Abstract
Viruses may play an important role in the evolution of human microbial communities. Clustered regularly interspaced short palindromic repeats (CRISPRs) provide bacteria and archaea with adaptive immunity to previously encountered viruses. Little is known about CRISPR composition in members of human microbial communities, the relative rate of CRISPR locus change, or how CRISPR loci differ between the microbiota of different individuals. We collected saliva from four periodontally healthy human subjects over an 11- to 17-mo time period and analyzed CRISPR sequences with corresponding streptococcal repeats in order to improve our understanding of the predominant features of oral streptococcal adaptive immune repertoires. We analyzed a total of 6859 CRISPR bearing reads and 427,917 bacterial 16S rRNA gene sequences. We found a core (ranging from 7% to 22%) of shared CRISPR spacers that remained stable over time within each subject, but nearly a third of CRISPR spacers varied between time points. We document high spacer diversity within each subject, suggesting constant addition of new CRISPR spacers. No greater than 2% of CRISPR spacers were shared between subjects, suggesting that each individual was exposed to different virus populations. We detect changes in CRISPR spacer sequence diversity over time that may be attributable to locus diversification or to changes in streptococcal population structure, yet the composition of the populations within subjects remained relatively stable. The individual-specific and traceable character of CRISPR spacer complements could potentially open the way for expansion of the domain of personalized medicine to the oral microbiome, where lineages may be tracked as a function of health and other factors.
Human microbial communities represent a vast and underexplored subset of our biosphere, and only recently have the depth and diversity of these communities begun to be elucidated (Eckburg et al. 2005; Ley et al. 2005, 2006; Gill et al. 2006; Gao et al. 2007; Huse et al. 2008; Costello et al. 2009). The primary tool for characterizing these communities is community-wide sequencing, as it provides a culture-independent method for examining aspects of community genomic content and variability. Both cellular life and viruses are subject to this type of analysis, with bacteria and archaea thus far as the primary focus, through exploration of microbial diversity based on analysis of 16S rRNA gene sequences (Angly et al. 2006; Huber et al. 2007; Pride and Schoenfeld 2008; Antonopoulos et al. 2009; Willner et al. 2009). There now have been numerous community-wide sequencing studies of microbes in the human oral cavity, vagina, gastrointestinal tract, and skin (Lepp et al. 2004; Eckburg et al. 2005; Jenkinson and Lamont 2005; Gao et al. 2007; Palmer et al. 2007; Costello et al. 2009; Bik et al. 2010; Ravel et al. 2010).
Bacteriophages (viruses of bacteria, henceforth referred to as viruses) represent the most abundant life forms on the planet, and are believed to inhabit every niche in which potential hosts exist. In contrast to the well-studied habitats in the environment (Breitbart et al. 2002; Rohwer and Thurber 2009; Rodriguez-Brito et al. 2010) and to the analysis of virus–host interactions in vitro (Roucourt et al. 2009), few studies have examined the diversity and potential impact of human bacteriophages (Breitbart et al. 2008; Willner et al. 2009). Because of their alternate lifestyles, in which they may be lytic and decimate their bacterial hosts or lysogenic and potentially confer new functional potential and selective advantage to their host (Canchaya et al. 2003), these viruses have a substantial capacity to alter human microbial communities (Weinbauer and Rassoulzadegan 2004; Kunin et al. 2008; Rodriguez-Valera et al. 2009). A few studies of virus communities in the human respiratory tract and feces have provided early insight into these microbial ecosystems (Breitbart et al. 2008; Nakamura et al. 2009; Willner et al. 2009). The viral communities found in hosts with cystic fibrosis differ greatly from those of healthy hosts, suggesting that these viruses might contribute to host pathology (Willner et al. 2009). However, as yet, there are limited data to suggest that viruses may be major sources of bacterial population control in the human oral cavity (Hitch et al. 2004).
