Abstract
Multiple signals, controlling both proliferation and differentiation, must be integrated in the reprogramming of intestinal epithelial cells during maturation along the crypt-luminal axis. The v-myb family member Mybl2, a molecule implicated in the development and maintenance of the stem cell phenotype, has been suggested to play an important role in proliferation and differentiation of several cell types and is a gene we have found is commonly regulated in several systems of colon cell maturation both in vitro and in vivo. Here we show that siRNA silencing of Mybl2 in proliferating Caco-2 cells increases expression of the cell-cycle regulators cdk2, cyclin D2, and c-myc and decreases expression of cdc25B and cyclin B2 with a consequent 10% increase of cells in G2/M and a complementary 10% decrease in G1. Mybl2 occupies sequences upstream of transcriptional start sites of cyclin D2, c-myc, cyclin B2, and cdc25B and regulates reporter activity driven by upstream regions of cdk2, cyclin D2, and c-myc. These data suggest that Mybl2 plays a subtle but key role in linking specific aspects of cell cycle progression with generation of signals for differentiation and may therefore be fundamental in commitment of intestinal epithelial cells to differentiation pathways during their maturation.
Keywords: Mybl2, differentiation, colon, commitment, cell cycle
INTRODUCTION
Epithelial cells lining the normal intestinal mucosa must simultaneously exert exquisite control over proliferation and differentiation. The proliferative compartment at the crypt base consists of stem and progenitor cells that not only robustly express cell cycle promoting genes to drive high levels of cell division but also simultaneously suppress expression of phenotypic markers of mature states. As these cells migrate upward along the crypt-luminal axis, they are reprogrammed by extrinsic and intrinsic signals to exit the cell cycle and undergo lineage specific differentiation. Although the distinction between factors that regulate intestinal cell proliferation versus differentiation is not always clear, pathways involved in these diverging cell phenotypes in the intestine, such as Wnt, K-Ras, Notch, and KLF4, have been identified. Nevertheless, the overlying coordination of these pathways in the transition from proliferating to maturing cell populations is not well characterized. Identification of molecules that integrate signals influencing both proliferation and differentiation will provide crucial insights into how stem and progenitor cell populations are reprogrammed to acquire mature states and to avoid acquisition of premalignant and malignant phenotypes.
Mechanisms that control transitions from proliferative to differentiated states can be elucidated by studying dividing cells induced to exit the cell cycle and adopt a mature phenotype, such as some colonic epithelial carcinoma cell lines reported to be reprogrammed from proliferative to mature states in culture (Fogh et al., 1977; Fogh, 1975). This reprogramming can be induced by, for example, the short chain fatty acid (SCFA) butyrate, a product of dietary fiber fermentation and a physiological regulator of colonic cell maturation in vivo (Augenlicht et al., 1995; Harig et al., 1989; Reddy, 1987; Tappenden et al., 1997). In other cases, contact inhibition of growth triggers lineage-specific differentiation along absorptive (Caco-2), goblet (HT29Cl16E), or secretory (HT29Cl19A) cell lineages. Discovering the requirements for reprogramming these highly proliferative tumor cells into growth-arrested, differentiating cells has shed light on many mechanisms that drive maturation of colon epithelial cells. We and others have profiled changes in gene expression in these maturing cell lineages (Fleet et al., 2003; Mariadason et al., 2002; Velcich et al., 2005), and we delineated RNA (Mariadason et al., 2005) and protein expression profiles (Chang et al., 2008) that characterize intestinal cell maturation in vivo from cells sequentially isolated along the crypt-villus axis of the mouse small intestine.
Here we show that Mybl2, which we initially identified from these data bases as a critical factor in several different lineages of differentiating colon epithelial cells, may play a distinct role in the transition of intestinal cells from the proliferative to the mature state. Mybl2 is a member of the v-myb family of transcription factors of which the founding member is the oncogene c-myb. Unlike A-myb, which is expressed primarily in B cells, developing central nervous system cells, mitotically active cells in the embryo, and reproductive tissues (Trauth et al., 1994), or c-myb, expressed mainly in hematopoietic precursors (Graf, 1992), Mybl2 is expressed in all proliferative cells examined (Sala and Watson, 1999; Sitzmann et al., 1996). In addition to its high expression in dividing cells, several lines of evidence link Mybl2 to a stem cell-like phenotype: 1) Mybl2 is one of 39 critical transcription factors commonly expressed in several different types of pluripotent stem cells (Muller et al., 2008); 2) Mybl2 maintains embryonic stem cells in an undifferentiated state and may be involved in early steps of differentiation, possibly by transcriptionally activating pluripotency-associated genes such as Oct4 (Tarasov et al., 2008a; Tarasov et al., 2008b); and 3) absence of functional Mybl2 is embryonic lethal, most likely because of the inability in these embryos to form an inner cell mass, the source of embryonic stem cells (Tanaka et al., 1999). Indeed, expression of Mybl2 has been proposed as one of several characteristics of the stem cell state (Boheler, 2009). Mybl2 regulates both proliferation and differentiation in several cell types: its expression can overcome G1 arrest in glioblastoma and osteosarcoma cells (Lin et al., 1994), and ectopic Mybl2 expression prevents full differentiation in leukemic (Bies et al., 1996) and neuroblastoma (Raschella et al., 1995) cells.
