Abstract
The Haemophilus influenzae HMW1 adhesin is an important virulence exoprotein that is secreted via the two-partner secretion pathway and is glycosylated at multiple asparagine residues in consensus N-linked sequons. Unlike the heavily branched glycans found in eukaryotic N-linked glycoproteins, the modifying glycan structures in HMW1 are mono-hexoses or di-hexoses. Recent work demonstrated that the H. influenzae HMW1C protein is the glycosyltransferase responsible for transferring glucose and galactose to the acceptor sites of HMW1. An Actinobacillus pleuropneumoniae protein designated ApHMW1C shares high-level homology with HMW1C and has been assigned to the GT41 family, which otherwise contains only O-glycosyltransferases. In this study, we demonstrated that ApHMW1C has N-glycosyltransferase activity and is able to transfer glucose and galactose to known asparagine sites in HMW1. In addition, we found that ApHMW1C is able to complement a deficiency of HMW1C and mediate HMW1 glycosylation and adhesive activity in whole bacteria. Initial structure-function studies suggested that ApHMW1C consists of two domains, including a 15-kDa N-terminal domain and a 55-kDa C-terminal domain harboring glycosyltransferase activity. These findings suggest a new subfamily of HMW1C-like glycosyltransferases distinct from other GT41 family O-glycosyltransferases.
Introduction
The Haemophilus influenzae HMW1 protein is a high-molecular weight non-pilus adhesin that mediates attachment to human epithelial cells, an essential step in the pathogenesis of H. influenzae disease [1]–[3]. HMW1 belongs to a family of proteins secreted via the two-partner secretion (TPS) pathway and requires a cognate outer membrane translocator protein called HMW1B for surface localization. HMW1 and HMW1B are prototype TPS proteins and are examples of TpsA and TpsB proteins, respectively. The HMW1 system requires an additional accessory protein called HMW1C for the fully functional system, a feature that is characteristic of a subset of TPS systems [3], [4]. In contrast to HMW1 and HMW1B, HMW1C lacks a signal sequence and remains in the cytoplasm.
In previous work, we established that HMW1 is a glycoprotein and undergoes glycosylation in the cytoplasm in a process that requires HMW1C [5]. Insertional inactivation of the hmw1C gene results in a change in apparent molecular mass of HMW1 (a 7–8 kDa decrease), partial degradation of HMW1, and a defect in tethering of HMW1 to the bacterial surface [5]. Examination of HMW1 proteolytic fragments by mass spectrometry identified 31 novel carbohydrate modification sites carrying 47 hexose units, corresponding to a molecular mass of ∼7.6 kDa [6]. All of the modified sites are asparagine residues, in all except one case in the conventional consensus sequence of N-linked glycans, namely Asn-X-Ser/Thr. Interestingly, the modifying carbohydrates at these sites are simple mono-hexose or di-hexose sugars rather than N-acetylated sugars, revealing an unusual carbohydrate modification and suggesting the presence of a glycosyltransferase with a novel enzymatic activity capable of transferring hexose moieties to asparagine residues [6]. Recently we established that HMW1C is the glycosyltransferase responsible for modifying HMW1 and is capable of transferring glucose and galactose from UDP-glucose and UDP-galactose to acceptor sites [7].
Carbohydrate modification of proteins is found in all domains of life and provides a mechanism for control of diverse cellular processes, including signal transduction, protein folding, sorting and stability [8], virus-cell interactions [9], and host immune responses [10]. In eukaryotes, N-linked protein glycosylation is the most common modification of secretory proteins and is coupled to protein translocation and folding. Since the realization that prokaryotes are able to glycosylate proteins, over 70 bacterial glycoproteins have been reported. The majority of these proteins are surface exposed and play a vital role in bacterial adhesion to host cells or evasion of host immunity. Studies of the glycan structures modifying bacterial glycoproteins have revealed that bacteria contain unusual and diverse carbohydrate units such as Pse and its derivatives [11]–[14]. The presence of simple mono-hexose and di-hexose structures at asparagine sites on HMW1 expands the recognized range of glycan structures on glycoproteins. Aside from the sugar structures of HMW1, the N-linked glycans on bacterial and non-bacterial glycoproteins are N-acetylated chitobiosyl core oligosaccharides attached to a well-established sequon of Asn-X-Ser/Thr. In contrast, O-linked glycans are either N-acetylated amino sugars or hexoses attached to Ser/Thr side chains, with no clear consensus sequence at the sites of attachment.
Analysis of sequenced genomes reveals a large number of predicted glycosyltransferases, amounting to ∼1–3% of ORFs in each genome [15]. The majority of these enzymes have low sequence similarity. Nonetheless, the CAZy database has provided very useful information on distinct groups of glycosyltransferases, classifying them into over 90 families [16]. Currently, HMW1C is classified into the GT41 family, which otherwise contains only O-linked GlcNAc transferases (OGT) [17], [18]. The OGT enzymes contain an N-terminal domain with so-called tetratricopeptide repeats (TPR) responsible for mediating recognition of a broad range of target proteins and a C-terminal glycosyltransferase domain responsible for binding and transferring UDP-GlcNAc to target proteins (Fig. 1A). With this information in mind, the observation that HMW1C lacks an N-terminal TPR domain and mediates N-linked glycosylation of HMW1 with simple hexoses raised questions about the specific structure and function of HMW1C-like proteins.
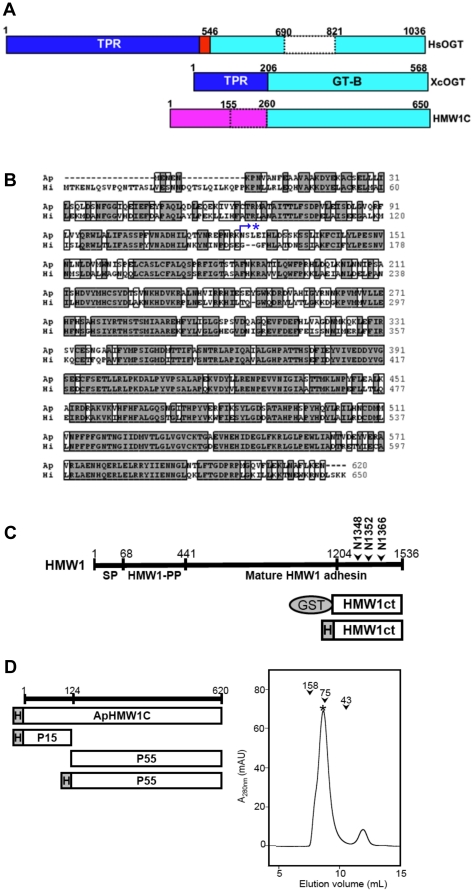
Figure 1. Representative GT41 members, HMW1C sequences, and schematics of recombinant proteins used in this study.