Clustered regularly interspaced short palindromic repeats (CRISPRs) represent a component of a CRISPR/Cas system that confers adaptive immunity against viruses and plasmids (Barrangou et al. 2007; Marraffini and Sontheimer 2008). The majority of bacteria and archaea possess at least one of these systems. As new viruses are encountered, a small portion of their genome is sampled and placed between often palindromic repeats at the end of the locus (Barrangou et al. 2007; Mojica et al. 2009). As the host is re-exposed to these same viruses, it resists predation through a mechanism of nucleic acid interference (Brouns et al. 2008; Hale et al. 2009). Analyses of CRISPR loci from bacteria and archaea in various environments have demonstrated substantial locus diversification, reflecting dynamic interactions among hosts and their viruses (Andersson and Banfield 2008; Deveau et al. 2008; Horvath et al. 2008; Tyson and Banfield 2008; Heidelberg et al. 2009; Semenova et al. 2009; van der Ploeg 2009). Others have used these loci to gather information about the history of virus exposures, and to type bacterial strains (Pourcel et al. 2005; Vergnaud et al. 2007; Zhang et al. 2009). However, CRISPRs have not been examined to any significant degree within human ecosystems. Given the nature of CRISPR systems, we believe that these genomic loci may serve as records of host–virus interactions in human environments and may reveal previously unrecognized mechanisms that underlie bacterial community evolution.
To improve our understanding of the dynamics between bacteria and viruses in the human oral cavity, we examined CRISPRs directly from members of the salivary microbiota of different human subjects over time. We exploited known CRISPR repeat sequences from laboratory streptococcal strains in order to determine (1) the presence and diversity of streptococcal CRISPRs in the human oral cavity, (2) whether the predominant features of individual CRISPR repertoires change over time, and (3) what these CRISPR sequences reveal about the nature of the viruses encountered by their streptococcal hosts.
Results
Recovery of streptococcal CRISPR repeat and spacer sequences from the human salivary microbiome
We recruited four subjects with good periodontal health and obtained saliva samples from February 2008 to July 2009 (Supplemental Table 1). No specific intervention took place during this 17-mo study period, and all subjects were sampled on Day 1, Day 30, Day 60, and Month 11. For subjects #1 and #2, additional samples had been collected in an identical manner 6 mo and 3 mo prior to the sampling on Day 1; these time points are denoted “Month −6” and “Month −3” for the sake of study consistency. We chose a conserved repeat sequence found in several streptococcal species, including Streptococcus mutans, Streptococcus thermophilus, Streptococcus pyogenes, and Streptococcus agalactiae, as the basis for a broad-range PCR, as the Streptococcus genus had been identified as a predominant community member in the oral cavity of many human subjects (Lazarevic et al. 2009; Nasidze et al. 2009a,b; Bik et al. 2010). For each subject and from each specimen, CRISPR spacers and repeats were amplified from salivary DNA using the conserved streptococcal repeat sequence–specific primers (Supplemental Fig. 1), and 384 clones were sequenced (Table 1; Supplemental Table 2).
Table 1.
Human subject CRISPR spacers
aContigs created using 100% identity over a minimum of 100 nucleotides.
bIncludes only unique spacers across all time points.
At least six different repeat sequences were identified from each subject (Supplemental Fig. 2), each with similar 3′-nucleotide sequences (Supplemental Table 3). Two such motifs were dominant and conserved among all four subjects and over the study time period (Supplemental Fig. 2). During the last sampling time point in subject #1, there was a more even distribution in the representation of repeat sequences.