In a screen for genes and pathways that are important in coordinating proliferation and differentiation in maturing colon cells, we found that Mybl2 is one of 14 genes commonly downregulated in expression during butyrate- and growth arrest-induced differentiation of Caco-2, HT29Cl16E, and HT29Cl19A cells in vitro and is downregulated as cells mature along the crypt-luminal axis of the mouse intestine in vivo (Papetti and Augenlicht, submitted). Mybl2 thus may also be important in the reprogramming of intestinal stem and progenitor cells as they mature. Data presented here demonstrate that Mybl2 exerts effects on both proliferation and differentiation pathways in colon epithelial cells. siRNA-mediated Mybl2 suppression alters the expression of several genes that control cell cycling, and the pattern of this regulation suggests that Mybl2 plays a role in priming cells for commitment to differentiation.
MATERIALS AND METHODS
Cell culture
The Caco-2 human adenocarcoma cell line was maintained as described (Mariadason et al., 2002) in medium containing 20% fetal bovine serum. For spontaneous differentiation, cells were grown to confluence (day 0) and harvested at various time points thereafter with medium changes every 2 days.
Western blot
Caco-2 cells scraped from 6-well plates were disrupted by sonication on ice for 2×10-second pulses with an intervening 30-second pause at setting 2 (7.5% output, Branson 450 Sonifier, Fisher Scientific). 20μg Caco-2 lysate was separated by SDS-PAGE and analyzed by Western blotting essentially as described (Papetti and Skoultchi, 2007) except protease inhibitor cocktail was from Sigma and blocking and antibody dilution buffer was 5% milk/TBST (0.05% Tween 20, 100mM Tris pH 7.5, 150mM NaCl). Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), detected using film or a Kodak 4000R Image Station, and analyzed with Kodak Molecular Imaging Software, version 4.0 (Eastman Kodak Company). Primary antibodies : 1:10 mouse anti-Mybl2 (R. Watson, Imperial College School of Medicine, London, UK), or 1:4000 mouse anti-α-tubulin (Sigma). Secondary antibody : 1:2000 goat anti-mouse IgG-HRP (Santa Cruz).
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Cells were collected in Trizol (Invitrogen) and total RNA prepared as described by the manufacturer. cDNA was synthesized from 1μg total RNA using the iScript cDNA Sythesis Kit (BioRad). qRT-PCR utilized SYBR Green PCR Master Mix (Applied Biosystems) in 20 or 30μl with 50ng cDNA and 100nM primers with a 7900HT Sequence Detection System (Applied Biosystems). Cycling conditions : 50°C for 2 min.; 95°C for 10 min.; 40 cycles of 95°C for 15 s; 60°C for 1 min. Specificity of amplification was confirmed by single peaks in dissociation curves and/or single bands on agarose gel electrophoresis. Analysis utilized SDS version 2.3 software (Applied Biosystems). GAPDH-normalized mRNA expression in differentiating cells was calculated relative to that in proliferating cells by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed as described (Choe et al.). Chromatin from 2.5×106 Caco-2 cells, fixed for 8 minutes with 1.1% formaldehyde and sonicated with a Branson 450 Sonifier for 24×15s pulses at setting 3 (7.5% output) with 45 s pauses, was immunoprecipitated with rabbit antisera to Mybl2 (N-19, Santa Cruz), c-myb (H-141, Santa Cruz), or control rabbit IgG (Santa Cruz). qRT-PCR was performed as described above using primers listed in supplemental table 2.
Reporter Assays
Subconfluent/proliferating Caco-2 cells grown in 12-well plates were transfected for 48 hours with 1μg/well luciferase reporter + 1μg/well pRL-TK renilla (Promega, Madison, WI) using Lipofectamine Plus (Invitrogen) as described by the manufacturer. For Mybl2 overexpression studies, proliferating Caco-2 cells were transfected in 24-well plates with 125ng/well of plasmids containing regions upstream of the transcriptional start sites of cdk2, cyclin D2, c-myc, cdc25B, or cyclin B2 genes cloned into the promoter region of luciferase (see supplementary Materials and Methods for details). Transfections consisted of 500ng pCMV control or Mybl2/pCMV (R. Watson) (Lin et al., 1994) + 125ng/well pRL-TK. For reporter assays with siRNA, Caco-2 cells were transfected as described below in 6-well plates (see siRNA Transfection) with 500ng/well luciferase reporter plasmid + 500ng/well pRL-TK, and 20μl lysate was assayed for luciferase and renilla activity on the LMaxII dual injection microplate reader (Molecular Devices). Data analysis utilized SoftMax Pro 5 software (Molecular Devices).
siRNA Transfection
Mybl2-specific or control nontargeting double-stranded siRNA oligos (ON-TARGETplus SMARTpool of 4 individual oligos, Thermo Scientific Dharmacon), contained 3' UU overhangs on both strands and a 5' phosphate on the antisense strand. Proliferating, subconfluent Caco-2 cells (~80,000 cells per well of 6-well plate) were transfected for 48 hours with 50nM siRNA using Oligofectamine (Invitrogen) as described (Papetti and Skoultchi, 2007).
Cell-Cycle Analysis
~106 Caco-2 cells were washed, scraped into ice-cold PBS, and resuspended in 0.5ml ice-cold PBS. 4.5ml 70% ethanol was added drop-wise with gentle mixing. Cells were fixed at −20°C overnight, washed 2× with 5ml PBS (pelleted at ~1830×g at 4°C), incubated in 1ml propidium iodide/RNAse staining buffer (BD Pharmingen) for 15 min at room temperature in the dark, filtered (70μ), and analyzed for DNA content on a FACScan flow cytometer (Becton Dickinson) with cell-cycle distribution determined using ModfFit LT software.
