Abstract
In January 2008, an outbreak of acute gastroenteritis at a waterpark was reported to the Bundang-gu Public Health Center in Seongnam, Korea. To determine the etiological agent and mode of transmission, a retrospective cohort study was done using structured questionnaires and stool samples from patients who had current gastrointestinal symptoms and three food handlers were tested. A total of 67 (31.0%) students and teachers developed acute gastroenteritis. No food items were associated with an increased risk of the illness. Norovirus was detected in 3 stool specimens collected from 6 patients who had severe diarrhea using semi-nested RT-PCR. All the specimens contained the genogroup I strains of the norovirus. Norovirus was also detected in the groundwater samples from the waterpark. In the nucleotide sequencing analysis, all the genogroup I noroviruses from the patients and groundwater samples were identified as the norovirus genotype I-4 strain. They were indistinguishable by DNA sequencing with a 97% homology. We conclude the outbreak of acute gastroenteritis caused by the norovirus was closely related to the contaminated groundwater.
Keywords: Norovirus, Gastroenteritis, Disease Outbreaks, Contaminated Groundwater, DNA Sequencing
INTRODUCTION
Noroviruses, which are single-stranded, non-enveloped RNA viruses, are grouped within the Caliciviridae family, and are currently divided into five genogroups. Of the five genogroups, group I, II and V usually infect humans (1). Patients who are infected by noroviruses usually show gastrointestinal manifestations including diarrhea, vomiting, abdominal pain, and low grade fever, and almost all of the infected cases resolve spontaneously (2). After the reverse transcription-PCR method for detecting noroviruses was established in the 1990s, noroviruses have been found to be one of the most common causes of food and waterborne gastroenteritis outbreaks in the world (3). However, there are few reports of norovirus-related epidemics with actual confirmed sources of the infection, especially ones using molecular methods to confirm the source (4, 5).
The decrease in waterborne outbreaks in Korea has been attributed to the improvement of treated water from munincipal water-treatment plant. However, with the exception of urban areas, groundwater may be the only drinking water available in many rural communities. Groundwater can be contaminated by a significant source of potential infection through improperly constructed and maintained water wells. Outbreaks of acute gastroenteritis caused by contaminated groundwater have been reported (4, 6). Therefore, groundwater contaminated by various pathogens is a potential source of waterborne outbreaks.
On January 14, 2008, 180 elementary and middle school students who had been admitted to an English Institute and 36 teachers visited a waterpark in Gyeonggi-do in the Republic of Korea. On January 16, 2008, several students from the group showed acute gastrointestinal symptoms including abdominal pain, diarrhea and vomiting, and visited a medical clinic in Bundang-gu, Gyeonggi-do, Korea. On January 17, 2008, the doctor of the clinic informed the Bundang-gu Public Health Center of a possible gastroenteritis outbreak. On January 18, 2008, a formal epidemiological study was initiated by an investigative team that consisted of staff from the Bundang-gu Public Health Center, Epidemic Intelligence Service officers from the provincial government of Gyeonggi-do and the Gyeonggi-do Institute of Health and Environment.
MATERIALS AND METHODS
Epidemiologic investigation
A retrospective cohort study was conducted among 180 students and 36 teachers from the English Institute on January 18 and 19, 2008. They were informed of the risk of acute gastroenteritis and were interviewed using a brief, standardized questionnaire. Data collected included age, sex, clinical information and pertinent details of food consumption at the waterpark. A case was defined as illness for any student or teacher who visited the waterpark and showed gastrointestinal manifestation such as abdominal pain, vomiting and diarrhea (two or more loose stool within a 24-hr period) on or after January 14, 2008. Three food handlers who worked at a restaurant in the waterpark were also interviewed about any current gastrointestinal manifestations, cooking procedures, and water sources. We also investigated the environment at a restaurant in the waterpark including cleavers, cutting boards, and dishcloths. Information about the management of the water supply and sewerage system was sought from the management of the waterpark. All the facilities in the waterpark were inspected carefully.
