Abstract
Because of the time and cost associated with Sanger sequencing of complete human mtDNA genomes, practically all evolutionary studies have screened samples first to define haplogroups and then either selected a few samples from each haplogroup, or many samples from a particular haplogroup of interest, for complete mtDNA genome sequencing. Such biased sampling precludes many analyses of interest. Here, we used high-throughput sequencing platforms to generate, rapidly and inexpensively, 109 complete mtDNA genome sequences from random samples of individuals from three Filipino groups, including one Negrito group, the Mamanwa. We obtained on average ∼55-fold coverage per sequence, with <1% missing data per sequence. Various analyses attest to the accuracy of the sequences, including comparison to sequences of the first hypervariable segment of the control region generated by Sanger sequencing; patterns of nucleotide substitution and the distribution of polymorphic sites across the genome; and the observed haplogroups. Bayesian skyline plots of population size change through time indicate similar patterns for all three Filipino groups, but sharply contrast with such plots previously constructed from biased sampling of complete mtDNA genomes, as well as with an artificially constructed sample of sequences that mimics the biased sampling. Our results clearly demonstrate that the high-throughput sequencing platforms are the methodology of choice for generating complete mtDNA genome sequences.
The increasing availability of complete mtDNA genome sequences from humans has greatly refined the human mtDNA phylogenetic tree and provided new insights into the phylogeography of particular haplogroups (Barnabas et al. 2006; Torroni et al. 2006; Abu-Amero et al. 2007; Derenko et al. 2007; Gonder et al. 2007; Fagundes et al. 2008; Soares et al. 2008; Perego et al. 2009). Such studies typically try to make inferences about population history based on the age of haplogroups (estimated from the number of mutations that have accumulated among mtDNA lineages belonging to the haplogroup) and their geographic distribution. However, making demographic inferences about populations (such as population size changes, population divergence times, migration/admixture events, etc.) from phylogeographic studies is problematic because different phylogenies can arise under the same demographic history, and vice versa (Nielsen and Beaumont 2009). Some studies equate ages of haplogroups with ages of populations, even though a haplogroup that arose a long time ago may have been introduced into a population only recently. Moreover, the method commonly employed to estimate the age of mtDNA haplogroups, namely, the “ρ” statistic, has been shown to often give misleading results for simulated data (Cox 2008).
Methods do exist for making demographic inferences from molecular genetic data (Drummond et al. 2002; Hey and Nielsen 2004), but a key requirement of such methods is that the genetic data should be from a random sample of individuals from the population. However, because of the expense and time needed to sequence complete mtDNA genomes with Sanger sequencing technology, previous studies of complete mtDNA genome sequences have generally either first screened samples by sequencing hypervariable segments of the mtDNA control region and/or genotyping coding region single nucleotide polymorphisms (SNPs) to classify haplogroups and then selecting one or two samples from each haplogroup for complete mtDNA genome sequencing, or have sequenced many samples from one particular haplogroup of interest in order to investigate the phylogeography of that haplogroup. Such sampling is biased and thus not suitable for demographic inference with existing methods.
Recently, methods have been developed for high-throughput, low-cost sequencing of many complete mtDNA genomes, using a parallel tagged sequencing approach and high-throughput (HT) sequencing platforms (Meyer et al. 2007, 2008b). Here, we have applied this approach and obtained 109 complete mtDNA genome sequences from random samples of individuals from three ethnolinguistic groups from the Philippines. Various analyses attest to the accuracy of the sequences generated by the high-throughput approach. Moreover, there are striking differences between Bayesian skyline plots (BSPs) of population size change through time constructed for our random samples of mtDNA genome sequences and previous such analyses based on biased samples (Atkinson et al. 2008), and we show that biased sampling can produce similar differences. Our results illustrate the value of random sampling of complete mtDNA genome sequences that can be obtained with the HT platforms and demonstrate that large-scale samples of complete mtDNA genome sequences can be obtained rapidly and efficiently with the HT platforms.
Results
GS and GS/FLX sequencing
The samples for this study come from three Filipino groups and include 26 Surigaonons, 44 Manobo, and 39 Mamanwa (a Negrito group). All samples were initially prepared for sequencing on the 454 Life Sciences (Roche) GS and GS FLX (hereafter referred to as GS/FLX) platforms, and sequences were obtained for 92 samples. For these, 95% of the resulting reads were assigned to tags, and 83.7% of the untagged reads mapped to the rCRS, with an average length of 222.4 bp. Any nucleotide position in a sequence with less than twofold coverage was automatically called an N, to denote missing data; our goal was to limit the number of such positions to <1% of the sequence for each individual. However, even though 113,045 reads were obtained in total with the GS/FLX, some individuals still did not have sufficient coverage. Furthermore, we observed numerous inconsistencies in homopolymer regions. Homopolymer regions are known to pose a problem with the GS/FLX sequencing technology as the exact number of bases in a run of three or more identical bases cannot be determined because of inaccuracy in the light signal intensity (Green et al. 2008).
Genome Analyzer II sequencing
Because of the abovementioned issues with the GS/FLX data, we switched to the Illumina Genome Analyzer II platform (hereafter referred to as GAII), since it has a much higher throughput and does not have problems sequencing homopolymers. Two GS/FLX pools were converted into GAII libraries and sequenced with the GAII. One pool was sequenced on one lane of the GAII with 51 cycles, with sequences read in one direction only. A total of 4,486,376 reads were obtained, of which 67% of reads were correctly tagged and 70% of the untagged reads mapped to the rCRS. The other pool was also sequenced on one lane with 51 cycles, except sequences were read in both directions. A total of 7,579,656 reads were obtained, of which 54% of the reads were correctly tagged and 43% of sequences mapped to the rCRS. The percentage of GAII reads that were correctly tagged and mapped is thus lower than that for the GS/FLX platform, which is to be expected since quality filtering was used before untagging and mapping for the GS/FLX data as described previously (Green et al. 2008) but not for the GAII data.