RESULTS
Mybl2 regulates expression of several genes that drive progression through specific phases of the cell cycle in colon cells
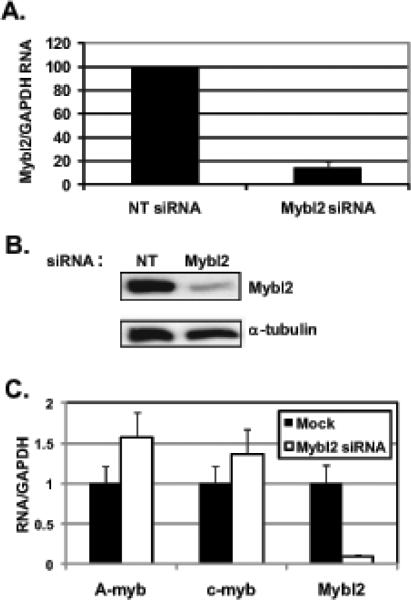
In cultured colon epithelial cells in vitro as well as in cells along the crypt-villus axis of the mouse small intestine in vivo, Mybl2 is highly expressed in proliferating cells and suppressed in those undergoing differentiation. (Papetti and Augenlicht, submitted), and Mybl2 is highly enriched in colon basal crypts versus sections adjacent to the lumen (Kosinski et al., 2007). To investigate Mybl2 function in colon epithelial cells, we silenced Mybl2 mRNA expression by ~86% in subconfluent, dividing Caco-2 cells using Mybl2-specific siRNA (Figure 1A,B). Importantly, this extent of Mybl2 mRNA suppression is similar to that in spontaneously differentiating Caco-2 cells (Papetti and Augenlicht, submitted) and therefore mimics the extent of physiological Mybl2 suppression in differentiating colon cells. Mybl2 suppression is not due to nonspecific effects of the oligo itself because a control non-targeting siRNA had no effect on Mybl2 expression (Figure 1A,B). Furthermore, this silencing is specific to Mybl2 as it does not suppress expression of the related family members A-myb or c-myb (Figure 1C).
Figure 1.
Subclonfluent, proliferating Caco-2 cells were transfected with Mybl2-specific or control, nontargeting (NT) siRNA for 48 hours. RNA (A) and protein (B) were measured as described in Materials and Methods. In (A), bar represents the average + standard deviation of 3 independent experiments, each performed in duplicate. (C) RNA from Mock- or Mybl2 siRNA-treated cells was measured for expression of A-myb, c-myb, and Mybl2 by qRT-PCR as described in Materials and Methods.
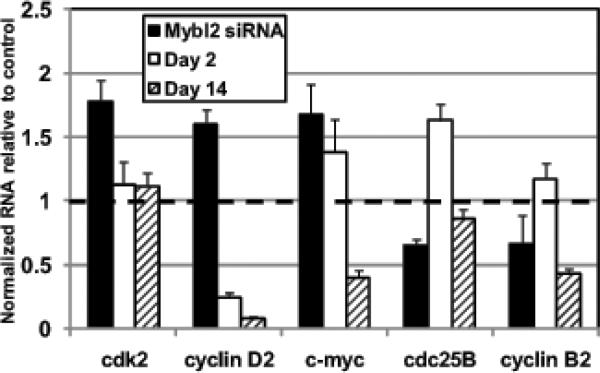
To distinguish whether the downregulation of Mybl2 during colon cell maturation either drives, or occurs as a result of, differentiation-specific gene expression, and to determine targets regulated by Mybl2, we compared gene expression patterns in Mybl2-suppressed proliferating Caco-2 cells to those in spontaneously differentiating Caco-2 cells at early (day 2) or intermediate (day 14) time points of induction. qRT-PCR was used to analyze expression of 45 genes (supplemental table 1) representing a spectrum of functional groups we previously identified as regulated during intestinal cell maturation in vitro and in vivo, including cell cycle-regulating genes, chromatin remodeling complexes, transcription factors, xenobiotic and drug resistance genes, and brush border enzymes (Mariadason et al., 2002).
Several characteristic markers of differentiated colon cells, including alkaline phosphatase (ALPi), dipeptidyl peptidase IV (DPPIV), E-cadherin, and epoxide hydrolase 2 (EPHX2), though substantially upregulated in spontaneously-differentiating Caco-2 cells, were not significantly elevated in Caco-2 cells in which Mybl2 was suppressed by siRNA (supplemental table 1). In addition, in contrast to spontaneously-differentiating Caco-2 cells, Mybl2 siRNA-treated Caco-2 cells did not growth arrest (see Figure 5) and did not acquire gross morphological changes, including acquisition of fluid-transporting domes, typical of differentiation along the absorptive cell lineage (data not shown). Importantly, two distinct subsets of genes were significantly regulated ≥1.5 fold by siRNA suppression of Mybl2 in colon epithelial cells. Mybl2 suppression upregulated expression of genes responsible for promoting progression through G1, including the cyclin dependent kinase cdk2, cyclin D2, and the proto-oncogene c-myc (Figure 2). However, except for cdk2, which remained unchanged during Caco-2 cell maturation, these cell cycle-promoting genes were downregulated during growth arrest and spontaneous differentiation of Caco-2 cells (Figure 2). In contrast, siRNA-mediated suppression of Mybl2 downregulated expression of genes that promote progression through G2/M, such as cdc25B and cyclin B2, although these genes were modestly upregulated early in Caco-2 differentiation (day 2) and downregulated later (day 14). Therefore, Mybl2 suppression, which is a characteristic of differentiating cells in vivo along the crypt-villus axis (Papetti and Augenlicht, submitted), does not directly drive colon cell differentiation as measured by differentiation markers, and Mybl2 may play a unique role in modulating the progression of cells through the cell cycle.