(A) Domain organization of three GT41 members, including the human OGT (HsOGT), the Xanthomonas campestris OGT (XcOGT), and Haemophilus influenzae HMW1C protein (HMW1C). The TPR and GT domains are indicated in blue and cyan, respectively. Based on the XcOGT structure [17], [18], domain boundaries of HsOGT were assigned (the nucleus localization signal in red). Fly and mammalian OGTs have a large insertion (in white) within the GT domain. In HMW1C, the N-terminal domain (in magenta) is different from the TPR domains in HsOGT and XcOGT. HMW1C residues 155 and 260 correspond to ApHMW1C residue 125 (limited proteolysis boundary) and XcOGT residue 203 (boundary for GT), respectively. (B) The sequence alignment of HMW1C (Hi) with its ortholog from A. pleuropneumoniae (Ap). The protease cleavage site is indicated with star. (C) Schematics of HMW1 and acceptor protein constructs. The known domain organization of HMW1 is shown: SP, the signal peptide (residues 1–68); HMW1-PP, the HMW1 pro-piece (residues 69–441) containing the secretion domain; and the mature adhesin (residues 442–1536). Several constructs representing different regions of HMW1 were generated as GST-fusion proteins to serve as acceptor proteins. Based on assessment of solubility and stability of each protein in solution, the best substrate was HMW1ct. For the substrate HMW1ct, a His-tagged version was also produced. The N-glycosylation sites, N1348, N1352, and N1366, within HMW1ct are indicated. (D) Schematics of enzyme constructs. ApHMW1C (GenBank: ABN74719.1) and its two sub-domains (P15 and P55) identified from the analysis of limited proteolysis were produced as His-tagged proteins. An analytical gel filtration profile of purified ApHMW1C (marked with star) revealed a calculated molecular weight of ∼70 kDa, consistent with a monomer. The peak positions of molecular standards are indicated as arrowheads (aldolase, 158 kDa; conalbumin, 75 kDa; and ovalbumin, 43 kDa).
To understand the structural basis for HMW1 glycosylation, we have pursued structure and function studies of HMW1C. Since HMW1C has been recalcitrant to crystallization, we elected to explore an Actinobacillus pleuropneumoniae HMW1C homolog called ApHMW1C, which is ∼65% identical to HMW1C (Fig. 1B). A. pleuropneumoniae is a gram-negative, non-motile organism that belongs to the Pasteurellaceae family (like H. influenzae) and is the etiological agent of porcine pleuropneumonia, a severe contagious pulmonary disease of pigs that causes important economic losses in industrialized pig production worldwide [19]. In this study, we found that purified ApHMW1C has N-glycosyltransferase activity and transfers glucose and galactose to known asparagine glycosylation sites in HMW1, analogous to HMW1C. In addition, we demonstrated that ApHMW1C is able to complement a deficiency of HMW1C and mediates HMW1 glycosylation and adhesive activity in whole bacteria. Our findings suggest a new subfamily of N-glycosyltransferases distinct from other GT41 family O-linked GlcNAc transferases.
Results
Identification and purification of ApHMW1C
BLAST analysis of the H. influenzae HMW1C protein revealed a sequence from Actinobacillus pleuropneumoniae as the closest homolog, with 65% identity and 85% similarity overall (Fig. 1B). This A. pleuropneumoniae sequence is encoded by orf APL_1635 and is hereafter referred to as ApHMW1C. The A. pleuropneumoniae genome does not contain a locus encoding a TPS system (like the hmw1 locus in H. influenzae that encodes HMW1, HMW1B, and HMW1C) or an isolated gene that encodes a predicted TpsA protein. Instead, the gene encoding ApHMW1C is adjacent to a gene cluster associated with sugar metabolism, namely APL_1631 (a homolog of glucosamine-fructose-6-phosphate aminotransferase), APL_1632 (a sugar metabolism transcriptional regulator), APL_1634 (putative glycosyltransferase), and APL_1635 (ApHMW1C). As a first step to examine the function of ApHMW1C, we overexpressed a His-tagged derivative using the pET45b vector backbone and E. coli BL21(DE3) (Fig. 1D). Subsequently we used a three-step purification protocol that included Ni2+-NTA affinity, anion-exchange, and gel filtration chromatography, yielding highly pure (>95%) recombinant protein. Based on the gel filtration profile of ApHMW1C (Fig. 1D), the calculated molecular weight was ∼70 kDa, consistent with the calculated molecular mass of 70,364 Da and the SDS-PAGE profile (Fig. 2A), indicating that ApHMW1C forms a monomer in solution.
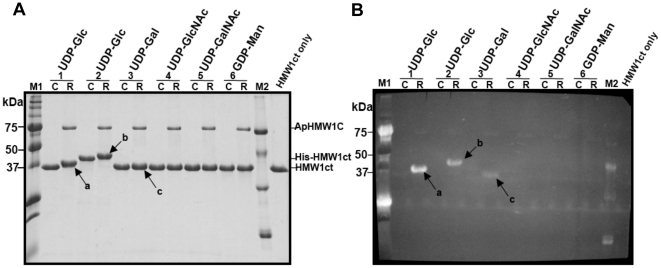
Figure 2. Glycosylation of HMW1ct by ApHMW1C.
To define the donor substrate specificity of ApHMW1C, glycosylation reactions were carried out in the reaction buffer with (R-lanes) or without (C-lanes) ApHMW1C using different UDP (or GDP) activated sugars. HMW1ct (without fusion tag) was used as the acceptor protein (lanes 1, and 3 to 6). As a control, His-tagged HMW1ct (His-HMW1ct) was also tested in a reaction with UDP-glucose as the donor sugar (lanes 2). (A) After the glycosylation reactions, samples were separated by SDS-PAGE, and the gel was stained with Coomassie Blue. (B) In parallel, a duplicate gel was transferred to a PVDF membrane and subjected to a detection reaction using the GlycoProfile III Fluorescent Glycoprotein Detection kit (Sigma). Glycosylated HMW1ct proteins are indicated by arrows: ‘a’ and ‘c’ are glycosylated HMW1ct reacted with UDP-glucose and UDP-galactose, respectively, and ‘b’ is glycosylated His-HMW1ct reacted with UDP-glucose. The lanes labeled “M1,” “M2,” and “HMW1ct only” indicate pre-staining protein markers (Precision Plus Protein Standards, Bio-Rad), glycosylated protein markers (ProteoProfile PTM Marker, Sigma), and HMW1ct only as a control, respectively.
Generation of HMW1ct acceptor protein
The high level of homology between ApHMW1C and HMW1C raised the possibility that ApHMW1C has glycosyltransferase activity and glycosylates an A. pleuropneumoniae acceptor protein. Given that A. pleuropneumoniae lacks an obvious TpsA protein, we examined the ability of ApHMW1C to glycosylate the H. influenzae HMW1 adhesin. The mature HMW1 adhesin is a large protein ∼125 kDa in size (Fig. 1C), and the solubility of the recombinant protein is limited (data not shown). Therefore, we overexpressed fragments of HMW1 as GST fusion proteins and then examined the solubility of these fragments after removal of the GST moiety. Three fragments were found to be soluble when the GST moiety was present, but two of these fragments formed protein precipitates after the GST moiety was removed. The fragment corresponding to amino acids 1205–1536 at the C-terminal end of HMW1 (referred to as HMW1ct; Fig. 1C) was soluble with and without the GST moiety. The same fragment of HMW1 with a His tag at the N terminus (His-HMW1ct) was also soluble. Purification of HMW1ct (33.7 kDa) and His-HMW1ct (36.8 kDa) resulted in highly pure (>90%) protein as assessed by SDS-PAGE (Fig. 2A), suitable for functional studies using these preparations as the acceptor protein.