Microbiological investigation of fecal samples
Fecal samples were collected on January 18, 2008 from six patients who had current gastrointestinal symptoms without antibiotic treatment. We additionally collected fecal samples from three food handlers. Fecal specimens were referred to Gyeonggi-do Institute of Health and Environment. Stool samples were tested for the presence of pathogenic bacteria including Salmonella, Shigella, Campylobacter and Vibrio vulnificus. Viral pathogens including adenovirus, rotavirus, astrovirus and norovirus were also tested with ELISA and semi-nested RT-PCR. Food leftovers were not available for microbiological investigation at the time of the initial investigation.
Water sample processing
On January 18, 2008, routine testing of the groundwater was done. The tests indicated positive results for general bacteria, total coliforms, and Escherichia coli. Following the notification of the results, water sampling and concentration procedures were additionally conducted. A total of 670 liters of groundwater samples were collected at the point of the reservoir. The collection of viruses from the water samples was performed according to the Information Collection Rule (ICR) method (USEPA1996) (7). Samples in the amount of 670 liters of groundwater were concentrated by filtration through a 1-MDS positively charged, 10-inch cartridge filter (CUNO Inc, Meriden, CT, USA). Briefly, the adsorbed viruses on the filter were eluted with 0.05 M glycine buffer, pH9.5, containing 1.5% beef extract. The elutions were immediately adjusted to pH 3.4-3.6 with 1M HCl and stirred for 30 min followed by centrifugation at 2,500 × g for 15 min. The resulting pellets were resuspended in 30 mL of 0.15 M sodium phosphate (pH 9.5) and incubated for 10 min, followed by centrifugation at 9,000 × g for 10 min. The supernatants were collected and then adjusted to pH 7.0 with 1 M NaOH. Finally, the concentrates were passed through a 0.2-µm-pore-size filter (Pall Gelman Laboratory, Ann Arbor, MI, USA). Viral RNA was extracted from 140 µL of the concentrated water samples using QIAamp microspin columns (viral RNA mini kit: QIAGEN, Valencia, CA, USA) according to the manufacturer's instruction and stored at -80℃ until analysis.
Viral RNA extraction in stool samples
Each one-gram clinical stool sample was added into 9 mL phosphate buffered saline (PBS) and mixed for 3 min. The samples were centrifuged at 3,000 rpm for 30 min at 4℃ to remove debris, and then the clarified supernatant was collected. According to the manufacturer's instructions, viral RNA was extracted from 140 µL of the samples by using QIAamp microspin columns (viral RNA mini kit: QIAGEN) and stored at -80℃ until analysis.
Norovirus detection by Semi-nested RT-PCR
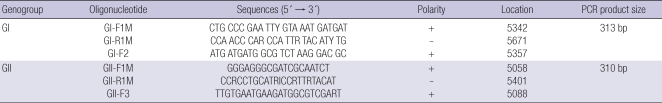
For stool samples and environmental water samples, semi-nested one step RT-PCR was performed as previously described (4, 8). Two primer sets (Table 1) for norovirus genogroup I and II which targeted the ORF1-ORF2 junction region of norovirus genome and amplified a 329 bp PCR product for norovirus GI and a 343 bp product for norovirus GII in the first one step RT-PCR respectively. First, the condition for one-step RT-PCR was consisted of RT for 40 min at 47℃ followed by Taq activation at 95℃ for 15 min; 35 PCR cycles of 94℃ for 30 sec, 54℃ for 30 sec, and 72℃ for 45 sec; and then a final soak at 72℃ for 7 min.
Table 1.
Semi-nested RT-PCR oligonucleotides used for Norovirus detection
Second, nested PCR was started at 94℃ for 3 min, followed by 25 cycles at 94℃ for 30 sec, 56℃ for 30 sec, and 72℃ for 45 sec, and then at 72℃ for 7 min. The final PCR products were 313 bp for GI and 310 bp for GII, which were further characterized by sequencing.