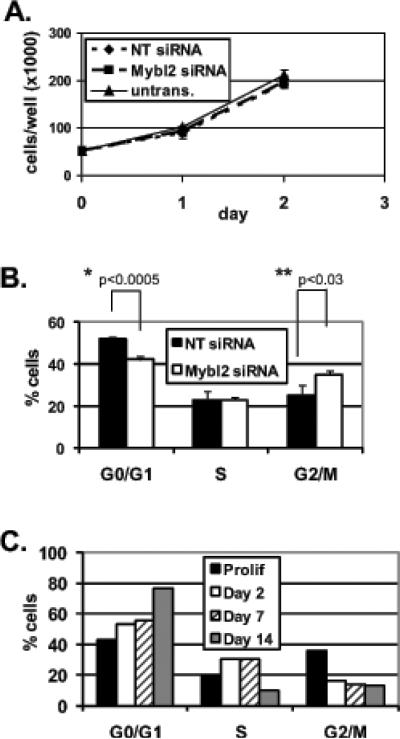
Figure 5.
(A) Proliferation of untransfected and Mybl2 or nontargeting (NT) siRNA-transfected Caco-2 cells was measured by counting cells at 24 hour intervals. (B) DNA content in siRNA-treated cells was measured by propidium iodide staining and flow cytometry as described in Materials and Methods, and cell-cycle distributions were calculated with ModFit software. Graph indicates quantitation of cells in each cell cycle phase. (C) Quantitation of cell cycle distributions in populations of proliferating and day 2, day 7, and day 14 spontaneously-differentiated Caco-2 cells. Asterisks indicate p values for Student's 2-tailed T-tests.
Figure 2.
RNA expression of genes regulated at least 1.5 fold in Mybl2-specific versus control nontargeting (NT) siRNA-treated Caco-2 cells compared to their expression in early (day 2) or late (day 14) spontaneously-differentiating cells as measured by qRT-PCR. Gene expression is normalized to GAPDH and plotted relative to controls (NT siRNA for Mybl2 siRNA or proliferating cells for day 2 and 14 differentiated cells) which are arbitrarily set to 1 (indicated by dashed line).
Mybl2 occupies and transcriptionally regulates sequences upstream of the transcriptional start sites of a subset of genes
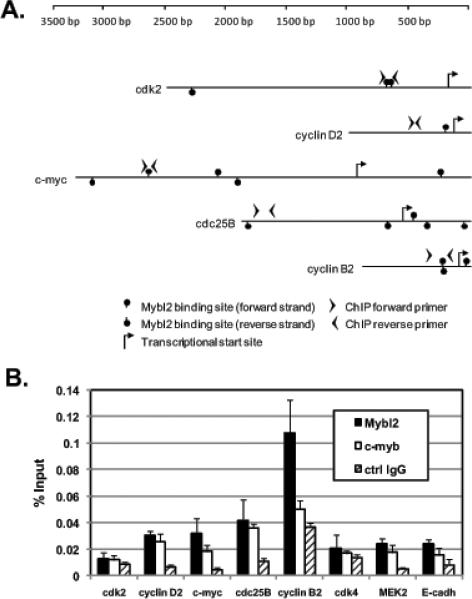
Interestingly, each of the genes significantly altered in expression by Mybl2 suppression harbors at least one consensus Myb binding site within 2 kb upstream of the transcriptional start site (Figure 3A). We therefore determined whether such regions bind Mybl2 and thereby possibly regulate Mybl2 gene transcription. Using chromatin immunoprecipitation (ChIP) with primers overlapping or flanking these potential Myb binding sites (Figure 3A), we found that Mybl2 occupies the regions upstream of the transcriptional start sites of cyclin B2, and, to a lesser extent, cyclin D2, c-myc and cdc25B (Figure 3B). For cyclin B2, this occupancy is specific to Mybl2 because the percentage of input chromatin bound by the related family member c-myb is less than that bound by Mybl2 and similar to that bound by a nonspecific control antibody (Figure 3B). Negligible enrichment for Mybl2 is seen upstream of the transcriptional start sites of cdk4, MEK2, and E-cadherin (Figure 3B), genes not altered in expression by Mybl2 knockdown (supplemental table 1).
Figure 3.
(A) Schematic of sequences upstream of the transcriptional start sites of cdk2, cyclin D2, c-myc, cyclin B2, and cdc25B indicating locations of primers for ChIP analysis. (B) Chromatin from proliferating Caco-2 cells was immunoprecipitated with antibodies to Mybl2, c-myb, and a nonspecific rabbit IgG (rIgG). The percent of input chromatin present in each immunoprecipitate was measured by qRT-PCR using the indicated primers. Each bar represents the mean + standard error of the mean of 3 independent experiments, each performed in duplicate PCR reactions.
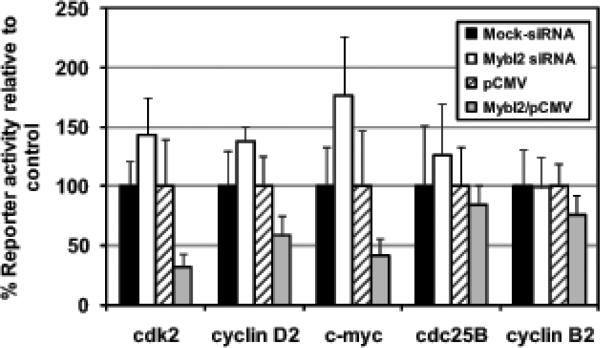
To determine whether Mybl2 can regulate transcriptional activity of these regions, we cloned each upstream of the luciferase coding region in pGL3 Basic and assayed the ability of either overexpressed Mybl2 or Mybl2 knockdown to affect reporter activity (see Materials and Methods for exact sequences). Consistent with the gene expression data (Figure 2), reporter gene activity driven by the upstream regions of cdk2, cyclin D2, and c-myc was reciprocal to Mybl2 expression: reporter activity was activated by Mybl2 knockdown (to 144%, 138%, and 176% control values, respectively) and repressed by Mybl2 overexpression (to 31.6%, 58.4%, and 41.9% control values, respectively) (Figure 4). Mybl2 knockdown or overexpression had negligible effects on transcriptional activity driven by upstream regions of cdc25B and cyclin B2. Thus, Mybl2 expression represses transcriptional activity of the upstream regions of cdk2, cyclin D2, and c-myc, therefore demonstrating that Mybl2 may regulate the expression of these genes by modulating the transcriptional activity of sequences upstream of the transcriptional start sites.