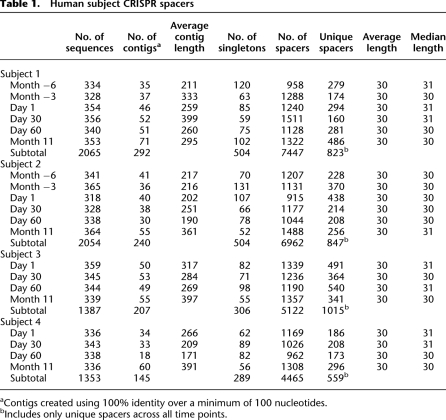
The richness of CRISPR spacer sequences varied between subjects and over time (Fig. 1; Table 1). For example, in subject #1, 7447 spacers were sampled over the 17-mo period, 823 of which were unique. As few as 174 (at Month −3), and as many as 486 (at Month 11) different spacers were identified at any given time point. Similar numbers of spacers were identified in subject #2 as for subject #1, more were identified in subject #3 (5122 spacers total, 1040 unique), and fewer in subject #4 (4465 spacers total, 571 unique). There was no clear, conserved, overall trend in spacer richness over time in any subject. Rarefaction analysis (Fig. 1) and Good's coverage (Supplemental Fig. 3) indicated variable degrees of completeness of sampling and coverage of the spacer population over time in each of the subjects. Variability in richness and coverage was most pronounced in subject #1. Good's coverage sampling estimate was >70 for all time points.
Figure 1.
Rarefaction analysis of CRISPR spacers in the saliva of human subjects at each sampled time point. Rarefaction curves were created using 10,000 random iterations based on spacer richness. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4. (Open circle) Month −6; (open square) Month −3; (closed triangle) Day 1; (closed square) Day 30; (closed circle) Day 60; (open triangle) Month 11.
For each time point in each subject, contigs of CRISPR locus sequences were created using stringent criteria (Table 1). The presence of singletons (sequences without significant homology with any other sequence recovered from that sample) reflects a high diversity of CRISPR loci present, as well as sampling effort.
Shared CRISPR spacers and beta diversity
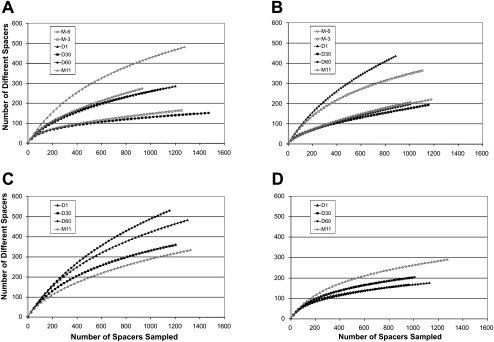
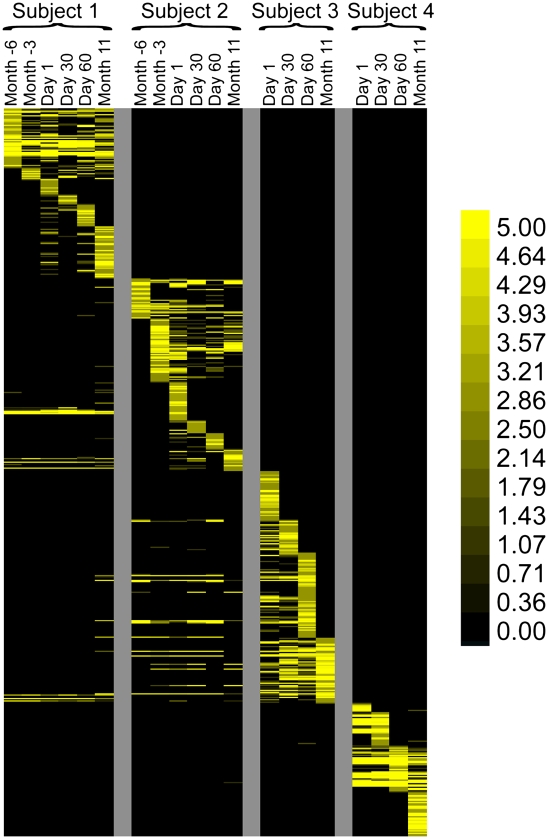
We analyzed CRISPR spacers in order to assess overlap in CRISPR spacer complements among the subjects and how CRISPR spacer diversity varied over time. As demonstrated in a heatmap, ∼7%–22% of the spacers were detected at all time points within each subject (Fig. 2). This core of shared spacers across time points within a subject suggests either selective pressure for conservation of certain spacers or the presence of relatively stable CRISPR loci within the streptococcal community. However, ∼15%–75% of spacers were detected only at single time points in each subject (Figs. 2, 3). Interestingly, the proportion of spacers that differed between Day 60 and Month 11 in each subject did not significantly exceed the proportion of spacers newly identified after shorter time intervals, with the exception of the samples from subject #4 (Fig. 3D).
