Abstract
Mass spectrometry and fluorescent probes have provided direct evidence that alkylating agents permeate the protein capsid of naked viruses and chemically inactivate the nucleic acid. N-acetyl-aziridine and a fluorescent alkylating agent, dansyl sulfonate aziridine, inactivated three different viruses, flock house virus, human rhinovirus-14, and foot and mouth disease virus. Mass spectral studies as well as fluorescent probes showed that alkylation of the genome was the mechanism of inactivation. Because particle integrity was not affected by selective alkylation (as shown by electron microscopy and sucrose gradient experiments), it was reasoned that the dynamic nature of the viral capsid acts as a conduit to the interior of the particle. Potential applications include fluorescent labeling for imaging viral genomes in living cells, the sterilization of blood products, vaccine development, and viral inactivation in vivo.
Antiviral agents usually attack the viral life cycle by inhibiting intracellular expression of viral enzymes or by interfering with extracellular steps such as interaction of the virus with the cellular receptor (1–5). Viral protease and replicase inhibitors are highly specific, but their efficacy can be significantly reduced by the emergence of viral mutants. A more general approach to disarming viruses is through chemical modification of the virus particles, such as with N-acetyl-aziridine (Fig. 1), as used in the production of killed-virus vaccines (6, 7). Here we report how alkylating agents inactivate viruses, and we introduce a versatile molecular design for viral inactivants.
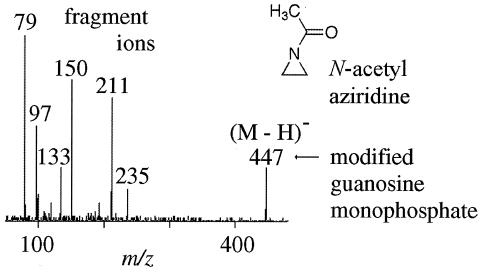
Figure 1.
Nanoelectrospray ionization tandem mass spectrometry (nanoESI-MS/MS) experiment performed on a modified RNA nucleotide. FHV RNA was extracted from N-acetyl-aziridine-treated and -untreated virus and exposed to RNase. The nanoESI-MS/MS results show that the treated virus had covalent N-acetyl-aziridine modification on approximately 0.4% of the nucleotides. The mass spectrometry analyses were performed on a Perkin–Elmer Sciex API III (Sciex, Thornhill, ON, Canada) modified with a nanoESI source from Protana A/S (Denmark). Palladium-coated, boro-silicate glass capillaries from Protana A/S were used for nanoelectrospray sample delivery.
Two possible mechanisms exist for viral inactivation with alkylating agents. One mechanism involves the modification of proteins, which would cause inhibition of viral cell entry or the release of the genome. The second mechanism allows the alkylating reagents direct access to the viral genome through a mobile protein capsid (8–12). The recent findings (8–11) that the protein capsids of viruses in solution have a much higher degree of dynamics than their crystallized counterparts suggested that the second mechanism might be the means of inactivation. Focusing on this latter idea, the ability of small alkylating agents to react with either the capsid or encapsidated nucleic acid was investigated initially by using flock house virus (FHV), an RNA-containing model virus used in previous studies (8, 9).
The alkylating agent N-acetyl-aziridine is a virus inactivant that has been used in vaccine preparation since the 1950s, yet no direct evidence for its mechanism of inactivation has been determined, making it a suitable starting point for this investigation. The chemistry of aziridines is dominated by ring strain, thus leading to enhanced reactivity in reactions where the strain is relieved. The tendency of aziridines to undergo ring-opening reactions with nucleophiles such as the nitrogen atoms in adenine and guanine make them natural alkylating agents of nucleotides.
Methods
Compounds.
The synthesis of N-acetyl-aziridine was performed according to standard synthetic procedures (13). Chlorambucil and cyclophosphamide were purchased from Sigma and used without further purification. N-dansylaziridine was synthesized from dansyl chloride dissolved in tetrahydrofuran (THF) and a solution of ethyleneimine and diisopropylethyl amine in THF was added dropwise to a final concentration of 1 and 2 equivalents, respectively. The temperature was maintained at 0°C during the addition. The solution was stirred for 5 h; after 3 h, the solution was allowed to warm to room temperature. The diisopropylethyl amine hydrochloride was filtered off, and the solution was evaporated to dryness. The crude product was recrystallized from benzene and cyclohexane. Purity was determined from MS and NMR.
Alkylation and MS.