Figure 4.
Proliferating Caco-2 cells were transfected with luciferase reporter plasmids driven by sequences upstream of the transcriptional start sites of the indicated genes (see Figure 3A) as well as either Mybl2 siRNA oligos or Mybl2/pCMV expression vector. Luciferase expression normalized to cotransfected Renilla activity is expressed relative to mock-treated cells (for siRNA experiments) or pCMV vector-transfected cells (for Mybl2 overexpression experiments). Each bar represents the mean + standard error of the mean of 3 independent experiments, each performed in triplicate.
Mybl2 knockdown does not alter colon cell proliferation but induces a modest accumulation of cells in G2/M with a concomitant decrease in G1
These data, and reports that Mybl2 expression is maximal at the G1/S boundary (Lam et al., 1992) and promotes progression from G1 to S phase (Lin et al., 1994), suggest that Mybl2 is a determinant of how colon cells progress through the cell cycle. We therefore assayed proliferation and cell-cycle distribution of Caco-2 cells transfected with Mybl2 versus control siRNA. Over 2 days, Caco-2 cell number was nearly identical in untreated as well as Mybl2 or nontargeting siRNA-treated cells (Figure 5A). Nevertheless, Mybl2 suppression induced a small but significant increase of 10% (p<0.03) of cells in G2/M phases of the cell cycle with a complementary 10% decrease (p<0.0005) of cells in G1 (Figure 5B). This modest shift in cell-cycle distribution is distinct from the accumulation of cells in G1 during spontaneous differentiation (Figure 5C) but consistent with functions of the cell cycle-associated gene expression changes induced by Mybl2 suppression: a more rapid progression of cells through G1 and a delay in progression through G2/M.
DISCUSSION
Similar to pluripotent stem cell differentiation, reprogramming of cells exiting the proliferative compartment in intestinal crypts establishes commitment to specific lineages of differentiation. Numerous genes and pathways, including c-myc, cdk2, cyclin B2, Rb, p27kip, Wnt signaling, and Notch signaling, regulate cell cycle entry and/or progression of intestinal epithelial cells. Correct functioning of the intestinal epithelium requires a link between progression through the cell cycle and the ability to respond to inducers of differentiation and commit to acquisition of a mature state. Therefore, integration of proliferative signals and their coupling to differentiation-specific pathways is critical for intestinal homeostasis and prevention of malignancy.
Mybl2 appears to be a key molecule at the crossroads of both proliferation and differentiation. We have analyzed the function of Mybl2 in colon epithelial cells by examining the effects of siRNA-mediated knockdown of Mybl2 on gene expression, cell cycle, and differentiation. We used siRNA oligos highly specific to Mybl2 that did not alter expression of the other myb family members, A-myb and c-myb (Figure 1). Moreover, although A-myb, Mybl2, and c-myb share high homology in their DNA binding domains (~75% at the amino acid level (Joaquin and Watson, 2003)) and can recognize the same DNA element, several observations indicate they have distinct functions and thus do not complement each other. First, expression patterns of the three proteins are distinct. A-myb is restricted to developing CNS, adult testis, breast ductal epithelium during pregnancy, and germinal center B-lymphocytes (Latham et al., 1996; Mettus et al., 1994; Trauth et al., 1994), and c-myb is expressed mainly in hematopoietic tissues. In contrast, Mybl2 is expressed in all proliferating cells (Sala, 2005; Sitzmann et al., 1996) and is enriched in stem cells (Muller et al., 2008; Tarasov et al., 2008b). Second, compromising the functions of A-Myb, Mybl2, or c-myb in transgenic mice produces very different results. A-Myb-targeted mice exhibit defects in spermatogenesis and mammary gland development (Toscani et al., 1997), whereas c-myb-deficient mice are embryonic lethal at around day E15 due to defects in fetal liver hematopoeisis (Mucenski et al., 1991). Mybl2 deficiency is embryonic lethal at E4.5–E6.5, likely due to a defect in inner cell mass formation (Tanaka et al., 1999). Third, in differentiating cells expressing both Mybl2 and c-myb (e.g. leukemia M1 cells induced by IL-6), the kinetics of downregulation for each molecule is distinct, with c-myb suppression preceding that of Mybl2 (Bies et al., 1996). Fourth, patterns of transcriptional regulation of promoters harboring Myb binding sites are distinct for c-myb versus Mybl2 (Watson et al., 1993). Thus, the functions of Mybl2 compromised by Mybl2 knockdown in our studies are likely not compensated for by A-myb or c-myb.