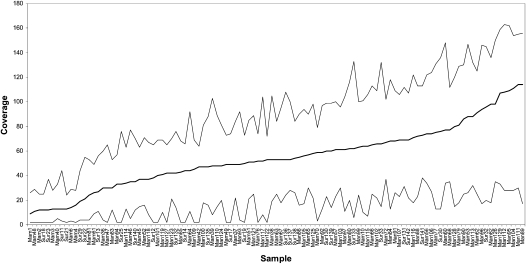
In order to call the base at a nucleotide position, we required a minimum of two reads for that position that would then have to agree. For positions with more than two reads, a scoring system (implemented in the MIA assembler) was used in which a consensus (majority) nucleotide was assigned to each position based on all of the reads covering that position from that sample. Reads that matched the consensus nucleotide received a score of +200, while reads that did not match the consensus nucleotide received a score of −600. These scores were then summed, and if the total score was less than 0, an N was assigned to that position. For the purposes of further analysis, sequences were required to have not more than 165 Ns (i.e., <1% missing data). In actuality, the number of Ns per sequence (Supplemental Table S2) ranged from 0 to 113, with an average of 9.9 Ns (i.e., 0.06% missing data) per sequence. The average coverage for the 109 sequences in the final data set was 54.6 (range 9–114), with average minimum coverage of 15.0 (range 2–38) and average maximum coverage of 91.4 (range 24–163), as shown in Figure 1 and Supplemental Table S2. Most positions were thus covered more than the minimum requirement of twofold; in total, there were 1424 positions in 23 sequences with only twofold coverage, or 0.4% of all positions in these 23 sequences (Supplemental Table S2).
Figure 1.
Average coverage (black line), and minimum and maximum coverage (gray lines) for the 109 mtDNA genome sequences in this study.
Verification of sequence authenticity
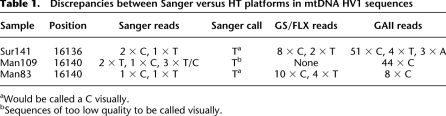
We compared the HV1 sequences obtained by Sanger sequencing and called with the SeqScape software to the HV1 sequences obtained from the HT platforms, and found three discrepancies (Table 1). For these discrepant positions, the coverage for the HT platforms was 22–68-fold, while the coverage from Sanger sequencing was twofold to sixfold. For two samples visual inspection of the Sanger sequencing trace files would lead to the same call as the HT platforms, while for the third sample the sequence quality was too poor at this position to call visually. Thus, the discrepancies for these three sequences can be attributed to problems with the SeqScape software. Overall, the HT platforms seem to give more reliable results, which is expected given the higher coverage that can be obtained with these platforms.
Table 1.
Discrepancies between Sanger versus HT platforms in mtDNA HV1 sequences
aWould be called a C visually.
bSequences of too low quality to be called visually.
Mutation analysis
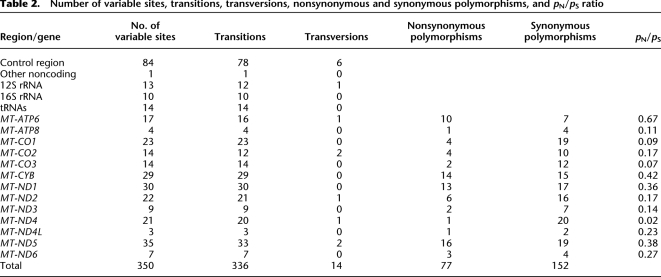
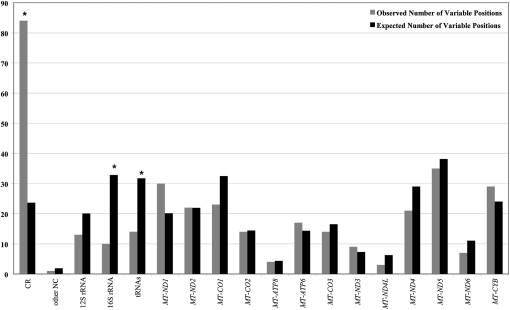
A total of 350 variable nucleotide positions were observed, all involving two nucleotides, of which 336 were transitions and 14 were transversions (Table 2). The ratio of transitions to transversions was higher for the coding region (32.3) than for the control region (13.0), but not significantly so (P = 0.09). There were significantly more variable positions in the control region and significantly fewer in the 16S rRNA gene and in the tRNA genes than expected, based on the length of each gene/region (Fig. 2). These are all familiar patterns in mtDNA genome sequences and further attest to the accuracy of the sequences (Pereira et al. 2009).
Table 2.
Number of variable sites, transitions, transversions, nonsynonymous and synonymous polymorphisms, and pN/pS ratio
Figure 2.
Observed and expected number of variable positions per mtDNA region/gene. (CR) Control region; (other NC) other noncoding; (asterisks) significant differences between the observed and expected numbers (P < 0.05, corrected for multiple comparisons).
Among the 13 protein-coding genes, there were 77 sites with nonsynonymous changes and 152 sites with synonymous changes (Table 2). The ratio of nonsynonymous polymorphisms per nonsynonymous site to synonymous polymorphisms per synonymous site (pN/pS) varied significantly among genes (P = 0.01). The most extreme values were observed for MT-ATP6, with 10 nonsynonymous and seven synonymous changes (pN/pS = 0.67), and MT-ND4, with one nonsynonymous and 20 synonymous changes (pN/pS = 0.02); when MT-ATP6 and MT-ND4 are removed from the analysis, the pN/pS ratios do not vary significantly among the remaining 11 genes (P = 0.23).