ApHMW1C glycosylation of HMW1ct
To assess whether ApHMW1C is capable of glycosylating HMW1, we performed in vitro glycosylation assays using purified ApHMW1C, HMW1ct, and either UDP-glucose, UDP-galactose, or GDP-mannose, analogous to previous experiments with H. influenzae HMW1C [7]. In addition, we performed an in vitro assay using purified ApHMW1C, His-HMW1ct, and UDP-glucose. As shown in Fig. 2, in assays using UDP-glucose and UDP-galactose, we observed retarded gel mobility of HMW1ct and His-HMW1ct, consistent with glycan modification. In assays using UDP-GlcNAc or UDP-GalNAc, we observed no evidence of glycosylation (Figs. 2A and 2B), indicating that N-acetylated sugars are not suitable substrate carbohydrates for ApHMW1C. In addition, we observed no evidence of glycosylation using GDP-mannose. In control experiments, heat inactivation of ApHMW1C eliminated glycosylation activity (data not shown). These results are consistent with the glycan structure of HMW1 [6], indicating that ApHMW1C has substrate specificity similar to the in vitro and in vivo activity of HMW1C.
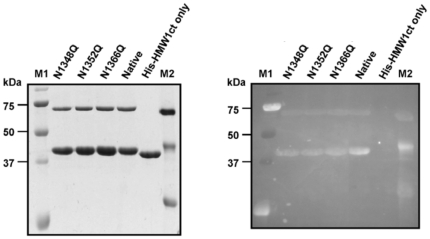
To determine whether ApHMW1C mimics HMW1C and modifies the same residues in HMW1ct, we used site-directed mutagenesis to convert Asn to Gln at the three known glycosylation sites in HMW1ct (N1348, N1352, and N1366) (Fig. 1C). The mutant proteins with a single mutation (N1348Q, N1352Q, or N1366Q) were over-expressed and purified and then analyzed for glycosylation intensity. As shown in Figure 3B, in in vitro glycosylation assays using the HMW1ct mutant proteins, ApHMW1C, and UDP-glucose, the band intensity of all HMW1ct mutant proteins was decreased compared to the intensity of wild type HMW1ct. These results establish that ApHMW1C mediates N-glycosylation of HMW1ct and functions analogously to HMW1C [7].
Figure 3. N-linked glycosylation of HMW1ct.
The glycosylation reactions were carried out in standard conditions using single (N1348Q, N1352Q, N1366Q) mutants of His-HMW1ct as acceptor proteins and UDP-glucose as donor substrate. (A) After the glycosylation reaction, the samples were separated by SDS-PAGE, and the gel was stained with Coomassie Blue. (B) A duplicate gel was transferred to a PVDF membrane and subjected to a detection reaction using the GlycoProfile III Fluorescent Glycoprotein Detection kit (Sigma). The lanes labeled “Native” and “His-HMW1ct only” are control reaction samples with and without ApHMW1C, respectively. M1 is a pre-staining protein marker (Precision Plus Protein Standards, Bio-Rad), and M2 is a glycosylated protein marker (ProteoProfile PTM Marker, Sigma).
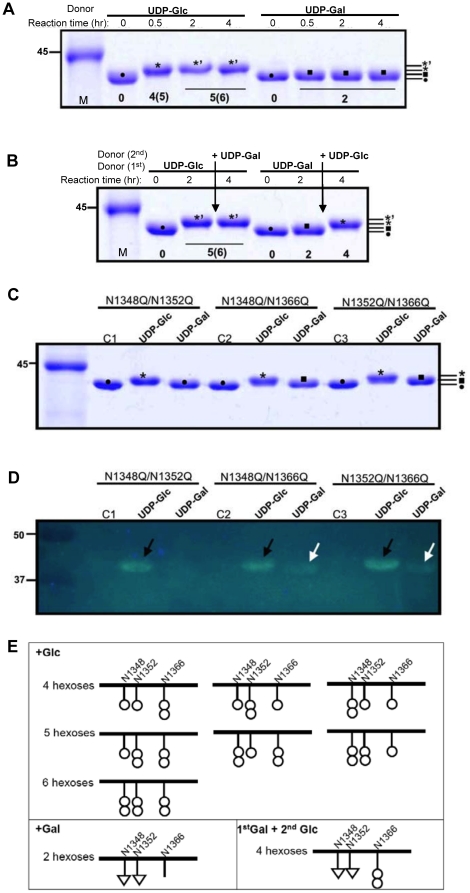
To investigate the apparent difference in gel shift and fluorescence intensity between the reaction with UDP-glucose and the reaction with UDP-galactose (Fig. 2), we monitored the progression of His-HMW1ct glycosylation by ApHMW1C. With both UDP-glucose and UDP-galactose, the modification reached a maximal gel shift within 2 hrs (Fig. 4A). Subsequently, we examined the effect of the order of addition of hexose sugars on glycosylation of HMW1ct. As shown in Fig. 4B, when adding UDP-galactose to the UDP-glucose saturated reaction, no change was observed, indicating that galactose is not further incorporated into the modified HMW1ct. On the other hand, adding UDP-glucose to the UDP-galactose saturated reaction resulted in a higher intermediate species of the modified protein, but did not restore to the maximal modification profile observed with UDP-glucose alone, suggesting that galactose may not be incorporated at all three Asn sites. To test this possibility, three HMW1ct variants with a double mutation (N1348Q/N1352Q, N1348Q/N1366Q, N1352Q/N1366Q) were over-expressed and purified and then analyzed for glycosylation intensity after incubation with ApHMW1C and either UPD-glucose or UDP-galactose. As shown in Fig. 4C, 4D, and 4E, glucose was incorporated at all three sites, whereas galactose was incorporated at Asn-1348 and Asn-1352 but not at Asn-1366. These results are consistent with a recent analysis of HMW1C glycosylation of HMW1 [7] and demonstrate that modification with galactose occurs at a restricted number of Asn sites.
Figure 4. Specificity of HMW1ct glycosylation.