All incubation reactions between the alkylating agents and the virus, as well as with free DNA or RNA, were performed at 25°C or 37°C for 3–5 h. The reaction solution typically contained 20 mM Tris buffer at pH 7.5. When N-dansylaziridine was incubated with the virus, a stock solution of the N-dansylaziridine alkylating agent was made up in acetonitrile and added to the virus until a final concentration of 5% acetonitrile was achieved. The 5% acetonitrile did not affect the infectivity of the virus.
The FHV was incubated with N-acetyl-aziridine in 20 mM Tris buffer for 4 h. N-acetyl-aziridine and reaction by-products such as hydrolyzed substrate were removed from the solution containing the intact virus with a 100-kDa molecular mass cutoff filter. The RNA was then extracted from the treated virus and exposed to nonspecific ribonucleases to generate individual nucleotides. In turn, these nucleotides were analyzed by nanoESI-MS/MS. The nanoESI experiments were performed on a Perkin–Elmer Sciex API III triple quadrupole mass spectrometer modified with a nanoESI source from Protana A/S (Denmark). The orifice and electrospray potential were set at −115 V and −650 V, respectively. A curtain gas of ultrapure nitrogen was pumped into the interface at a rate of 0.6 liter/min to aid evaporation of solvent droplets and prevent particulate matter from entering the analyzer region. Desolvated ions entered the analyzer via the vacuum interface and were guided by entrance optics. Palladium-coated, boro-silicate glass capillaries from Protana A/S were used for sample delivery. The collision-induced dissociation experiments were performed with ultra-pure argon as a collision gas. The precursor ion spectra were acquired by scanning the first quadrupole, whereas collisions with argon (target thickness of 30 × 1014 atom/cm2) in the second quadrupole produced ion dissociation. The third quadrupole was used to mass-select the fragment ion. Spectra were the result of averaging from 50 to 200 scans depending on the number of scans necessary to obtain a signal-to-noise ratio greater than 50.
The viral proteolysis ms experiments were conducted by using a PerSeptive Biosystems (Framingham, MA) Voyager Elite matrix-assisted laser desorption/ionization (MALDI) time-of-flight reflectron mass spectrometer. All MALDI analyses were conducted by using 3,5-dimethoxy-4-hydroxycinnamic acid matrix (Aldrich) dissolved in a 50% acetonitrile/water solution. Proteolytic digests were performed at room temperature by using modified trypsin (Promega). All digests were performed in 20 mM Tris⋅HCl (pH 7.5); total volume was 25 μl. Aliquots of 0.5 μl were withdrawn from the reaction vessel at time points of 1, 3, 5, 10, and 20 min, and the digest reactions were quenched on addition of the acidic matrix. Modified trypsin (Promega) was used at a 1:100 (wt/wt) enzyme-to-virus ratio.
Cells and Virus.
Drosophila melanogaster cells (Schneider's line 1) were grown as monolayers in Schneider's insect medium supplemented with 15% heat-inactivated FBS, as described (14). Infections were carried out in suspension at a cell density of 4 × 107 cells/ml. Specifically, 2.5 ml of such a suspension was inoculated with FHV at a multiplicity of 1–5 plaque-forming units per cell. After 1 h at room temperature with gentle agitation, the cell suspension was transferred to a 100-mm tissue culture dish containing 12.5 ml of cell growth medium. Plates were further incubated at 27°C without agitation.
Two days after infection, cells were dislodged into the growth medium and lysed by addition of Nonidet P-40 to a final concentration of 0.5% (vol/vol). After 10 min on ice, cell debris was removed by low-speed centrifugation at 13,888 × g for 10 min at 4°C. FHV particles in the supernatant were pelleted through a 30% (wt/wt) sucrose cushion in 50 mM Hepes (pH 7)/5 mM CaCl2/0.1% (wt/vol) BSA/0.1% (vol/vol) 2-mercaptoethanol at 274,000 × g for 2.5 h at 11°C. The pellet was resuspended in 50 mM Hepes (pH 7)/5 mM CaCl2/0.1% (vol/vol) 2-mercaptoethanol and layered on a 10–40% (wt/wt) sucrose gradient in the same buffer. Particles were sedimented at 274,000 × g for 1.5 h at 11°C, and the virus band was harvested by needle puncture of the gradient tube.
FHV titers were determined by plaque assay on monolayers of D. melanogaster cells, as described (15).
RNA Extraction and Agarose Gel Electrophoresis.