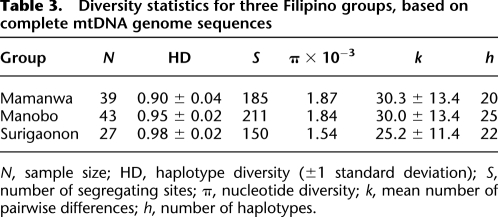
Some basic statistics describing variation in the sequences are presented in Table 3. The Mamanwa have a lower haplotype diversity than the other two Filipino groups, indicating a greater proportion of shared haplotypes. Otherwise, levels of genetic variation in the Mamanwa are comparable to the other groups. Pairwise FST values indicate a higher level of differentiation between the Mamanwa and Manobos (FST = 0.11) or Surigaonons (FST = 0.12) than between Manobos and Surigaonons (FST = 0.03).
Table 3.
Diversity statistics for three Filipino groups, based on complete mtDNA genome sequences
N, sample size; HD, haplotype diversity (±1 standard deviation); S, number of segregating sites; π, nucleotide diversity; k, mean number of pairwise differences; h, number of haplotypes.
Haplogroup affiliation and dating of novel haplogroups
A total of 22 different haplogroups were observed (Fig. 3; Table 4), 11 belonging to macrohaplogroup M (Fig. 4) and 11 belonging to macrohaplogroup N (Fig. 5). It should be noted that only rarely did a sequence in this study match the sequence for the assigned haplogroup exactly, so sequences were assigned to the closest haplogroup for which the sequence contained all mutations that define the haplogroup. Interestingly, many of the new lineages within a haplogroup show population specificity. For example, sequences from all three populations were assigned to haplogroup E1a1a1 (Fig. 4). However, only one Mamanwa sequence actually matched haplogroup E1a1a1 exactly. The other sequences had additional mutations that defined five new lineages within haplogroup E1a1a1: three were found exclusively in nine Manobo, one was found exclusively in four Mamanwa, and one (with two sub-branches) was found exclusively in three Surigaonon. Thus, even though haplogroup E1a1a1 is found in all three populations, the novel lineages within this haplogroup are completely population-specific. This same pattern (i.e., a haplogroup shared between populations, but lineages within the haplogroup exhibiting population specificity) is exhibited by several haplogroups.
Figure 3.
Map of sampling locations, and mtDNA haplogroup frequencies, for the three Filipino groups in this study.
Table 4.
Haplogroup frequencies in the three Filipino groups
Figure 4.
Nearest haplogroup affiliation of the mtDNA genome sequences obtained in this study that belong to macrohaplogroup M. The colors of the ID labels indicate population affiliation; (blue) Mamanwa; (red) Manobo; (green) Surigaonon.
Figure 5.
Nearest haplogroup affiliation of the mtDNA genome sequences obtained in this study that belong to macrohaplogroup N. The colors of the ID labels indicate population affiliation; (blue) Mamanwa; (red) Manobo; (green) Surigaonon.
Most of the haplogroups observed in this study have already been reported in the Philippines (Tabbada et al. 2010) and/or elsewhere in Southeast Asia (Trejaut et al. 2005; Pierson et al. 2006; Soares et al. 2008), further supporting the accuracy of the sequences. However, we found two novel haplogroups, designated provisionally here as M* and N*. Sequences assigned to haplogroup M* (Fig. 4) share some mutations with haplogroup M4, but do not have the basal mutation (12007) defining that haplogroup; therefore, the sequences were not assigned to M4. These M* sequences were observed in two Mamanwa and one Surigaonon. Haplogroup N* branches directly from the base of macrohaplogroup N (Fig. 5) and was observed in one Manobo and, strikingly, 14 Mamanwa. This novel N* haplogroup thus accounts for 36% of Mamanwa mtDNA sequences.
In order to estimate the divergence time of the novel M* and N* haplogroups, we carried out a phylogenetic analysis in BEAST. The topology for macrohaplogroup N (Fig. 5) involves a trifurcation leading to haplogroups N*, Y2a, and R, and different runs in BEAST gave different branching orders for these three branches. However, the Kishino-Hasegawa and Shimodaira-Hasegawa tests in PAUP* (Kishino and Hasegawa 1989; Shimodaira and Hasegawa 2001) indicate that there are no significant differences among all possible topologies for these three branches. The resulting dates for the divergence of N* are from 55,000 to 60,000 yr ago, with a 95% HPD (the lower and upper bound of the 95% highest posterior density interval) range of ∼44,000–72,500 yr ago. The age of the divergence of the M* haplogroup was similarly estimated from the age of macrohaplogroup M to be 47,862 yr ago (95% HPD = 39,907–59,236 yr). The analysis with a normally distributed prior for the mutation rate and the root of the tree resulted in an estimated mutation rate of 1.637 × 10−8 (95% HPD = 9.9 × 10−9–2.41 × 10−8), similar to previous estimates (Atkinson et al. 2008). In this analysis, the root of the tree was estimated to be 150,277 yr old (95% HPD = 90,716–217,514), in accordance with previous estimates (Endicott et al. 2009).