The glycosylation reactions were carried out in standard conditions using His-HMW1ct as acceptor protein and UDP-glucose or UDP-galactose as donor substrate. (A) At each time point, an aliquot of the reaction was stopped by adding an equal volume of 2X SDS-PAGE sample buffer and followed by heating at 95°C for 4 min. (B) At each time point, SDS-PAGE samples were prepared as in A. However, two hrs after reaction with the first donor substrate, the second donor substrate was added to the reaction, as indicated. All samples were separated by 12% SDS-PAGE, and the gel was stained with Coomassie blue. The distinct shifts due to incorporated sugars are indicated by symbols (•, 0 hexose; ▪, 2 hexoses; ⋆, 4 or 5 hexoses; and ⋆’, 5 or 6 hexoses). (C) The glycosylation reactions were carried out in standard conditions using double mutants of His-HMW1ct (N1348Q/N1352Q, N1348Q/N1366Q, and N1352Q/N1366Q) by ApHMW1C using UDP-glucose or UDP-galactose as donor substrates. C1, C2, and C3 indicate control reactions without ApHMW1C. Samples were separated by 12% SDS-PAGE and were stained with Coomassie blue. (D) In parallel, a duplicated gel was transferred to a PVDF membrane and subjected to a detection reaction using the GlycoProfile III Fluorescent Glycoprotein Detection kit (Sigma). The glycosylated proteins by UDP-glucose or by UDP-galactose are indicated by arrows. (E) Model of hexose modifications at Asn-1348, Asn-1352, and Asn-1366.
ApHMW1C glycosylation of HMW1
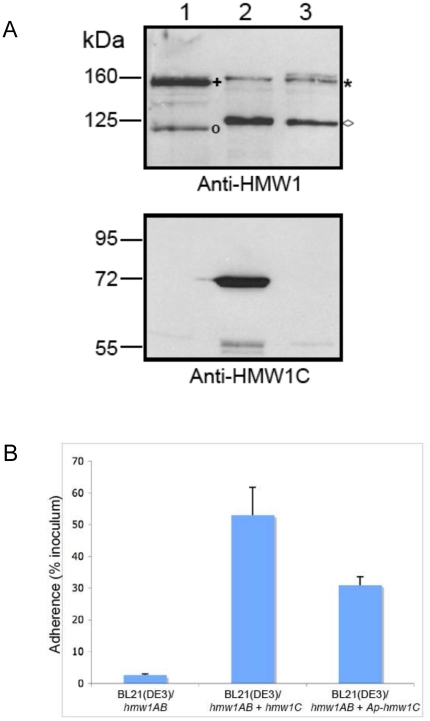
To extend our in vitro results and examine whether ApHMW1C is capable of glycosylating full-length HMW1, we introduced ApHMW1C into E. coli BL21(DE3) harboring the hmw1 locus with a deletion of hmw1C (hmw1AB). As controls, we examined E. coli BL21(DE3)/hmw1AB and E. coli BL21(DE3)/hmw1AB + hmw1C. As shown in Figure 5A, ApHMW1C was capable of complementing a deletion of hmw1C, restoring normal glycosylation, stability, and processing of HMW1. In addition, ApHMW1C was able to restore HMW1 adhesive activity, as measured by bacterial adherence to cultured epithelial cells (Fig. 5B).
Figure 5. Ability of ApHMW1C to complement a deficiency in HMW1C.
Panel A shows Western immunoblots of whole cell sonicates of E. coli BL21(DE3)/pACYC-HMW1ΔC (lane 1), E. coli BL21(DE3)/pACYC-HMW1ΔC + pET45b-HMW1C (lane 2), and E. coli BL21(DE3)/pACYC-HMW1ΔC + pET45b-ApHMW1C (lane 3). Lane 1 contains twice as much protein as loaded in lanes 2 and 3 to increase the visibility of the non-glycosylated HMW1 species. The blot in the upper panel was performed with a guinea pig antiserum reactive with HMW1, and the blot in the lower panel was performed with a guinea pig antiserum reactive with H. influenzae HMW1C. The asterisk indicates the glycosylated HMW1 pro-protein, and the plus sign indicates the non-glycosylated HMW1 pro-protein. The diamond indicates the glycosylated HMW1 mature protein, and the circle indicates the non-glycosylated HMW1 mature protein. Panel B shows in vitro adherence results comparing adherence by E. coli BL21(DE3)/pACYC-HMW1ΔC (hmw1AB), E. coli BL21(DE3)/pACYC-HMW1ΔC + pET45b-HMW1C (hmw1AB + hmw1C), and E. coli BL21(DE3)/pACYC-HMW1ΔC + pET45b-ApHMW1C (hmw1AB + Aphmw1C) to Chang epithelial cells. Bars and error bars represent mean and standard error measurements from a representative assay with measurements performed in triplicate.
Catalytic properties of ApHMW1C
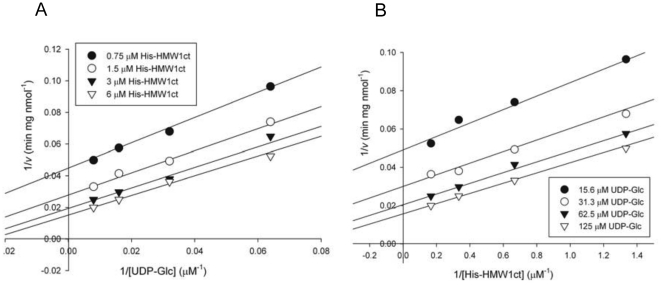
To delineate the catalytic properties of ApHMW1C, the kinetics of the glycosyltransferase reaction were monitored using a continuous coupled spectrophotometric assay. In this assay, formation of UDP was measured by oxidation of NADH using pyruvate kinase and lactate dehydrogenase as coupling enzymes [20]. His-HMW1ct was used as the acceptor protein, and UDP-glucose and UDP-galactose were used as donor substrates. ApHMW1C activity was not influenced by the addition of Mg2+ and Mn2+ or by the presence of excess EDTA according to gel shift assays (data not shown), suggesting that activity is independent of exogenous metal ions. Despite these results, we included K+ and Mg2+ in standard assay conditions because of their importance as metal cofactors for the coupling enzymes. The apparent Km and Vmax values for reactions with UDP-glucose and with UDP-galactose were determined by fitting initial rate data to the Michaelis-Menten equation (eq. 1), as summarized in Table 1. This analysis demonstrated that UDP-glucose is preferred over UDP-galactose as a donor substrate (kcat/Km = 3.3×103 M−1s−1 vs. kcat/Km = 5.2×102 M−1s−1), consistent with our results using the glycan detection method. For the glucose transfer reaction, the true Km values of two substrates (sugar and protein) were determined by measuring initial rates at concentrations ranging from 15.6 to 125 µM for UDP-glucose and from 0.75 to 6 µM for HMW1ct. The pattern of double-reciprocal Lineweaver-Burk plots for initial rates suggests a sequential mechanism, since all lines were linear and converged at the left side of the y-axis (Fig. 6). A ping-pong mechanism can be ruled out, since it would be expected to generate parallel lines in Lineweaver-Burk plots. Moreover, ApHMW1C could not auto-hydrolyze the UDP moiety from UDP-glucose in the absence of an acceptor protein (data not shown), suggesting that ApHMW1C may adopt the ordered or random sequential mechanism.