SDS and NaCl were added to gradient-purified FHV particles at a final concentration of 1% (wt/vol) and 0.2 M, respectively. RNA was extracted with an equal volume of acidic phenol-chloroform and precipitated with 3 volumes of ethanol in the presence of 0.3 M sodium acetate and 20 μg of glycogen. After several hours at −20°C, the RNA was pelleted, washed with 70% ethanol, dried, and dissolved in nuclease-free water. Electrophoretic analysis was performed on nondenaturing 1% agarose gels in Tris-acetate–EDTA buffer. At the end of the run, the gel was incubated in the presence of ethidium bromide to visualize nucleic acids.
Results and Discussion
Initially, FHV was incubated with 5 mM N-acetyl-aziridine at 37° C for 2 h when a slight alkylation of the proteins was observed by using MALDI-MS. However, at lower concentrations of the aziridine (1 mM), no alkylation of the capsid proteins was observed, although the virus infectivity (as indicated by plaque assay) had dropped by a factor of 1,000. Sucrose gradient analysis and electron microscopy confirmed that the N-acetyl-aziridine was not affecting the integrity of the virion. These observations suggested that inactivation of FHV occurred through N-acetyl-aziridine modification of the viral genome. To test this hypothesis, nonreacted N-acetyl-aziridine and reaction byproducts were removed from the solution containing the intact virus with a 100-kDa molecular mass cutoff filter. The RNA was then extracted from the treated virus and exposed to nonspecific ribonucleases to generate individual nucleotides. These nucleotides were then analyzed by nanoESI-MS/MS. The nanoESI-MS/MS results showed the presence of N-acetyl-aziridine-modified nucleotides (Fig. 1), and precursor ion scanning (16) with nanoESI-MS/MS revealed that ≈0.4% of the RNA had been alkylated. These results demonstrated that alkylating agents can pass through the viral capsid and react with the genome without affecting the proteins.
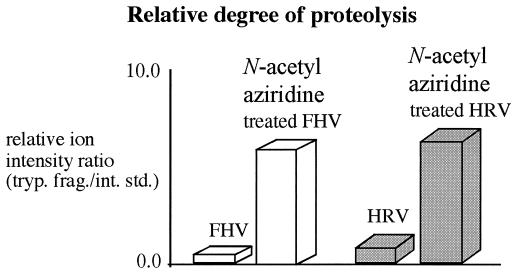
Further evidence of genome alkylation was obtained by proteolysis experiments of the intact virus. FHV and human rhinovirus-14 particles that had been incubated with 1 mM N-acetyl-aziridine were significantly more susceptible to proteolytic cleavage by trypsin than the nonalkylated viruses (Fig. 2), as determined by liquid chromatography ms (LC/MS) and MALDI MS. Interestingly, no chemical modification to the proteolytic fragments was observed in these experiments. These results were reminiscent of previous studies in which an increase in the rate of proteolysis was observed for FHV particles containing nonviral RNA (9). The rate increase was interpreted as resulting from an increase in particle dynamics caused by the disruption of protein–RNA interactions normally established between coat protein and the authentic viral genome. Similarly, alkylation of the RNA in the presence of N-acetyl-aziridine may have led to the disruption of protein-RNA interactions, thereby leading to increased susceptibility to proteolytic cleavage.
Figure 2.
Relative degree of proteolysis of capsid proteins of N-acetyl-aziridine-treated and -untreated FHV and human rhinovirus-14. The analysis was performed with two different mass spectrometry techniques, liquid chromatography (LC)/MS and MALDI MS), which provided the same results. The LC/MS and MALDI experiments were done with a Hewlett-Packard 1100 electrospray system and a PerSeptive STR Voyager mass spectrometer, respectively.
To test this model further, other alkylating agents were used to modify/inactivate the viral genome, and the proteolytic digestion analysis was applied as a screen. The alkylating agents tested included ethyleneimine, β-propiolactone, dimethyl sulfate, and N-nitroso ethyl urea. All of the above compounds increased the rate of proteolysis in the intact virus, but βpropiolactone also reacted with the viral capsid proteins. This property diminishes the potential value of β-propiolactone as a viral inactivant. Because of their very toxic nature, no further experiments were done with dimethyl sulfate and N-nitroso ethyl urea.