Comparison of HV1 versus coding sequence variation
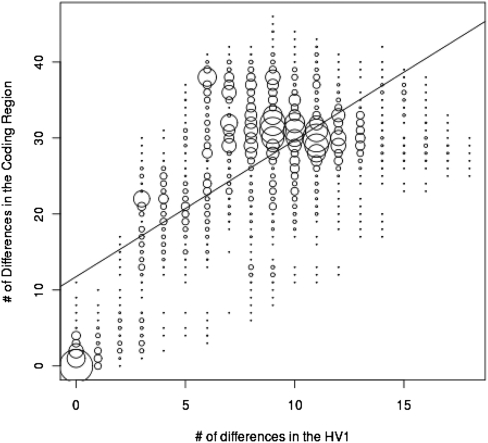
A strategy that is frequently used to select samples for complete mtDNA genome sequencing is to first sequence HV1, and then select individuals with different HV1 sequences for complete mtDNA genome sequencing (Derenko et al. 2007; Friedlaender et al. 2007). The underlying assumption is that individuals with identical HV1 sequences will also have identical, or nearly identical, coding region sequences. To investigate this assumption, we plotted the number of differences in HV1 sequences versus the number of differences in the coding sequences between each pair of individuals (Fig. 6). Of the pairwise comparisons with no differences in the HV1 sequences, 62.5% of these had one or more differences in the coding region, up to a maximum of 11 differences (Fig. 6). Thus, there can be appreciable coding region variation among individuals with identical HV1 sequences.
Figure 6.
Plot of the number of differences in the HV1 sequences versus the number of differences in the coding region sequences for each pair of individuals. The best-fit line is indicated.
Bayesian skyline plots
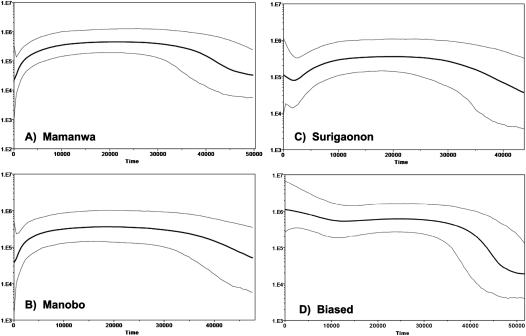
In order to estimate population size change through time, we carried out a Bayesian analysis (Drummond et al. 2005). The results of this analysis are depicted as a plot of population size change throughout time, termed a Bayesian skyline plot (BSP). The BSPs for the Mamanwas, Manobos, and Surigaonons are generally similar (Fig. 7A–C), and indicate population growth from 50 thousand yr ago (kya) until ∼30–35 kya, followed by population stasis until ∼6–8 kya, at which point population size decreases. The Surigaonons differ from the other groups in showing another signal of population growth, beginning ∼2–3 kya. Assuming a generation time of 25 yr, the current estimates of effective population size would be about 500 for the Mamanwa and Manobo, and 4000 for the Surigaonon.
Figure 7.
Bayesian skyline plots. The y-axis for each plot is the product of the effective population size and the generation time. (A) Mamanwa; (B) Manobo; (C) Surigaonon; (D) biased sample consisting of 28 sequences, each from a different haplogroup or lineage within a haplogroup.
The BPSs for these three Filipino groups differ markedly from those for other human populations that were also based on complete mtDNA genome sequences, and which tend to show strong signals of population growth throughout the past 50,000 yr or so (Atkinson et al. 2008). A possible reason for this discrepancy is that previous studies of complete mtDNA genome sequences suffer from biased sampling, as described above. To investigate if such biased sampling could influence the BSP analysis, we mimicked this sort of sampling by selecting 28 sequences, each from a different haplogroup (or lineage within a haplogroup) from our data and carrying out the BSP analysis. The resulting BSP (Fig. 7D) differs dramatically from the BSPs for the individual Filipino populations (Fig. 7A–C): there is not only a much stronger signal of initial population growth extending from 50 kya to 35 kya, but another signal of growth beginning around 10 kya and no subsequent signal of population decrease. Moreover, the current estimated effective population size for the biased sample is about 40,000, which is 10–100 times that of the corresponding estimates for the individual populations. Thus, biased sampling can give spurious signals of population growth and incorrect estimates of effective population size in the BSP analysis.
Discussion
We have used HT sequencing platforms to generate complete mtDNA genome sequences rapidly and efficiently. Although we began the study with the GS FLX platform, we switched to the Illumina GA platform as it provides higher coverage and fewer problems with sequences in homopolymer regions. Ongoing improvements in HT sequencing technologies will undoubtedly increase throughput and sequence accuracy and lower costs. A potential concern with HT platforms is that the error rate per base pair is much higher than with traditional Sanger sequencing (Bentley et al. 2008; Johnson and Slatkin 2008). However, because of the much higher coverage per position obtained with the HT platforms (an average of ∼55-fold in this study), the accuracy of the resulting sequences is expected to be higher than that for sequences obtained via Sanger sequencing. This was borne out in our study; a comparison of the HV1 sequences obtained via Sanger sequencing versus the HT platforms revealed five discrepancies in 109 individuals (thus, the concordance rate = 99.99%), all of which could be attributed to problems with the base-calling software for the Sanger sequencing.
With the average of ∼55-fold coverage obtained in this study, no sequence had more than 1% missing data, and on average each sequence had only 0.06% missing data. This level of missing data does not influence any of the analyses carried out here, such as haplogroup assignment, diversity statistics, BEAST and BSP analyses, and the like. However, it is possible that for other applications, such as in forensic casework or disease studies, even less missing data would be desirable. This can readily be achieved by increasing the amount of coverage; for example, sequencing just one sample on one lane of the Illumina GA platform can produce up to 16,000-fold coverage of the mtDNA genome (He et al. 2010). However, because of heteroplasmy in mtDNA genomes (Li et al. 2010), some Ns will always be present.