Sequencing and phylogenetic analysis
The PCR products from stool and groundwater samples were further characterized by sequencing and phylogenetic analysis. Sequence alignment analysis of the viruses present in the water and stool samples was carried out with the known proto-type Norovirus sequences (9) using MegAlign, version 5.03 (DNA-Star, Madison, WI, USA).
Nucleotide sequence accession number
The sequence of the norovirus GI-4 strain reported in this article has been deposited in GenBank under the accession number AF145896.
Ethics statement
This study was approved by the institutional review board of Hanyang University Hospital (August 5, 2010). Informed consent was exempted. All of the data of patients were processed anonymously.
Statistical analysis
Statistical analyses were performed using the chi-square test or Fisher's exact test for categorical variables. Statistical significance was determined using a P value < 0.05. All analyses were carried out using the software package SPSS for Windows ver. 12.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Epidemiological investigation
The waterpark is located in the suburbs of Gyeonggi-do, Korea. The waterpark provides both indoor and outdoor spas and swimming pools and subsidiary facilities including a restaurant. Disinfected groundwater is the only source of drinking water and boiled water is not usually supplied to visitors.
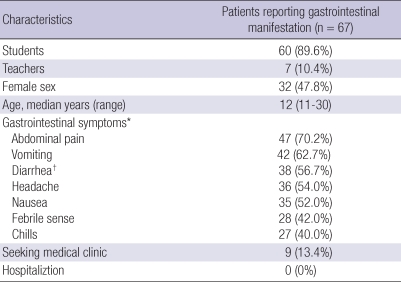
A total of 60 students and 7 teachers among 216 visitors from the Institute were identified as having gastrointestinal manifestations. Of the 67 patients, 35 (52.2%) were male and 32 (47.8%) were female. The median age of the patients was 12 yr (range, 11 to 30 yr). They had gastrointestinal manifestations such as abdominal pain (70.2%), vomiting (62.7%), and diarrhea (56.7%). No patients were hospitalized due to severe illnesses, and almost all patients spontaneously recovered from the illness with fluid or symptomatic management (Table 2). The median incubation time from exposure was 36 hours. (range 4 to 96 hr) However, no food items were associated with a statistically significant increased risk of the illness. After norovirus had been detected in groundwater samples, exposure to drinking water was investigated by telephone calls. However, water consumption was not associated with increased odds of becoming ill (data not shown).
Table 2.
Demographic and clinical characteristics of outbreak caused by norovirus at a waterpark
*The symptoms based on interviews; †Defined as two or more loose stools within a 24 hr.
Microbiological investigation
We collected 9 stool samples from 6 patients and 3 food handlers on January 18, 2008. None of the stool specimens from the patients or food handlers were positive for bacterial culture. Noroviruses were detected from 3 of the 6 stool samples taken from patients using norovirus RT-PCR, but no other viruses were detected in the fecal samples from the patients and the three food handlers. All specimens contained the norovirus genogroup I (GI) strain. However, all environmental samples from the restaurant were negative for bacterial and virological tests.
Prior to this outbreak, water analysis and periodical inspections of the groundwater and water supply system had not shown any abnormal results. However, routine testing of the groundwater collected on January 18 showed positive results for general bacteria, total coliforms and E. coli. Groundwater samples were collected to detect noroviruses at the point of the reservoir. Norovirus GI strain was detected in the groundwater samples.
Norovirus genotyping
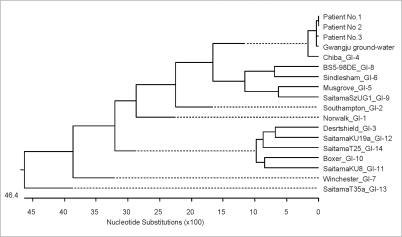
The amplified PCR products from stool and groundwater samples were sequenced, and a phylogenetic analysis was performed. The noroviruses detected in the fecal specimens from the outbreak cases were typed as GI-4, as was the norovirus isolated from the groundwater samples (Fig. 1). They were indistinguishable by DNA sequencing with a 97% homology.