Table 1. Kinetic parameters of ApHMWC and its derivative proteins.
| Parameter a | Km | Vmax | kcat | kcat/Km |
| (µM) | (nmol min−1 mg−1) | (s−1) | (M−1s−1) | |
| UDP-Glc | ||||
| ApHMW1C | 38.9±5.8 | 108.8±4.0 | 0.130±0.005 | (3.3±0.8) ×103 |
| P55 | 67.3±3.8 | 141.0±2.4 | 0.132±0.002 | (2.0±0.6) ×103 |
| P15 | ND | ND | ND | ND |
| UDP-Gal | ||||
| ApHMW1C | 147.5±24.8 | 64.3±4.3 | 0.077±0.005 | (5.2±2.1) ×102 |
| P55 | 206.8±50.8 | 44.2±4.9 | 0.041±0.005 | (2.0±0.9) ×102 |
| P15 | ND | ND | ND | ND |
These are apparent values, determined by varying the concentration of one substrate (sugar donor substrate) at a fixed concentration of the second (protein acceptor).
Figure 6. Initial velocity of ApHMW1C.
(A) Double reciprocal plots of the initial velocity of ApHMW1C as a function of UDP-glucose concentration at the fixed His-HMW1ct concentrations, as indicated (inset). (B) Double reciprocal plots of the initial velocity of ApHMW1C as a function of His-HMW1ct concentration at the fixed UDP-glucose concentrations, as indicated (inset). The true Km values of UDP-glucose and His-HMW1ct corresponded to 54.5 µM and 2.3 µM, respectively, as obtained by eq. 2.
Probing the domain structure of ApHMW1C
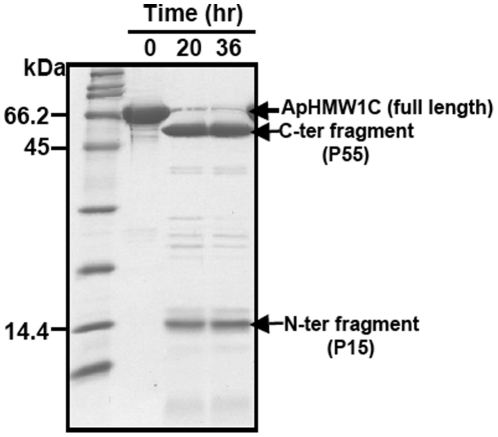
To experimentally determine the domain structure of ApHMW1C and gain general insights into the architecture of HMW1C-like proteins, we performed limited proteolysis of ApHMW1C (Fig. 7). Using 1∶100 and 1∶50 molar ratios of trypsin to ApHMW1C, reaction mixtures were monitored and revealed a gradual disappearance of full-length ApHMW1C and appearance of two major fragments. N-terminal sequencing established that the small fragment (referred to as P15; Fig. 1D) corresponds to the N-terminal domain of ApHMW1C and that the large fragment (referred to as P55; Fig. 1D) corresponds to the C-terminal domain of ApHMW1C starting with Asn-125. To investigate the functional roles of these domains, we produced each domain as a recombinant His-tagged protein. The P15 domain could be purified as a soluble protein, but the P55 domain was completely insoluble. Interestingly, P15 and P55 could be co-purified when we performed scale-up preparative limited proteolysis followed by chromatography. Based on these observations, the two domains appear to be associated and the N-terminal P15 domain appears to be important for the stability and/or solubility of the recombinant ApHMW1C. Next, we investigated the glycosylation activity of P15 and P55 (Fig. 1D). The P15 fragment was purified by affinity and gel filtration chromatography and had no detectable activity with either donor substrate. Given that His-tagged P55 was insoluble, the P55 fragment was obtained by preparative limited trypsin proteolysis followed by purification using ion exchange and gel filtration chromatography. Purified P55 showed 60% catalytic efficiency (kcat/Km = 2.0×103 M−1s−1) relative to native ApHMW1C (kcat/Km = 3.3×103 M−1s−1) using UDP-glucose and 38% catalytic efficiency (kcat/Km = 2.0×102 M−1s−1) relative to native ApHMW1C (kcat/Km = 5.2×102 M−1s−1) using UDP-galactose (Table 1). These results demonstrate that P55 (i.e. the fragment generated by deleting the P15 region from ApHMWC) has reduced enzyme activity.
Figure 7. Probing functional domains of ApHMW1C.
SDS-PAGE of limited proteolysis identified two stable fragments from ApHMW1C. Proteolysis reactions were carried out as described in experimental procedures. Shown is the reaction with trypsin:ApHMW1C (w/w) ratio of 1∶100 at 6°C.
Discussion
In this study, we characterized the enzyme properties of an A. pleuropneumoniae HMW1C homolog designated ApHMW1C. Both HMW1C and ApHMW1C have been assigned to the GT41 family of glycosyltransferases in the CAZy database. The GT41 family includes both bacterial and eukaryotic proteins and is characterized by large O-linked GlcNAc transferases with a long N-terminal extension containing so-called tetratricopeptide repeats (the TPR domain). To date the only member of the GT41 family that has been functionally characterized is the conserved mammalian O-linked GlcNAc transferase referred to as OGT. This enzyme modifies the side chains of serine and threonine residues of nuclear and cytosolic proteins with GlcNAc [21]–[24]. The N-terminal TPR domain of OGT mediates the recognition of a broad range of target acceptor proteins, and the C-terminal region contains the glycosyltransferase catalytic domain [17], [18], [25]. OGT-mediated glycosylation is a dynamic process in which GlcNAc is rapidly added and removed compared with the lifetime of the acceptor protein, analogous to protein phosphorylation. Given that HMW1C and ApHMW1C catalyze N-glycosylation and transfer simple hexose sugars to multiple asparagine sites of the HMW1 adhesin [6], HMW1C-like proteins clearly differ from previously reported enzymes in the GT41 family.
Historically, glycosyltransferases have been very difficult to study biochemically. These enzymes are notoriously unstable, often containing flexible loops and domains and commonly associating with other interacting proteins [26]. Thus far, we have been unable to obtain sufficiently high concentrations of HMW1C in solution for crystallography studies. As an alternative approach, we turned to ApHMW1C and found that we were able to purify high quantities that remained soluble. At the amino acid sequence level, ApHMW1C and HMW1C are ∼65% identical and ∼85% similar. However, the A. pleuropneumoniae genome does not have a locus corresponding to a TPS system (e.g., the hmw1 locus in H. influenzae encoding HMW1, HMW1B, and HMW1C) or an isolated gene that encodes a predicted TpsA protein. Instead, the gene encoding ApHMW1C is adjacent to a gene cluster with orfs involved in sugar metabolism. This observation suggests the possibility that HMW1C-like proteins have evolved in certain organisms to transfer sugars to protein acceptors that are not necessarily TpsA proteins in TPS systems.
In the absence of a known acceptor protein for ApHMW1C in A. pleuropneumoniae, we addressed the specific function of ApHMW1C using HMW1 as an acceptor protein. In in vitro assays containing ApHMW1C, a C-terminal fragment of HMW1 (HMW1ct), and either UDP-glucose or UDP-galactose, we observed evidence of glycosylation of HMW1ct via both the Glycoprofile III Fluorescent Glycoprotein Detection kit and a noticeable shift in mobility on SDS-PAGE gels. To extend these findings, we examined the specificity of ApHMW1C glycosylation activity by mutagenesis, focusing on the three Asn residues in HMW1ct that are known to be glycosylated by HMW1C [6]. Individual mutations of each Asn site (N1348Q, N1352Q, N1366Q) resulted in a clear reduction in fluorescence intensity (Fig. 3).