There are two potential limitations of the approach used here to generate complete mtDNA genome sequences. The first is the use of long-range PCR products as the sequencing template, which requires high-quality DNA. However, capture-based methods can be used to generate sequencing libraries enriched for mtDNA sequences (He et al. 2010), even from low-quality DNA. The second potential limitation is the higher error rate per base pair, as discussed above. Although increasing coverage will in general result in more accurate sequences, there are other steps that should be taken to ensure accurate sequences, especially when coverage happens to be low; these include requiring reads from both strands, as discussed in more detail elsewhere (Li et al. 2010).
A further advantage of the HT platforms is that they enable unbiased, population-based sequencing of complete mtDNA genomes, as compared to the biased sampling that characterizes practically all previous studies of complete mtDNA genome sequences. In sum, these advantages clearly establish HT platforms as the methodology of choice for generating complete mtDNA genome sequences.
Patterns of mtDNA variation
Overall, the patterns of mtDNA variation revealed in this study are similar to those observed in previous studies of complete mtDNA genome sequences (Ingman and Gyllensten 2001; Kivisild et al. 2006; Pereira et al. 2009). In particular, we observed a significant excess of variable positions in the control region (Fig. 2), which can be attributed to weaker functional constraints, as it is the major noncoding region of the mtDNA genome. There was also a significant deficiency of variable positions in the 16S rRNA and tRNA genes (Fig. 4), consistent with the view that the mitochondrial rRNA and tRNA genes are subject to strong functional constraints and hence exhibit reduced variation. In addition, the transition:transversion ratio was quite high (Table 2); observing these familiar patterns in the HT-generated sequences further enhances confidence in the accuracy of these sequences.
There were a total of 77 nonsynonymous polymorphisms among the 13 protein-coding genes (Table 2). Of particular interest is the significant excess of nonsynonymous polymorphisms (relative to the other mtDNA protein-coding genes) in the MT-ATP6 gene (Table 2). Previously, an excess of nonsynonymous polymorphisms in the MT-ATP6 gene was observed in Siberian populations, and was hypothesized to reflect positive selection for cold adaptation (Mishmar et al. 2003). Subsequent analyses cast doubt on this interpretation, suggesting instead that flaws in the statistical analyses (Kivisild et al. 2006) and/or relaxation of functional constraints (Ingman and Gyllensten 2007) were better explanations for the observed variation in MT-ATP6. While more sophisticated analyses would be required to investigate any putative signal of selection, our observation of an excess of nonsynonymous polymorphisms in MT-ATP6 in Filipino populations further argues against the cold adaptation hypothesis and in favor of relaxation of functional constraints (Ingman and Gyllensten 2007).
Demographic inferences and biased versus unbiased sampling
Because of the cost and time associated with the traditional Sanger sequencing approach, practically all previous studies of complete human mtDNA genome sequences have been forced to rely on a biased sampling approach. That is, mtDNA variation is first assayed in samples of interest by some other method (typically, obtaining HV1 sequences), and then a limited set of samples (typically, either one from each haplogroup, or several samples from one haplogroup of particular interest) is selected for complete mtDNA genome sequencing. The limitation of this approach is shown by the fact that individuals with identical HV1 sequences can harbor appreciable coding sequence variation (Fig. 6). Thus, a significant advantage of the HT platforms is that they permit unbiased, population-based sampling of complete mtDNA genome sequences.
The importance of such unbiased sampling is amply demonstrated in the BSP analyses of population size changes through time (Fig. 7). Previously, BSP analyses of complete mtDNA genome sequences, obtained via biased sampling, indicated a general pattern of overall population growth that varied in intensity across geographic regions (Atkinson et al. 2008). The BSPs for each of the three individual Filipino populations differ dramatically from this overall pattern, and instead indicate an initial phase of moderate population growth, followed by a long period of constant population size, and then a decline in population size starting 6–8 kya (Fig. 7A–C). In contrast, the BSP for an artificially constructed biased sample (Fig. 7D) differs from those for the individual populations and strongly resembles previously published BPSs (Atkinson et al. 2008). Moreover, the final estimate of effective population size is 10–100 times larger for the biased than for the unbiased samples. Thus, biased sampling can have a dramatic impact on this type of demographic analysis, and the conclusions of previous BSP analyses (Atkinson et al. 2008) should be viewed with caution.
History of Filipino populations
The complete mtDNA genome sequences provide new insights into the genetic history and relationships of these three Filipino groups. Of particular interest are the Mamanwa, as they are a Negrito group. Mainly because of their physical characteristics (including short stature, frizzy hair, and dark skin pigmentation), it is generally thought that Filipino Negrito groups are descended from a separate, early migration from Africa to Asia (Omoto 1984; Bellwood 1997; Hill et al. 2006; Thangaraj et al. 2006). However, there is very little in the way of available genetic data to address this issue. In fact, a recent study of 50,000 autosomal SNP data in Asian populations concluded that Filipino Negrito and non-Negrito groups are descended from the same single primary wave of colonization to East Asia (Abdulla et al. 2009).
The complete mtDNA genome sequences reveal that the Mamanwa possess a novel haplogroup, designated here as N*, that branches from the base of the macrohaplogroup N phylogeny. This haplogroup is at high frequency (36%) in the Mamanwa but absent or nearly so in the other two Filipino groups (Fig. 3; Table 4). The estimated divergence time of this haplogroup is ∼55,000–60,000 yr ago, implying that the ancestors of the Mamanwa may have become isolated from the ancestors of the other Filipino groups at about this time. These results may seem at variance with the 50,000 SNP data, which does not indicate a separate history of Negrito and non-Negrito Filipino groups (Abdulla et al. 2009). However, a possible scenario that reconciles the mtDNA genome sequences with the 50,000 autosomal SNP data involves would-be early isolation of the ancestors of Negrito groups from non-Negrito groups, followed by more recent gene flow from non-Negrito groups into Negrito groups, for which there is evidence in both the mtDNA data (as shown by the ∼60% frequency of mtDNA haplogroups in the Mamanwa that are characteristic of other southeast Asian groups) and the 50,000 SNP data (Abdulla et al. 2009), as well as in Y-chromosome data (Delfin et al. 2010). We are currently obtaining additional data and exploring other analyses to investigate this further. But in any event, the results of this study amply demonstrate the utility and validity of HT platforms for rapid and efficient sequencing of complete human mtDNA genomes, in particular, for providing the random samples of mtDNA genome sequences needed for demographic analyses.