Fig. 1.
Phylogenetic analysis of norovirus genogroup-I at a waterpark in Gyeonggi-do. Analysis was conducted using MegAlign, version 5.03 (DNAStar, Madison, Wis). The noroviruses detected in the fecal specimens from the patients of this outbreak were typed as GI-4 (most similar to GI-4 Chiba the GenBank accession number AF145896), as was the norovirus isolated from the groundwater.
DISCUSSION
This study reports an outbreak involving 67 patients who had acute gastrointestinal manifestations at a waterpark in Gyeonggi-do, Korea. Norovirus GI-4 strain was identified in both fecal specimens from patients and groundwater samples using sequence analysis. These finding provided clear evidence of the causative agent and route of transmission. In addition to this, epidemiological findings provided even more evidence. The median incubation time of this outbreak was 36 hr, which is similar to previously reported outbreaks caused by noroviruses (10, 11). Vomiting and abdominal pain were the predominant features, which corresponds with the characteristics of illnesses caused by noroviruses among school-age children and adolescents (12). Based on these results, we concluded that the norovirus-contaminated groundwater was the source of the outbreak.
It is interesting that norovirus GI strain was the causative agent. GII norovirus is more commonly detected in Korea. Of the 114 positive samples of norovirus among children from 2005 to 2006 in Korea, noroviruses belonged to the GI and GII genogroups, and these showed 10.5% and 89.5% respectively (13). Likewise, the GII norovirus has become the pandemic strain in the United States (14). However, in a previous large waterborne outbreak caused by noroviruses in Korea, GI-4 was detected in four fecal samples although GI-4 was not identified in groundwater samples at that time (4). In addition, some cases infected with GI norovirus have been reported during winter in Korea (13). Furthermore, GI-4 was identified from raspberries and patients in a recent gastroenteritis outbreak (15). These results suggest that GI-4 norovirus can cause large outbreaks of gastroenteritis.
Groundwater is a common source of infection in waterborne outbreak. Groundwater can be easily contaminated by potential pathogens through poorly constructed or maintained water wells (16). Contamination of groundwater may cause outbreaks of gastroenteritis (4). Periodic monitoring with indicator bacteria such as total coliforms and E. coli has been performed to identify groundwater contamination. However, epidemiologic surveillance data on viral pathogens in groundwater are scarce in Korea. Recently, wastewater surveillance by molecular methods has been suggested as a useful tool for rapid monitoring and characterization of various groups of viral pathogens (17). Therefore, molecular techniques for groundwater surveillance could provide additional benefits for the quality control of groundwater and outbreak prevention.
In conclusion, we described an outbreak of gastroenteritis caused by GI-4 norovirus among the visitors of the waterpark, which was closely associated with the contaminated groundwater.
AUTHOR SUMMARY
An Outbreak of Gastroenteritis Caused by Norovirus-Contaminated Groundwater at a Waterpark in Korea
Seong-Joon Koh, Han Gil Cho, Bo Hyun Kim, and Bo Youl Choi
In the present study, we described an outbreak of acute gastroenteritis among students and teachers who had visited a waterpark in Gyeonggi-do in January 2008. An epidemiological study was conducted to determine the etiological agent and mode of transmission. A total 67 (31.0%) students and teachers developed acute gastroenteritis. No food items were associated with an increased risk of the illness. Norovirus was detected in 3 stool samples. Norovirus was also found in the groundwater samples of the waterpark. All the noroviruses from patients and groundwater samples were identified as the norovirus genotype I-4 strain. They were indistinguishable by nucleotide sequence analysis with a 97% homology. In conclusion, this outbreak caused by norovirus was closely associated with the contaminated groundwater.
