clustering analysis of the 249 ERα-positive breast tumors. Classification performance of discriminating gene sets identified from the 19 significantly deregulated genes in tumors with PIK3CA mutations (18 up-regulated genes + NKAIN1). Each gene set separates the 157 tumors without PIK3CA mutation (N) and the 92 tumors with PIK3CA mutations (M) in two main clusters (cluster 1 and 2).
(PDF)