Coupled with the fact that the highest expression of Mybl2 occurs in proliferating, rather than differentiating, colon epithelial cells (Papetti and Augenlicht, submitted), our data suggest that Mybl2 may function in regulating commitment to differentiation in cycling cells, thus linking cell cycle modulation to lineage-specific maturation. This is reflected in the phenotype of Mybl2-suppressed cells and the known functions of the cell cycle genes modulated in expression. While spontaneously-differentiated Caco-2 cells - in which Mybl2 is downregulated - clearly arrest in G1 (Figure 5C), a premature decrease in Mybl2 induced by siRNA biases cells towards accumulation in G2/M, with a complementary reduction in G1 (Figure 5B), consistent with the fact that three of the genes elevated by siRNA downregulation of Mybl2 (cdk2, cyclinD2 and c-myc) promote progression through G1, and thus their upregulation may promote transit out of G1. In contrast, downregulation of cdc25B and cyclin B2 by Mybl2 siRNA likely retards cells in G2/M. Similar effects of Mybl2 knockdown (i.e. accumulation of cells in G2/M) are reported in megakaryocytes and murine stem cells (Garcia and Frampton, 2006; Tarasov et al., 2008a).
Our data suggest that Mybl2 binds regulatory elements of, and regulates the promotion of transcription by, sequences upstream of the transcriptional start site of a subset of these genes. In accord with the Mybl2 knockdown-induced changes in gene expression (Figure 2), Mybl2 silencing activated and Mybl2 overexpression repressed reporter activity driven by upstream sequences in the G1 regulators cdk2, cyclin D2, and c-myc (Figure 4), and Mybl2 indeed occupied the upstream sequences of c-myc, cyclin D2, cyclin B2 and cdc25B (Figure 3). The absence of detectable Mybl2 binding to cdk2 sequences by ChIP and Mybl2 regulation of cyclin B2 and cdc25B by reporter assay may be due to more complex transcription factor interactions, such as an indirect recruitment of Mybl2 by E2F (Zhu et al., 2004) or Sp1 (Cicchillitti et al., 2004; Sala et al., 1999) family members, and lack of relevant cis-regulatory elements. In many cases, such necessary sequences may be located many kilobases upstream or downstream of the transcriptional start site (Ohler and Wassarman) and thus may have been absent in the reporter constructs and/or DNA fragments defined by primers used in the ChIP assay. Furthermore, activation of exogenously-expressed Mybl2 targets is complex and has been reported to be dependent on possible structural changes in Mybl2 due to phosphorylation by cdk2/cyclin A (Johnson et al., 1999; Lane et al., 1997), interactions with coregulators (Schubert et al., 2004), and removal of a C-terminal inhibitory domain (Lane et al., 1997). Therefore, in Caco-2 cells, which already robustly express Mybl2, overexpressed Mybl2 may not adopt the proper conformation to activate transcription from target promoters. Regardless, however, of the detailed mechanism employed by each locus, the data demonstrate that Mybl2 expression represses a subset of G1 regulators (eg cdk2, c-myc and cyclin D2) and upregulates expression of a subset of G2/M regulators (eg cyclin B2 and cdc25B) which can cooperate to gate progression of cells through the cell cycle.
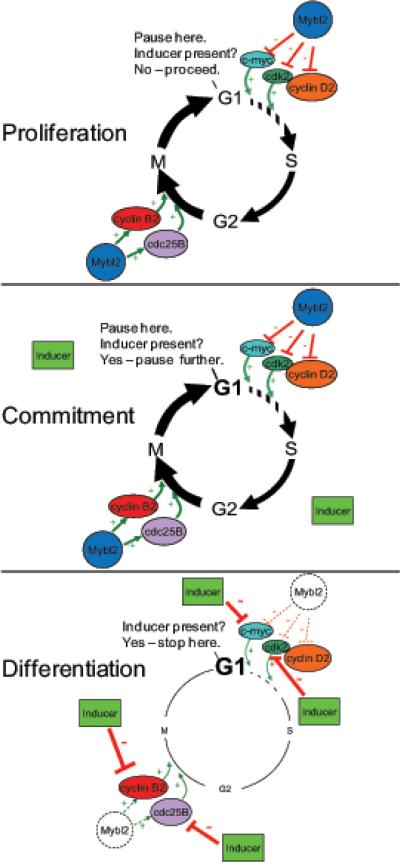
Therefore, our working hypothesis is that Mybl2 downregulation does not drive colon cell differentiation, but that by modulating both G1 and G2/M regulators, Mybl2 may contribute to preparing cells for the regulatory influence of other transcription factors (e.g., Hes1 and Hath1 of the Notch pathway, or KLF4) that directly drive commitment to different lineages. The modest, though significant, changes induced by Mybl2 knockdown or overexpression suggest that Mybl2 may induce a transient pause in G1, sufficient to ensure that the cell will commit to differentiation induced by additional signals, yet without inducing cell-cycle arrest, as the cell progresses through the transit amplifying compartment, as depicted in the model in Figure 7. Once commitment has occurred, Mybl2 downregulation then allows other differentiation-specific factors to completely arrest cells in G1 and fully express the differentiation program.
This model predicts changes in types and levels of transcription factors, including Mybl2, that associate with genes involved in lineage-specific commitment and expression of lineage-specific markers, at different stages of the cell cycle. Reported data are consistent with this: not only does Mybl2 regulate cell-cycle promoting genes, including cdc2 (Sala et al., 1997; Zhu et al., 2004), cyclins D1 and A1 (Bartusel et al., 2005), and cyclin B1 (Knight et al., 2009; Zhu et al., 2004), but also genes involved in acquiring differentiated states, such as elastin (Hofmann et al., 2005), collagen (Cicchillitti et al., 2004), and adenosine receptor (St Hilaire et al., 2008). In this regard, we have found that Mybl2 may also modulate the expression of differentiation-specific genes, including sucrase isomaltase and the glucose transporter Glut1, both of which contain Myb binding sites in their promoter regions, in colon epithelial cells (data not shown). Thus, our data emphasize that there may be an integrated kinetic relationship between Mybl2 and cell-cycle regulation that prepares intestinal epithelial cells for additional key signals coordinating intestinal cell maturation along the crypt-luminal axis (Mariadason et al., 2002; Mariadason et al., 2005; Velcich et al., 2005).