Methods
DNA samples
Saliva samples were collected with informed consent, and with the permission and assistance of the Philippines National Commission on Indigenous People, from three groups from northern Mindanao in the Philippines (Fig. 3). Samples were obtained from 39 Mamanwas (a Negrito group) from three villages (Tabasinga, Mabuhay, and Urbistondo); 44 Manobos (a non-Negrito group) from two villages (Talacogon and Sabang Gibong) along the Agusan del Sur River; and 26 Surigaonons (an urban group) from Sitio and Surigao. Two milliliters of saliva was collected from each individual and stored in 2 mL of lysis buffer; DNA was extracted as described previously (Quinque et al. 2006).
Sequencing HV1
The hypervariable region 1 (HV1) of the mtDNA control region was amplified with primers L15926 and H10029 as described elsewhere (Pakendorf et al. 2003), and amplicons were purified using a Millipore Manu03050 Filter plate. Cycle sequencing was performed with the nested primers L16001 (Cordaux et al. 2003) and H16401 (Vigilant et al. 1989) and sequenced in both directions with the BigDye Terminator Kit v3.1 (Applied Biosystems) on an ABI 3700 sequencer. Samples with 16189C, resulting in the “C-stretch,” were sequenced twice in both directions, to ensure at least twofold coverage of each position. Sequences were assembled with SeqScape v2.1.1 (Applied Biosystems) and compared to the Revised Cambridge Reference Sequence (Andrews et al. 1999).
Sequencing complete mtDNA genomes
The entire mtDNA genome was amplified in two overlapping products of ∼8338 and 8647 bp, using primer pairs L644/H8982 and L8789/H877 (Supplemental Table S1). Long-range PCR was carried out using the Expand Long Range dNTP pack (Roche) and 3 ng of template DNA in a 50-μL volume, using the protocol provided by the manufacturer. The annealing temperature was 68.5°C for product 1 and 66°C for product 2. PCR products were purified using SPRI beads (Agencourt) using the manufacturer's instructions. The two PCR products for each individual were mixed in equimolar ratios and nebulized using nebulizers and reagents from the 454 Life Sciences (Roche) GS or GS FLX Library Preparation kit following the manufacturer's instructions. MinElute spin columns (QIAGEN) were used to purify the nebulized DNA, which was then eluted in 20 μL of elution buffer. About 400 ng of DNA was used for tagging nebulized PCR products with an individual-specific tag sequence, as described previously (Meyer et al. 2008b). The GS and GS FLX libraries were prepared according to the standard manufacturer's protocol, with two modifications that enable higher library yields. The first modification decreases the need to perform titration runs of libraries (Meyer et al. 2008a), and the second allows more DNA to be retrieved at the last step of the protocol (Maricic and Paabo 2009).
All samples were initially prepared for the GS or GS FLX platform in three pools, consisting of the tagged, nebulized PCR products. Two pools were subsequently converted into libraries suitable for sequencing on the Illumina Genome Analyzer II platform, as described elsewhere (Krause et al. 2010). These libraries were each sequenced on one lane on the Illumina Genome Analyzer II, one with single reads and one with paired end reads; the sequences of the primers used for sequencing are provided in Supplemental Table S1.
mtDNA sequence assembly
All reads were sorted according to tags, and reads that did not contain a correct tag were removed. Complete mtDNA genome sequences were assembled with MIA, an in-house assembler described previously (Briggs et al. 2009), using the rCRS as a reference to which all reads were mapped. A multiple alignment of the consensus sequences obtained with MIA was performed with mafft v6.708b (Katoh et al. 2009). The mtDNA genome sequences have been deposited in GenBank (accession numbers GU733718–GU733826).
Haplogroup assignment
Sequences were assigned to haplogroups according to Phylotree.org Build 6 (van Oven and Kayser 2009), using a custom Perl script. Sequences were assigned to the closest matching haplogroup for which all mutations that define the haplogroup were observed in that sequence. As in Phylotree, positions 309.1C(C), 16182C, 16183C, 16193.1C(C), and 16519 were not used for haplogroup assignment since these are subject to highly recurrent mutations.
Data analysis
Basic descriptive diversity statistics were calculated with dnaSP. MEGA 4 (Kumar et al. 2008) was used to calculate the mean number of nonsynonymous and synonymous sites in each protein-coding gene, using the standard mtDNA amino acid codon table, while mtGENESYN (Pereira et al. 2009) was used to calculate the number of nonsynonymous and synonymous mutations in the protein-coding genes, and the number of mutations in the rRNA genes, tRNA genes, and noncoding regions. The pN/pS ratio for each protein-coding gene was obtained by dividing the number of nonsynonymous mutations per nonsynonymous site by the number of synonymous mutations per synonymous site.
The comparison of differences in the hypervariable region 1 and the coding region between pairs of sequences was done with a custom Perl script, available upon request. The number of pairwise differences in the HVRI (positions 16,001–16,568) were plotted against the number of pairwise differences in the coding region (positions 577–16,000) with a regression line.