Based on the observed differences in the gel shift and the fluorescence intensity between the reaction with UDP-glucose and the reaction with UDP-galactose, two hexoses might be added at different sites in the acceptor protein and/or in different forms such as mono- or di-hexoses [6]. These possibilities were addressed by monitoring glycosylation as a function of time. Using our assay conditions, the 2hr reaction time appears to be sufficient for the maximal modification with both donor substrates. Interestingly, we observed only one species of the modified protein with UDP-galactose but two species with UDP-glucose in the course of the reaction (Fig. 4A), indicating that the transfer of glucose is more complex than the transfer of galactose. Indeed, the results of the reaction order of hexose sugars indicated that glucose can be incorporated at site(s) where galactose cannot and that glucose can be further incorporated to produce di-hexoses (Fig. 4B), as originally observed with HMW1C glycosylation of HMW1 [7]. Using HMW1ct derivatives with double mutations, we established that ApHMW1C can transfer the glucose moiety from UDP-glucose to all three of the documented glycosylation Asn sites within HMW1ct but can transfer the galactose moiety from UDP-galactose to only two of the three Asn sites. These observations demonstrate that UDP-glucose is the major donor substrate for ApHMW1C, resulting in transfer of glucose to either an Asn residue or to another glucose modifying an Asn residue, consistent with findings with HMW1C/HMW1.
HMW1C and ApHMW1C share 42–65% identity and 58–85% similarity with proteins in a number of other gram-negative bacterial pathogens, including the enterotoxigenic E. coli EtpC protein and predicted proteins in H. ducreyi, Yersinia spp., Mannheimia spp., and Burkholderia spp, among others [7]. Considering the results with ApHMW1C and HMW1C, HMW1C-like proteins catalyze N-glycosylation of an acceptor protein and appear to have acquired a secondary O-glycosylation activity that involves transferring a donor sugar to an acceptor sugar, forming di-hexoses on glycoproteins. The capacity to generate di-hexose O-glycosyidic bonds may explain why HMW1C-like proteins share similarity with GT41 O-glycosyltransferases within the C-terminal domain (Fig. 1A). However, the fact that OGTs transfer GlcNAc from UDP-GlcNAc to the side chain hydroxyl group of an amino acid (Ser or Thr) rather than transferring glucose from UDP-Glc to the hydroxyl group of glucose as in HMW1C-like proteins may explain why the sequence identities between HMW1C-like enzymes and OGT-like enzymes are marginal. One obvious common feature of these two groups of GT41 proteins is that both bind sugars donors that are activated by UDP, requiring a binding pocket that is formed by the C-terminal half of the GT-B domain (Fig. 1A) [17], [18].
Using a continuous coupled spectrophotometric assay to assess enzymatic activity, we observed that ApHMW1C is an efficient N-glycosyltransferase capable of transferring glucose and galactose moieties to multiple asparagine residues of HMW1 with kcat/Km values of 3.3×103 M−1s−1 and 5.2×102 M−1s−1, respectively. Like the majority of enzymes in the GT-B superfamily of glycosyltransferases [27]–[32], ApHMW1C showed metal-independent behavior (data not shown), suggesting that ApHMW1C has a GT-B fold, consistent with assignment to the GT41 family. Based on the results of limited proteolysis, ApHMW1C appears to contain two domains, namely a minor N-terminal domain (P15) and a major C-terminal domain (P55). Regardless of donor substrates, the P55 domain had lower catalytic efficiency. At this point, it is unclear whether or not the small N-terminal domain contributes to enzyme efficiency. In order to understand the structural basis for the enzyme mechanisms and specificities of O-linked GlcNAc transferase and HMW1C-like proteins, obtaining the crystal structure of an HMW1C-like glycosyltransferase is highly desirable.
In bacteria, only limited information is available about the structures of protein-linked glycans, the detailed process of protein glycosylation, and the functional implications of glycosylation. However, given that nearly all of the known glycoproteins are surface-exposed, glycosylation appears to have an important role in pathogenesis. Recently characterized functions of bacterial protein glycosylation include adhesion, protection against proteases, antigenic variation, and evasion of immunity [5], [12], [33]–[38]. Among bacterial glycoproteins, H. influenzae HMW1 and diarrheagenic E. coli AIDA-I are particularly noteworthy, representing two branches of the type V secretion system. HMW1 is a TpsA protein secreted by the TPS pathway, and AIDA-I is an autotransporter protein. It is known that glycosylation of HMW1 protects against premature degradation and is required for HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence. Similarly, glycosylation of AIDA-I is required for normal conformation and for AIDA-I mediated adhesion to epithelial cells [39]. In considering the secretion pathway, glycosylation might play a role in secretion of exoproteins, reminiscent of the eukaryotic secretory pathway. Of note, HMW1 contains N-linked hexoses at over 30 sites along the mature protein, whereas AIDA-I contains O-linked heptoses at ∼16 sites along the protein [39]. The enzyme responsible for glycosylation of AIDA-I has not been studied. Seemingly, these bacterial glycoproteins secreted by the type V secretion pathway share common features such as simple glycan structures (hexoses in HMW1 vs. heptoses in AIDA-I) and multiple site modification. Yet, these proteins are distinct in that HMW1 has N-linked glycans and AIDA-I has O-linked glycan. In order to further our knowledge of the mechanisms and functional implications of glycosylation of proteins secreted via the type V secretion pathway, structural and functional studies on the corresponding enzyme systems are critical. Along these lines, our functional characterization of ApHMW1C is an important advance.
In summary, HMW1C and ApHMW1C add simple hexose sugars to asparagine sites of acceptor proteins and appear to represent a new subfamily of bifunctional glycosyltransferases (N-linkage to protein acceptor and O-linkage to glucose acceptor), distinct from GT41 family O-glycosyltransferases. Determination of the structures of HMW1C and ApHMW1C will provide a major advance in understanding how members of this family glycosylate HMW1 and other acceptor proteins.
Materials and Methods
Materials
Restriction enzymes were purchased from New England Biolabs. Pfu DNA polymerase and T4 DNA ligase were purchased from Stratagene and Promega, respectively. Primers used for PCR were synthesized by IDT. Unless indicated otherwise, chemicals were purchased from Sigma, including UDP- and GDP-sugars, β-NADH and phosphoenolpyruvate (PEP), and coupling enzymes (LDH/PK).
Cloning and expression
The gene encoding ApHMW1C (GenBank: ABN74719.1) was amplified by PCR from genomic DNA of Actinobacillus pleuropneumoniae (purchased from ATCC: Cat no. 27088D-5) using primers 1 and 2 (Table 2) and was inserted into KpnI/SacI-digested pET45b (Novagen) to express an N-terminal His-tag protein. The gene encoding H. influenzae HMW1C was amplified by PCR from pHMW1-15 [40] using primers 3 and 4 (Table 2) and was inserted into KpnI/SacI-digested pET45b. The DNA fragment encoding the C-terminal end of Haemophilus influenzae HMW1 corresponding to amino acids 1205–1536 (HMW1ct, Fig. 1C) was amplified by PCR from pHMW1-15 [40] using primers 5 and 6 (Table 2) and was inserted into BamHI/XhoI-digested pGEX-6p-1 (GE Healthcare) and BamHI/XhoI-digested pET28a (Novagen). Mutants of HMW1ct were generated using the QuikChange II site-directed mutagenesis kit (Stratagene) and mutagenic primer sets (Table 2) according to the manufacturer's instructions.