A growing body of recent evidence indicates that robust Mybl2 expression is an important characteristic of stem cells (Boheler, 2009; Muller et al., 2008; Tarasov et al., 2008a; Tarasov et al., 2008b), but the role that Mybl2 plays in these pluripotent cells is not clear. Tarasov et al. reported that Mybl2 is required for chromosomal integrity and cellular euploidy and that, by binding and regulating promoter activity of the Pou5f1(Oct4) gene, promotes maintenance of the undifferentiated stem cell state (Tarasov et al., 2008a). Stem cells must not only be dedicated to self renewal and prevention of differentiation but must also be poised to adopt differentiation programs in response to induction signals. No mechanism has yet been proposed to account for the ability of progenitor cells to transition from a proliferative, undifferentiated population to one committed to the acquisition of a differentiated state. It will be interesting to explore whether the involvement of Mybl2 in preparing undifferentiated intestinal cells for commitment to maturation, suggested by our data, may also be part of the mechanism whereby continually cycling stem cells are poised to commit to differentiation pathways.
Supplementary Material
Figure 6.
Model of Mybl2's role in coordinating expression of cell cycle regulatory genes to prime cells for commitment to lineage-specific differentiation. In proliferating cells, Mybl2 may cause cells to pause in G1 long enough to receive and process differentiation induction signals by modestly activating expression of genes, such as cyclin B2 and cdc25B, that promote progression through G2/M and suppressing expression of genes, including cdk2, cyclin D2, and c-myc, that drive progression through G1. Once cells are induced to differentiate, Mybl2 is downregulated, and other factors completely arrest cells in G1 and robustly activate maturation-specific gene expression.
ACKNOWLEDGEMENTS
*We wish to thank Georgia Corner for assisting with gene functional group classifications and Michele Houston for FACScan assistance.
This work was supported by grants U54 CA100926, CA114265, CA123473, and P013330 from the National Cancer Institute.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Augenlicht L, Velcich A, Heerdt BG. Short-chain fatty acids and molecular and cellular mechanisms of colonic cell differentiation and transformation. Adv Exp Med Biol. 1995;375:137–148. doi: 10.1007/978-1-4899-0949-7_12. [DOI] [PubMed] [Google Scholar]
- Bartusel T, Schubert S, Klempnauer KH. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene. 2005;351:171–180. doi: 10.1016/j.gene.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Bies J, Hoffman B, Amanullah A, Giese T, Wolff L. B-Myb prevents growth arrest associated with terminal differentiation of monocytic cells. Oncogene. 1996;12(2):355–363. [PubMed] [Google Scholar]
- Boheler KR. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J Cell Physiol. 2009;221(1):10–17. doi: 10.1002/jcp.21866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Chance MR, Nicholas C, Ahmed N, Guilmeau S, Flandez M, Wang D, Byun DS, Nasser S, Albanese JM, Corner GA, Heerdt BG, Wilson AJ, Augenlicht LH, Mariadason JM. Proteomic changes during intestinal cell maturation in vivo. J Proteomics. 2008;71(5):530–546. doi: 10.1016/j.jprot.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates cdk6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. J Biol Chem. 285(5):3044–3052. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillitti L, Jimenez SA, Sala A, Saitta B. B-Myb acts as a repressor of human COL1A1 collagen gene expression by interacting with Sp1 and CBF factors in scleroderma fibroblasts. The Biochemical journal. 2004;378(Pt 2):609–616. doi: 10.1042/BJ20031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet JC, Wang L, Vitek O, Craig BA, Edenberg HJ. Gene expression profiling of Caco-2 BBe cells suggests a role for specific signaling pathways during intestinal differentiation. Physiol Genomics. 2003;13(1):57–68. doi: 10.1152/physiolgenomics.00152.2002. [DOI] [PubMed] [Google Scholar]
- Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59(1):221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- Fogh JaT, G. New Human Tumor Cell LInes. In: Fogh J, editor. Human Tumor Cell LInes in vitro. Plenum Press; New York: 1975. pp. 115–141. [Google Scholar]
- Garcia P, Frampton J. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J Cell Sci. 2006;119(Pt 8):1483–1493. doi: 10.1242/jcs.02870. [DOI] [PubMed] [Google Scholar]
- Graf T. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr Opin Genet Dev. 1992;2(2):249–255. doi: 10.1016/s0959-437x(05)80281-3. [DOI] [PubMed] [Google Scholar]
- Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320(1):23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- Hofmann CS, Wang X, Sullivan CP, Toselli P, Stone PJ, McLean SE, Mecham RP, Schreiber BM, Sonenshein GE. B-Myb represses elastin gene expression in aortic smooth muscle cells. J Biol Chem. 2005;280(9):7694–7701. doi: 10.1074/jbc.M412501200. [DOI] [PubMed] [Google Scholar]
- Joaquin M, Watson RJ. Cell cycle regulation by the B-Myb transcription factor. Cell Mol Life Sci. 2003;60(11):2389–2401. doi: 10.1007/s00018-003-3037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TK, Schweppe RE, Septer J, Lewis RE. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J Biol Chem. 1999;274(51):36741–36749. doi: 10.1074/jbc.274.51.36741. [DOI] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28(15):1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, Chen X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104(39):15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam EW, Robinson C, Watson RJ. Characterization and cell cycle-regulated expression of mouse B-myb. Oncogene. 1992;7(9):1885–1890. [PubMed] [Google Scholar]