Bayesian skyline plots were produced from the coding region sequences (positions 577–16,023) using MCMC sampling in the program BEAST (version 5.1) (Drummond et al. 2002; Drummond and Rambaut 2007). The plots were obtained with a piecewise linear model, and ancestral gene trees were based on the Tamura-Nei substitution model (Tamura and Nei 1993) with invariant sites and a gamma-distributed rate (TrN + I + G). To select a model of nucleotide substitution, PAUP* portable version 4.0d105 (Swofford 2003) was used to generate likelihood scores of different competing models, and MODELTEST version 3.7 (Posada and Crandall 1998) was used to choose the best-fit model. A Bayes factor computed via importance sampling (Newton et al. 1994) indicated that the strict molecular clock could not be rejected and was therefore used for the analysis. We allowed 20 discrete changes in the population history using a coalescent-based tree prior with the linear model in which population size grows and declines between changing points. Each MCMC sample was based on a run of 40,000,000 generations sampled every 4000 steps with the first 4,000,000 generations regarded as burn-in. Three independent runs were made for each population, and a mutation rate of 1.691 × 10−8 (Atkinson et al. 2008) was used. Each run was analyzed using the program Tracer (http://tree.bio.ed.ac.uk/software/tracer/) for independence of parameter estimation and stability of MCMC chains (Drummond and Rambaut 2007).
Phylogenetic trees giving a date for the divergence time of the new N* and M* haplogroups were generated in BEAST for the coding region under the same conditions as described above, but with a constant population size model that was supported by a Bayes factor analysis. The tree was based on seven independent runs of 20,000,000 generations each, sampled every 2000 steps, with the first 2,000,000 generations regarded as burn-in. For this analysis one African Mbenzele sequence (GenBank accession no. AF346996) was used to root the tree. All log files were reviewed in Tracer (http://tree.bio.ed.ac.uk/software/tracer/), and all tree files from the independent runs were combined with a custom Python script and with TreeAnnotator v1.5.1, which is a part of the BEAST package (Drummond and Rambaut 2007). Since there are many reported mutation rates based on external and internal calibrations and different methodologies (Mishmar et al. 2003; Atkinson et al. 2008; Endicott and Ho 2008; Fagundes et al. 2008; Ho and Endicott 2008; Endicott et al. 2009; Soares et al. 2009), phylogenetic trees were also analyzed with a normally distributed prior range for the mutation rate with a mean of 1.5 × 10−8 and a standard deviation of 5.0 × 10−9, and a normally distributed prior range for the age of the root of the tree with a mean of 150,000 yr and a standard deviation of 50,000 yr, which incorporates all TMRCA dates of modern humans reported previously (Endicott et al. 2009).
Acknowledgments
We thank all of the individuals who donated their samples. For valuable assistance with the sample collection, we thank Irinetta C. Montinola, Wilfredo Sinco, and Fernando A. Almeda Jr., all from the Surigao Heritage Center; Girlie Patagan from the National Council of Indigenous People, Surigao; Elizabeth S. Larase and Juliet P. Erazo from the Office of Non Formal Education, Surigao; and the Rotary Club of Surigao. We thank Matthias Meyer, Johannes Krause, Tomislav Maricic, Tillmann Fünfstück, Hernán Burbano, Frederick Delfin, Irina Pugach, and Janet Kelso for technical assistance and valuable discussion. This research was funded by the Max Planck Society.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession nos. GU733718–GU733826. The raw reads have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under accession no. ERP000381.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.107615.110.
References
- Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, Chaurasia A, Chen CH, Chen JM, Chen YT, et al. 2009. Mapping human genetic diversity in Asia. Science 326: 1541–1545 [DOI] [PubMed] [Google Scholar]
- Abu-Amero KK, Gonzalez AM, Larruga JM, Bosley TM, Cabrera VM 2007. Eurasian and African mitochondrial DNA influences in the Saudi Arabian population. BMC Evol Biol 7: 32 doi: 10.1186/1471-2148-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147 doi: 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ 2008. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol 25: 468–474 [DOI] [PubMed] [Google Scholar]
- Barnabas S, Shouche Y, Suresh CG 2006. High-resolution mtDNA studies of the Indian population: Implications for palaeolithic settlement of the Indian subcontinent. Ann Hum Genet 70: 42–58 [DOI] [PubMed] [Google Scholar]
- Bellwood P 1997. Prehistory of the Indo-Malaysian archipelago. University of Hawaii Press, Honolulu, HI [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z, et al. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325: 318–321 [DOI] [PubMed] [Google Scholar]
- Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin SM, Stoneking M 2003. Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet 11: 253–264 [DOI] [PubMed] [Google Scholar]
- Cox MP 2008. Accuracy of molecular dating with the Rho statistic: Deviations from coalescent expectations under a range of demographic models. Hum Biol 80: 335–357 [DOI] [PubMed] [Google Scholar]
- Delfin F, Salvador JM, Calacal GC, Perdigon HB, Tabbada KA, Villamor LP, Halos SC, Gunnarsdottir E, Myles S, Hughes DA, et al. 2010. The Y-chromosome landscape of the Philippines: Extensive heterogeneity and varying genetic affinities of Negrito and non-Negrito groups. Eur J Hum Genet. doi: 10.1038/ejhg.2010.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, et al. 2007. Phylogeographic analysis of mitochondrial DNA in Northern Asian populations. Am J Hum Genet 81: 1025–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 doi: 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161: 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192 [DOI] [PubMed] [Google Scholar]
- Endicott P, Ho SY 2008. A Bayesian evaluation of human mitochondrial substitution rates. Am J Hum Genet 82: 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott P, Ho SYW, Metspalu M, Stringer C 2009. Evaluating the mitochondrial timescale of human evolution. Trends Ecol Evol 24: 515–521 [DOI] [PubMed] [Google Scholar]