Table 2. Primers used for cloning, site-directed mutagenesis, and sequence analysis.
| Protein names | Primer # | Primer sequences (5′ → 3′) |
| ApHMW1C | 1 | cgg ggt acc atg gaa aac gaa aat aaa ccg aat |
| 2 | cgc gag ctc tta att ttc ttt tag gaa cgc att t | |
| HMW1C | 3 | gtg ggt acc atg aca aaa gaa aat tta caa agt g |
| 4 | gtg gag ctc tta ttt ttt actcaagtgcttccg | |
| HMW1ct | 5 | gcg gga ttc gca aat agc ggt gca tta acc |
| 6 | ggc ctc gag cta ccg ccc gtt atc agc gat | |
| HMW1ct variants | ||
| HMW1ctN1348Q | 7 | gca gga agt att aat gcc gcc cag gtg aca cta aat act aca ggc |
| HMW1ctN1352Q | 8 | aat gcc gcc aat gtg aca cta cag act aca ggc act tta act acc |
| HMW1ctN1348Q/N1352Q | 9 | aat gcc gcc cag gtg aca cta cag act aca ggc act tta act acc |
| HMW1ctN1366Q | 10 | acc gtg aag ggt tca aac att cag gca acc agc ggt acc ttg gtt |
Protein purification
To purify His-tagged proteins, the pET-based plasmids containing the genes encoding ApHMW1C, ApHMW1C-HMW1C hybrids, and HMW1ct were expressed in E. coli BL21(DE3) (Stratagene). Bacteria were grown to an OD600 of 0.8, and expression was induced with 0.2 mM IPTG at room temperature for 2 hrs. Subsequently, bacterial cells were harvested, resuspended in buffer-A (20 mM Tris-HCl, pH 8.0, 250 mM NaCl, 5 mM β-mercaptoethanol) containing 10 mM imidazole, and disrupted using sonication. After centrifugation at 40,000xg for 20 min (Sorvall), supernatants were subjected to a binding reaction with Ni2+-NTA (Qiagen) resin for 30 min in a batch procedure. Protein-resin complexes were then packed onto a column and washed with buffer-A containing 20 mM imidazole. The proteins were eluted using a step gradient method including 40, 60, 100, 300 and 500 mM imidazole in buffer-A. Fractions containing target proteins were diluted five times with buffer-B (20 mM Tris-HCl pH 8.0), loaded onto a HiTrap-Q column (GE Healthcare), and eluted with a linear gradient of 50 mM–1M NaCl in buffer-B using an AKTA FPLC system (GE Healthcare). The peak fractions were combined, concentrated, and further purified using a Superdex 75 10/300 GL gel filtration column (GE Healthcare) equilibrated with buffer-C containing 50 mM HEPES, pH 7.0, 200 mM NaCl, 0.1 mM EDTA, and 5% glycerol.
To purify GST fusion proteins, constructs were expressed in E. coli BL21 (Stratagene). Bacteria were grown to an OD600 of 0.8, and expression was induced with 0.4 mM IPTG at room temperature for 2 hrs. Bacterial cells were harvested and resuspended in PBS containing 1 mM DTT. Following cell lysis using sonication, cell lysates were centrifuged at 40,000xg for 20 min, and the supernatant was added to GSH-Sepharose 4 fast flow beads (GE Healthcare). After binding for 2 hrs at 4°C and washing with PBS, GST-fusion proteins were eluted with 10 mM GSH in 50 mM Tris-HCl, pH 8.0. The GST moiety was cleaved via on-column digestion with 80 units PreScission protease (GE Healthcare) in 4 mL of cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT) overnight at 4°C [41]. Eluted proteins were further purified using a HiTrap-Q column as described above. The protein concentration was determined by the Bradford assay following the manufacturer's instructions (Bio-Rad).
Glycan detection
To detect glycosylation of the HMW1ct acceptor protein in vitro, ApHMW1C in a final concentration of 1.6 µM was added to a 100 µL reaction mixture containing 50 mM HEPES, pH 8.0, 50 mM KCl, 5 mM MgCl2, 0.5 mM UDP- or GDP-sugar, and 6.3 µM HMW1ct. The mixtures were incubated at room temperature for 2 hrs and were then added to an equal volume of 2X SDS-PAGE sample buffer and incubated at 95°C for 4 min. The samples were separated on a 15% SDS-PAGE gel, transferred to a PVDF membrane using a Semi-Dry Transfer Cell (Bio-Rad), and detected by a GlycoProfile III Fluorescent Glycoprotein Detection kit (Sigma) according to the manufacturer's instructions.
Enzyme assays and data analysis
Glycosyltransferase activity was measured using a continuous coupled spectrophotometric assay for UDP formation in standard assay conditions [20]. Reaction mixtures containing 50 mM HEPES pH 8.0, 50 mM KCl, 5 mM MgCl2, 0.5 mM UDP-glucose, 3.2 µM His-HMW1ct, 0.2 mM β-NADH, 2 mM phosphoenolpyruvate (PEP), 2.5 U lactate dehydrogenase (LDH), and 1.75 U pyruvate kinase (PK) were pre-incubated at room temperature for 10 min. Reactions were started by adding ApHMW1C to a final concentration of 0.9 µM, and the initial glycosylation rates were measured by monitoring the decrease of absorbance at 340 nm for 1 min. One unit was defined as the amount of enzyme that produced 1 nmol of UDP per minute in standard assay conditions. Initial rates (v) from the kinetic studies were plotted as 1/v vs. 1/[substrate concentration]. The data were fit to the appropriate rate equations using the Enzyme Kinetic Module version 1.3 integrated into SigmaPlot ver 10.0.1 (Systat). The apparent KM and Vmax values were determined by fitting the initial rates to the Michaelis-Menten equation (eq. 1). Data conforming to a sequential mechanism were fit to the Random Bi-Bi equation (eq. 2).
| (1) |
| (2) |
In eqs 1–2, A and B are the concentration of the substrates, v and V represent initial and maximum rates, respectively, Ka and Kb are Michaelis constants for A and B, respectively, and Kia is the inhibition constant for substrate A.
Adherence assays
Adherence assays were performed with Chang epithelial cells (human conjunctiva; ATCC CCL 20.2) (Wong-Kilbourne derivative clone 1-5c-4) as described previously [42]. Bacteria were prepared by inoculating LB broth containing antibiotics to select for the relevant plasmids and incubating overnight, then diluting 1∶10 in fresh LB broth with antibiotics and incubating for 90 min, then adding IPTG to a final concentration of 0.03 mM and incubating for 2 hrs to induce expression of ApHMWC or HMW1C. Percent adherence was calculated by dividing the number of adherent colony-forming units by the number of inoculated colony-forming units. All strains were examined in triplicate, and assays were repeated three times.