- Lane S, Farlie P, Watson R. B-Myb function can be markedly enhanced by cyclin A-dependent kinase and protein truncation. Oncogene. 1997;14(20):2445–2453. doi: 10.1038/sj.onc.1201086. [DOI] [PubMed] [Google Scholar]
- Latham KE, Litvin J, Orth JM, Patel B, Mettus R, Reddy EP. Temporal patterns of A-myb and B-myb gene expression during testis development. Oncogene. 1996;13(6):1161–1168. [PubMed] [Google Scholar]
- Lin D, Fiscella M, O'Connor PM, Jackman J, Chen M, Luo LL, Sala A, Travali S, Appella E, Mercer WE. Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1-mediated G1 arrest. Proc Natl Acad Sci U S A. 1994;91(21):10079–10083. doi: 10.1073/pnas.91.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Arango D, Corner GA, Aranes MJ, Hotchkiss KA, Yang W, Augenlicht LH. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62(16):4791–4804. [PubMed] [Google Scholar]
- Mariadason JM, Nicholas C, L'Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, Arango D, Augenlicht LH. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128(4):1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Mettus RV, Litvin J, Wali A, Toscani A, Latham K, Hatton K, Reddy EP. Murine A-myb: evidence for differential splicing and tissue-specific expression. Oncogene. 1994;9(10):3077–3086. [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr., Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65(4):677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, Schmidt NO, Loring JF. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455(7211):401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Wassarman DA. Promoting developmental transcription. Development. 137(1):15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papetti M, Skoultchi AI. Reprogramming leukemia cells to terminal differentiation and growth arrest by RNA interference of PU.1. Mol Cancer Res. 2007;5(10):1053–1062. doi: 10.1158/1541-7786.MCR-07-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschella G, Negroni A, Sala A, Pucci S, Romeo A, Calabretta B. Requirement of b-myb function for survival and differentiative potential of human neuroblastoma cells. J Biol Chem. 1995;270(15):8540–8545. doi: 10.1074/jbc.270.15.8540. [DOI] [PubMed] [Google Scholar]
- Reddy BS. Dietary fiber and colon cancer: animal model studies. Prev Med. 1987;16(4):559–565. doi: 10.1016/0091-7435(87)90072-7. [DOI] [PubMed] [Google Scholar]
- Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 2005;41(16):2479–2484. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Sala A, Kundu M, Casella I, Engelhard A, Calabretta B, Grasso L, Paggi MG, Giordano A, Watson RJ, Khalili K, Peschle C. Activation of human B-MYB by cyclins. Proc Natl Acad Sci U S A. 1997;94(2):532–536. doi: 10.1073/pnas.94.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, Saitta B, De Luca P, Cervellera MN, Casella I, Lewis RE, Watson R, Peschle C. B-MYB transactivates its own promoter through SP1-binding sites. Oncogene. 1999;18(6):1333–1339. doi: 10.1038/sj.onc.1202421. [DOI] [PubMed] [Google Scholar]
- Sala A, Watson R. B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments. J Cell Physiol. 1999;179(3):245–250. doi: 10.1002/(SICI)1097-4652(199906)179:3<245::AID-JCP1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Schubert S, Horstmann S, Bartusel T, Klempnauer KH. The cooperation of B-Myb with the coactivator p300 is orchestrated by cyclins A and D1. Oncogene. 2004;23(7):1392–1404. doi: 10.1038/sj.onc.1207255. [DOI] [PubMed] [Google Scholar]
- Sitzmann J, Noben-Trauth K, Kamano H, Klempnauer KH. Expression of B-Myb during mouse embryogenesis. Oncogene. 1996;12(9):1889–1894. [PubMed] [Google Scholar]
- St Hilaire C, Yang D, Schreiber BM, Ravid K. B-Myb regulates the A(2B) adenosine receptor in vascular smooth muscle cells. Journal of cellular biochemistry. 2008;103(6):1962–1974. doi: 10.1002/jcb.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem. 1999;274(40):28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- Tappenden KA, Thomson AB, Wild GE, McBurney MI. Short-chain fatty acid-supplemented total parenteral nutrition enhances functional adaptation to intestinal resection in rats. Gastroenterology. 1997;112(3):792–802. doi: 10.1053/gast.1997.v112.pm9041241. [DOI] [PubMed] [Google Scholar]
- Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, Li RA, Lim B, Stewart CL, Liu Y, Van Eyk JE, Wersto RP, Wobus AM, Boheler KR. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS ONE. 2008a;3(6):e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov KV, Testa G, Tarasova YS, Kania G, Riordon DR, Volkova M, Anisimov SV, Wobus AM, Boheler KR. Linkage of pluripotent stem cell-associated transcripts to regulatory gene networks. Cells, tissues, organs. 2008b;188(1–2):31–45. doi: 10.1159/000118787. [DOI] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, Hatton KS, Reddy EP. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997;386(6626):713–717. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- Trauth K, Mutschler B, Jenkins NA, Gilbert DJ, Copeland NG, Klempnauer KH. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. Embo J. 1994;13(24):5994–6005. doi: 10.1002/j.1460-2075.1994.tb06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velcich A, Corner G, Paul D, Zhuang M, Mariadason JM, Laboisse C, Augenlicht L. Quantitative rather than qualitative differences in gene expression predominate in intestinal cell maturation along distinct cell lineages. Experimental cell research. 2005;304(1):28–39. doi: 10.1016/j.yexcr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Watson RJ, Robinson C, Lam EW. Transcription regulation by murine B-myb is distinct from that by c-myb. Nucleic acids research. 1993;21(2):267–272. doi: 10.1093/nar/21.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. Embo J. 2004;23(23):4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.