- Fagundes NJ, Kanitz R, Eckert R, Valls AC, Bogo MR, Salzano FM, Smith DG, Silva WA Jr, Zago MA, Ribeiro-Dos-Santos AK, et al. 2008. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am J Hum Genet 82: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlaender JS, Friedlaender FR, Hodgson JA, Stoltz M, Koki G, Horvat G, Zhadanov S, Schurr TG, Merriwether DA 2007. Melanesian mtDNA complexity. PLoS ONE 2: e248 doi: 10.1371/journal.pone.0000248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA 2007. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol 24: 757–768 [DOI] [PubMed] [Google Scholar]
- Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. 2008. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134: 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YP, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Kinzler KW, Vogelstein B, Papadopoulos N 2010. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Soares P, Mormina M, Macaulay V, Meehan W, Blackburn J, Clarke D, Raja JM, Ismail P, Bulbeck D, et al. 2006. Phylogeography and ethnogenesis of aboriginal Southeast Asians. Mol Biol Evol 23: 2480–2491 [DOI] [PubMed] [Google Scholar]
- Ho SY, Endicott P 2008. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am J Hum Genet 83: 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U 2001. Analysis of the complete human mtDNA genome: Methodology and inferences for human evolution. J Hered 92: 454–461 [DOI] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U 2007. Rate variation between mitochondrial domains and adaptive evolution in humans. Hum Mol Genet 16: 2281–2287 [DOI] [PubMed] [Google Scholar]
- Johnson PLF, Slatkin M 2008. Accounting for bias from sequencing error in population genetic estimates. Mol Biol Evol 25: 199–206 [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537: 39–64 [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M 1989. Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA-sequence data, and the branching order in Hominoidea. J Mol Evol 29: 170–179 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al. 2006. The role of selection in the evolution of human mitochondrial genomes. Genetics 172: 373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, Paabo S 2010. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr Biol 20: 231–236 [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K 2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M 2010. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet 87: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Paabo S 2009. Optimization of 454 sequencing library preparation from small amounts of DNA permits sequence determination of both DNA strands. Biotechniques 46: 51–57 [DOI] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M 2007. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res 35: e97 doi: 10.1093/nar/gkm566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, Krause J, Weihmann A, Paabo S, Hofreiter M 2008a. From micrograms to picograms: Quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res 36: e5 doi: 10.1093/nar/gkm1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Hofreiter M 2008b. Parallel tagged sequencing on the 454 platform. Nat Protoc 3: 267–278 [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci 100: 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton MA, Raftery AE, Davison AC, Bacha M, Celeux G, Carlin BP, Clifford P, Lu C, Sherman M, Tanner MA, et al. 1994. Approximate Bayesian-inference with the weighted likelihood bootstrap. J R Stat Soc Ser B Methodol 56: 3–48 [Google Scholar]
- Nielsen R, Beaumont MA 2009. Statistical inferences in phylogeography. Mol Ecol 18: 1034–1047 [DOI] [PubMed] [Google Scholar]
- Omoto K 1984. The Negritos: Genetic origins and microevolution. Acta Anthropogenet 8: 137–147 [PubMed] [Google Scholar]
- Pakendorf B, Wiebe V, Tarskaia LA, Spitsyn VA, Soodyall H, Rodewald A, Stoneking M 2003. Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am J Phys Anthropol 120: 211–224 [DOI] [PubMed] [Google Scholar]
- Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, Olivieri A, Kashani BH, Ritchie KH, Scozzari R, Kong QP, et al. 2009. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol 19: 1–8 [DOI] [PubMed] [Google Scholar]
- Pereira L, Freitas F, Fernandes V, Pereira JB, Costa MD, Costa S, Maximo V, Macaulay V, Rocha R, Samuels DC 2009. The diversity present in 5140 human mitochondrial genomes. Am J Hum Genet 84: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson MJ, Martinez-Arias R, Holland BR, Gemmell NJ, Hurles ME, Penny D 2006. Deciphering past human population movements in Oceania: Provably optimal trees of 127 mtDNA genomes. Mol Biol Evol 23: 1966–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA 1998. MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I 2006. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem 353: 272–277 [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M 2001. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247 [DOI] [PubMed] [Google Scholar]
- Soares P, Trejaut JA, Loo JH, Hill C, Mormina M, Lee CL, Chen YM, Hudjashov G, Forster P, Macaulay V, et al. 2008. Climate change and postglacial human dispersals in Southeast Asia. Mol Biol Evol 25: 1209–1218 [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Rohl A, Salas A, Oppenheimer S, Macaulay V, Richards MB 2009. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet 84: 740–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- Tabbada KA, Trejaut J, Loo JH, Chen YM, Lin M, Mirazon-Lahr M, Kivisild T, De Ungria MCA 2010. Philippine mitochondrial DNA diversity: A populated viaduct between Taiwan and Indonesia? Mol Biol Evol 27: 21–31 [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526 [DOI] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Reddy AG, Singh VK, Singh L 2006. Unique origin of Andaman Islanders: Insight from autosomal loci. J Hum Genet 51: 800–804 [DOI] [PubMed] [Google Scholar]
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ 2006. Harvesting the fruit of the human mtDNA tree. Trends Genet 22: 339–345 [DOI] [PubMed] [Google Scholar]
- Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Lee ZY, Lin M 2005. Traces of archaic mitochondrial lineages persist in Austronesian-speaking Formosan populations. PLoS Biol 3: e247 doi: 10.1371/journal.pbio.0030247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M 2009. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30: E386–E394 [DOI] [PubMed] [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC 1989. Mitochondrial-DNA sequences in single hairs from a Southern African population. Proc Natl Acad Sci 86: 9350–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]