Another class of compounds investigated was nitrogen mustard derivatives such as chlorambucil and cyclophosphamide. These compounds consist of two chloroethyl groups that are covalently bound to a nitrogen; an aziridinium ion is then formed by a ring-closing reaction between the nitrogen and the ethyl group, with subsequent loss of the chloro ion. Although these molecules are potent alkylating agents, no increase in proteolytic digestion could be detected. Because both compounds are significantly larger than N-acetyl-aziridine, we initially assumed that this was because of their inability to diffuse into the viral capsid. However, when the N,N-dimethyl aziridinium ion, whose size is similar to that of N-acetyl-aziridine, was tested, it also did not increase the proteolysis rate of the viral capsid. These results indicated that perhaps the positive charge on the molecule prevented the diffusion of the alkylating agent into the viral capsid. Plaque assay experiments confirmed that nitrogen mustard derivatives did not inactivate FHV. The observation that hydrophobic alkylating agents penetrated the capsid more efficiently than charged compounds suggested that the access route to the viral interior might be lined by hydrophobic residues. The crystal structure of the FHV particle does not provide much information regarding the pathway that the compounds employed in this study used to gain access to the encapsidated nucleic acid. There are no holes or crevices that extend from the capsid surface to the interior of the particle. In fact, on the basis of the x-ray structure, the capsid should be impermeable even to water molecules.
Our data suggested that capsid permeability could be exploited further by the design of molecules that would be small enough to pass through the protein shell, yet large enough to retain activity. The relative ease of synthesizing aziridines with various functional groups makes this class of molecules readily available. For some aziridines to be effective alkylating agents in aqueous solution, the ring nitrogen atom must be protonated and will thus have a positive charge. Two examples of aziridines that require protonation to be reactive are N-(2-hydroxyethyl) aziridine and N-ethylaziridine, and both of these molecules were investigated. Under mildly acidic conditions, when the nitrogen atom is protonated, they were inactive against viruses. These results, combined with the previous observations that aziridinium ions did not inactivate virus further, suggested that positively charged molecules cannot penetrate the capsid effectively. To overcome this problem, more reactive aziridines that alkylate nucleophiles without being protonated were investigated. The most effective way to achieve this increased reactivity was to use electron-withdrawing groups on the nitrogen atom such as acyl, carboalkoxy, sulfonyl, and diphenylphosphinyl groups.
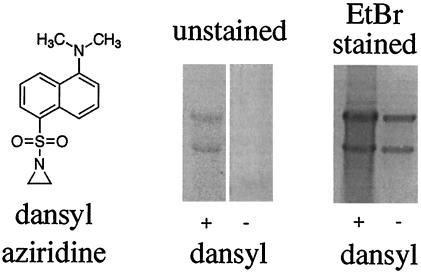
The sulfonyl-substituted aziridines were particularly attractive for selective reactivity because the sulfonyl does not undergo undesired side reactions with proteins such as the acetylation obtained with N-acetyl-aziridine. One particular sulfonyl-substituted aziridine, N-dansylaziridine (Fig. 3), was an effective viral inactivant. The N-dansylaziridine penetrated the capsid of FHV and reacted with the intact viral genome, thus inactivating the virus and rendering the genome fluorescent because of covalent attachment of the dansyl group (Fig. 3).
Figure 3.
Agarose gel electrophoresis of N-dansylaziridine-modified and -unmodified viral RNA. RNA was extracted from N-dansylaziridine-treated FHV and an untreated control sample by using phenol-chloroform. The extracted material was ethanol-precipitated and electrophoresed through a nondenaturing 1% agarose gel. (Left) Structure of N-dansylaziridine. (Center) After electrophoresis, the gel was placed on a transilluminator and viewed by using a 300-nm light source. Nucleic acids representing FHV RNA1 (3.1 kb, upper band) and RNA2 (1.4 kb, lower band) were visible for the sample treated with N-dansylaziridine, but not the control. (Right) The gel was stained with ethidium bromide for 30 min and viewed again with a transilluminator. RNA in both lanes is now visible.
Ms experiments showed that the covalent attachment of N-dansylaziridine to FHV was accompanied by an increased rate of proteolysis of the viral capsid, although both negative staining and sucrose gradient centrifugation showed no evidence of loss of particle structural integrity. Because alkylation of proteins was not detected in these mass spectral studies of the N-dansylaziridine-exposed FHV, the results indicated that the capsid mobility was enhanced because of the disruption of protein–RNA interactions in the native virus. This effect was presumably caused by chemical modification of the nucleic acid, similar to the observations made with N-acetyl-aziridine.
The selectivity of N-dansylaziridine to reaction with the genome rather than the capsid protein was tested by exposing it to equimolar mixtures of a small RNA oligonucleotide and soluble FHV capsid protein. Even after prolonged exposure (6 h), no N-dansylaziridine modification of the protein was detected by MALDI MS, yet significant RNA modification was observed. Additional control experiments were performed in which FHV was treated with nonreactive compounds (at 5 mM), including dansyl sulfonate, rhodamine, and ethidium bromide. These compounds did not increase or decrease the rate of proteolysis, suggesting that molecules that do not covalently modify the nucleotides do not disrupt the viral protein–RNA interactions.