Strains and protein analysis for in vivo functional assays
To test whether ApHMW1C is active in vivo, the following strains were used: BL21(DE3) expressing pACYC-HMW1ΔC (hmw1AB) [5], BL21(DE3) expressing pACYC-HMW1ΔC + pET45b-ApHMW1C, and BL21(DE3) expressing pACYC-HMW1ΔC + pET45b-HMW1C. Whole cell sonicates were prepared by suspending bacterial pellets in 10mM HEPES, pH 7.4 and sonicating to clarity. Proteins were resolved by SDS-PAGE using 7.5% polyacrylamide gels. Western blots were performed using antiserum GP96, a guinea pig polyclonal antiserum against the HMW1 and HMW2 proteins [43].
Limited proteolysis of ApHMW1C
Protease digestions were performed as described previously [44] with the following modifications. For each 100 µl reaction, 100 µg of a purified ApHMW1C sample was incubated with trypsin (Roche Dignostics) in 20 mM Tris-HCl (pH 7.5), NaCl 100 mM, and 1 mM EDTA (pH 8). Reactions were performed at trypsin:ApHMW1C (w/w) ratios of 1∶50, 1∶100, and 1∶500 by incubating in a heat block at 37°C for up to 3 hrs or at 6°C for up to 36 hrs. At each time point, aliquots of the reactions were stopped by the addition of 2X SDS-PAGE loading buffer followed by boiling for 4 min. The samples were analyzed by SDS-PAGE, transferred to a PVDF membrane, and submitted for N-terminal sequencing (Midwest Analytical Inc., St. Louis).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Institutes of Health (NIH) grant RO1-AI068943 and Grant E-1616 from the Welch Foundation to HJY and by NIH grant RO1-DC02873 to JWS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barenkamp SJ, Leininger E. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect Immun. 1992;60:1302–1313. doi: 10.1128/iai.60.4.1302-1313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Geme JW, 3rd, Falkow S, Barenkamp SJ. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci U S A. 1993;90:2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Geme JW, 3rd, Kumar VV, Cutter D, Barenkamp SJ. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Geme JW, 3rd, Yeo HJ. A prototype two-partner secretion pathway: the Haemophilus influenzae HMW1 and HMW2 adhesin systems. Trends Microbiol. 2009;17:355–360. doi: 10.1016/j.tim.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Grass S, Buscher AZ, Swords WE, Apicella MA, Barenkamp SJ, et al. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol Microbiol. 2003;48:737–751. doi: 10.1046/j.1365-2958.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- 6.Gross J, Grass S, Davis AE, Gilmore-Erdmann P, Townsend RR, et al. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J Biol Chem. 2008;283:26010–26015. doi: 10.1074/jbc.M801819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW., 3rd The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 2010;6(5):e1000919. doi: 10.1371/journal.ppat.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 9.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 11.Guerry P, Szymanski CM. Campylobacter sugars sticking out. Trends Microbiol. 2008;16:428–435. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt MA, Riley LW, Benz I. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 2003;11:554–561. doi: 10.1016/j.tim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155:317–327. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
- 15.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 16.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke AJ, Hurtado-Guerrero R, Pathak S, Schüttelkopf AW, Borodkin V, et al. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 19.Sebunya TN, Saunders JR. Haemophilus pleuropneumoniae infection in swine: a review. J Am Vet Med Assoc. 1983;182:1331–1337. [PubMed] [Google Scholar]
- 20.Gosselin S, Alhussaini M, Streiff MB, Takabayashi K, Palcic MM. A continuous spectrophotometric assay for glycosyltransferases. Anal Biochem. 1994;220:92–97. doi: 10.1006/abio.1994.1303. [DOI] [PubMed] [Google Scholar]
- 21.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 22.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 23.Iyer SP, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci U S A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, Chen YW, Clausen H, Cohen SM, Conti E. Structural insights into the Notch-modifying glycosyltransferase Fringe. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb1144. [DOI] [PubMed] [Google Scholar]
- 26.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 27.Moréra S, Imberty A, Aschke-Sonnenborn U, Rüger W, Freemont PS. T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism. J Mol Biol. 1999;292:717–730. doi: 10.1006/jmbi.1999.3094. [DOI] [PubMed] [Google Scholar]
- 28.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 29.Grizot S, Salem M, Vongsouthi V, Durand L, Moreau F, et al. Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. J Mol Biol. 2006;363:383–394. doi: 10.1016/j.jmb.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 30.Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci U S A. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y, Chen L, Ha S, Gross B, Falcone B, et al. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 33.Kuo C, Takahashi N, Swanson AF, Ozeki Y, Hakomori S. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J Clin Invest. 1996;98:2813–2818. doi: 10.1172/JCI119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenthal C, Elsinghorst EA. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect Immun. 1999;67:4084–4091. doi: 10.1128/iai.67.8.4084-4091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann JL, O'Gaora P, Gallagher A, Thole JE, Young DB. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 1996;15:3547–3554. [PMC free article] [PubMed] [Google Scholar]
- 37.Marceau M, Nassif X. Role of glycosylation at Ser63 in production of soluble pilin in pathogenic Neisseria. J Bacteriol. 1999;181:656–661. doi: 10.1128/jb.181.2.656-661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romain F, Horn C, Pescher P, Namane A, Riviere M, et al. Deglycosylation of the 45/47-kilodalton antigen complex of Mycobacterium tuberculosis decreases its capacity to elicit in vivo or in vitro cellular immune responses. Infect Immun. 1999;67:5567–5572. doi: 10.1128/iai.67.11.5567-5572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charbonneau ME, Girard V, Nikolakakis A, Campos M, Berthiaume F, et al. O-linked glycosylation ensures the normal conformation of the autotransporter adhesin involved in diffuse adherence. J Bacteriol. 2007;189:8880–8889. doi: 10.1128/JB.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barenkamp SJ, St Geme JW., 3rd Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect Immun. 1994;62:3320–3328. doi: 10.1128/iai.62.8.3320-3328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkpatrick AS, Yokoyama T, Choi KJ, Yeo HJ. Campylobacter jejuni Fatty Acid Synthase II: Structural and functional analysis of b-hydroxyacyl-ACP dehydratase (FabZ). Biochem Biophys Res Commun. 2009;380:407–412. doi: 10.1016/j.bbrc.2009.01.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buscher AZ, Burmeister K, Barenkamp SJ, St Geme JW., 3rd Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J Bacteriol. 2004;186:4209–4217. doi: 10.1128/JB.186.13.4209-4217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buscher AZ, Grass S, Heuser J, Roth R, St Geme JW., 3rd Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol Microbiol. 2006;61:470–483. doi: 10.1111/j.1365-2958.2006.05236.x. [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama T, Paek S, Ewing CP, Guerry P, Yeo HJ. Structure of a sigma28-regulated nonflagellar virulence protein from Campylobacter jejuni. J Mol Biol. 2008;384:364–376. doi: 10.1016/j.jmb.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]