References
- 1.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Said MA, Perl TM, Sears CL. Healthcare epidemiology: gastrointestinal flu: norovirus in health care and long-term care facilities. Clin Infect Dis. 2008;47:1202–1208. doi: 10.1086/592299. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Norovirus activity--United States, 2006-2007. MMWR Morb Mortal Wkly Rep. 2007;56:842–846. [PubMed] [Google Scholar]
- 4.Kim SH, Cheon DS, Kim JH, Lee DH, Jheong WH, Heo YJ, Chung HM, Jee Y, Lee JS. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J Clin Microbiol. 2005;43:4836–4839. doi: 10.1128/JCM.43.9.4836-4839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewitt J, Bell D, Simmons GC, Rivera-Aban M, Wolf S, Greening GE. Gastroenteritis outbreak caused by waterborne norovirus at a New Zealand ski resort. Appl Environ Microbiol. 2007;73:7853–7857. doi: 10.1128/AEM.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallay A, De Valk H, Cournot M, Ladeuil B, Hemery C, Castor C, Bon F, Megraud F, Le Cann P, Desenclos JC Outbreak Investigation Team. A large multi-pathogen waterborne community outbreak linked to faecal contamination of a groundwater system, France, 2000. Clin Microbiol Infect. 2006;12:561–570. doi: 10.1111/j.1469-0691.2006.01441.x. [DOI] [PubMed] [Google Scholar]
- 7.Fout G, Schaefer F, 3rd, Messer J, Dahling D, Stetler R. ICR microbial laboratory manual. Washington, DC: US Environmental Protection Agency; 1996. Publication no. EPA/600/R-95/178. [Google Scholar]
- 8.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 9.Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Takai R, Oka T, Takeda N, Katayama K. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J Clin Microbiol. 2004;42:2988–2995. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podewils LJ, Zanardi Blevins L, Hagenbuch M, Itani D, Burns A, Otto C, Blanton L, Adams S, Monroe SS, Beach MJ, Widdowson M. Outbreak of norovirus illness associated with a swimming pool. Epidemiol Infect. 2007;135:827–833. doi: 10.1017/S0950268806007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werber D, Lausević D, Mugosa B, Vratnica Z, Ivanović-Nikolić L, Zizić L, Alexandre-Bird A, Fiore L, Ruggeri FM, Di Bartolo I, Battistone A, Gassilloud B, Perelle S, Nitzan Kaluski D, Kivi M, Andraghetti R, Pollock KG. Massive outbreak of viral gastroenteritis associated with consumption of municipal drinking water in a European capital city. Epidemiol Infect. 2009;137:1713–1720. doi: 10.1017/S095026880999015X. [DOI] [PubMed] [Google Scholar]
- 12.Arias C, Sala MR, Dominguez A, Torner N, Ruiz L, Martinez A, Bartolomé R, de Simón M, Buesa J. Epidemiological and clinical features of norovirus gastroenteritis in outbreaks: a population-based study. Clin Microbiol Infect. 2010;16:39–44. doi: 10.1111/j.1469-0691.2009.02831.x. [DOI] [PubMed] [Google Scholar]
- 13.Yoon JS, Lee SG, Hong SK, Lee SA, Jheong WH, Oh SS, Oh MH, Ko GP, Lee CH, Paik SY. Molecular epidemiology of norovirus infections in children with acute gastroenteritis in South Korea in November 2005 through November 2006. J Clin Microbiol. 2008;46:1474–1477. doi: 10.1128/JCM.02282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maunula L, Roivainen M, Keranen M, Makela S, Soderberg K, Summa M, von Bonsdorff CH, Lappalainen M, Korhonen T, Kuusi M, Niskanen T. Detection of human norovirus from frozen raspberries in a cluster of gastroenteritis outbreaks. Euro Surveill. 2009;14:pii: 19435. [PubMed] [Google Scholar]
- 16.Goss MJ, Barry DA, Rudolph DL. Contamination in Ontario farmstead domestic wells and its association with agriculture: 1. Results from drinking water wells. J Contam Hydrol. 1998;32:267–293. [Google Scholar]
- 17.Aw TG, Gin KY. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J Appl Microbiol. 2010;109:716–730. doi: 10.1111/j.1365-2672.2010.04701.x. [DOI] [PubMed] [Google Scholar]