Other nonenveloped viruses such as human rhinovirus-14 and foot and mouth disease virus were also inactivated by N-dansylaziridine. In the case of foot and mouth disease virus, inactivation occurred at a N-dansylaziridine concentration of less than 100 μM, a concentration far lower than that required for inactivation by using N-acetyl-aziridine (1 mM). Interestingly, inactivation by N-dansylaziridine of all of the viruses tested was found to be highly temperature-dependent. The viruses were not inactivated at 25°C but were inactivated at 37°C, suggesting that at the higher temperature the capsid is more mobile, thus making the viral genome more accessible to the alkylating agents. Control experiments measuring the susceptibility of an RNA oligonucleotide at 25°C and 37°C showed no appreciable difference in the dansyl sulfonate–aziridine reactivity as measured by MALDI MS.
Overall, our results show clearly that viral capsid mobility allows for selective modification of the viral genome. The activity of fluorescent N-dansylaziridine suggests further that even more sophisticated alkylating agents can be used to modify viral genetic material with several potential applications: (i) Fluorescent anchors that can be used to label the encapsidated RNA and monitor its release from the virion during the uncoating process, or to visualize its movement within the cells; (ii) dendrimer-based anchors that could be advantageous as multivalent antiviral agents in drug or vaccine development; and (iii) anchors such as sialic acid that could bind to viral glycoproteins of enveloped viruses, e.g., influenza virus. Alternatively, solid support anchors could effectively extract viruses from body fluids such as blood, and virus identification could be achieved through enzymatic digestion of the capsid proteins with mass analysis and proteomic database searching. The goal of this multidisciplinary effort in mass spectrometry, virology, and organic chemistry is to improve our understanding of structural virology and design molecules that selectively attack the viral genome for the inactivation of viruses in vivo.
Acknowledgments
We gratefully acknowledge Jennifer Boydston and Sebastian Wendeborn for their many helpful comments and suggestions. We also thank Elaine Chase for the human rhinovirus preparation. K.B. thanks the Knut and Alice Wallenberg Foundation for a postdoctoral fellowship. We are grateful for support from National Institutes of Health Grants GM55775, GM53491, and GM10704 (to G.S., A.S., and T.J.S., respectively).
Abbreviations
- FHV
flock house virus
- nanoESI-MS/MS
nanoelectrospray ionization tandem MS
- MALDI
matrix-assisted laser desorption/ionization
- LC/MS
liquid chromatography MS
References
- 1.Balfour H H., Jr N Engl J Med. 1999;340:1255–1268. doi: 10.1056/NEJM199904223401608. [DOI] [PubMed] [Google Scholar]
- 2.Hodinka R L. Infect Dis Clin North Am. 1997;11:945–967. doi: 10.1016/S0891-5520(05)70399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patick A K, Potts K E. Clin Microbiol Rev. 1998;11:614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith T J, Kremer M J, Luo M, Vriend G, Arnold E, Kamer G, Rossmann M G, McKinlay M A, Diana G D, Otto M J. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 5.Wade R C. Structure (London) 1997;5:1139–1145. doi: 10.1016/s0969-2126(97)00265-7. [DOI] [PubMed] [Google Scholar]
- 6.Brown F, Crick J. J Immunol. 1959;82:444–448. [PubMed] [Google Scholar]
- 7.Brown F, Meyer R F, Law M, Kramer E, Newman J F E. Biologicals. 1998;26:39–47. doi: 10.1006/biol.1998.0122. [DOI] [PubMed] [Google Scholar]
- 8.Bothner B, Dong X F, Bibbs L, Johnson J E, Siuzdak G. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- 9.Bothner B, Schneemann A, Marshall D, Reddy V, Johnson J E, Siuzdak G. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 10.Lewis J K, Bothner B, Smith T J, Siuzdak G. Proc Natl Acad Sci USA. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Yafal A G, Lee Y, Hogle J, Chow M. J Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer B, Fraenkel-Conrat H. Biochemistry. 1969;8:3266–3269. doi: 10.1021/bi00836a020. [DOI] [PubMed] [Google Scholar]
- 13.Dermer O C, Ham G E. Ethyleneimine and Other Aziridines. New York: Academic; 1969. p. 592. [Google Scholar]
- 14.Friesen P D, Rueckert R R. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneemann A, Zhong W, Gallagher T M, Rueckert R R. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatman K, Hollenbeck T, Hagey L, Vallee M, Purdy R, Weiss F, Siuzdak G. Anal Chem. 1999;71:2358–2363. doi: 10.1021/ac9806411. [DOI] [PubMed] [Google Scholar]