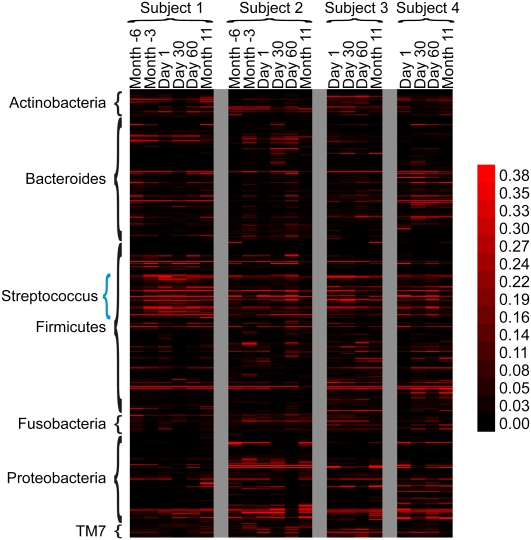
Figure 2.
Heatmap of unique spacers present in each subject at all time points. Each row represents a unique spacer sequence. The intensity scale bar is located to the right.
Figure 3.
Shared CRISPR spacers in the saliva of individual human subjects at each time point. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4. (Gray) The proportion of spacers shared with other time points within each subject; (black) spacers that are unique to each time point within each subject.
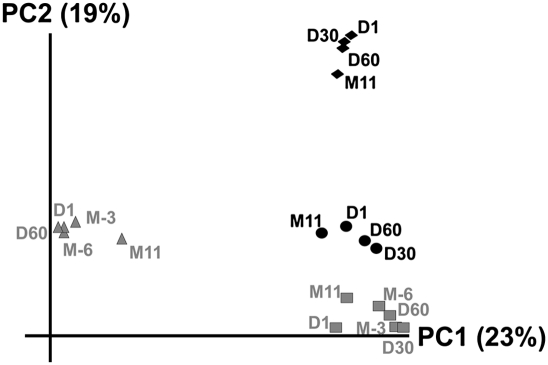
Fewer spacers were shared among subjects than were shared within a subject over time (Supplemental Fig. 4A). We examined differences in spacer composition between subjects using a measurement of beta diversity (Supplemental Fig. 4B). Interestingly, the highest levels of beta diversity were seen between subjects #1 and #2, who share a household. When beta diversity was analyzed using principal coordinates analysis, spacer composition was found to be highly specific to each subject (Fig. 4).
Figure 4.
Principal coordinates analysis of CRISPR spacer composition from human saliva based on beta diversity. (Gray triangles) Subject #1; (gray squares) subject #2; (black circles) subject #3; (black diamonds) subject #4.
Relationships between bacterial community composition and CRISPR spacer population
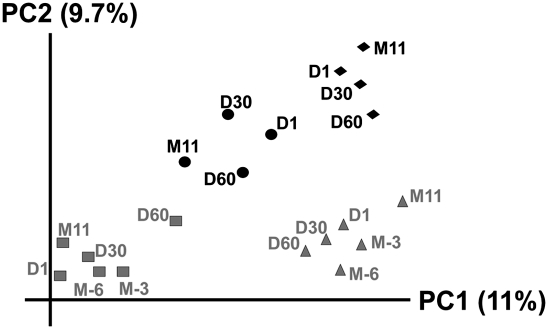
We analyzed the composition of the bacterial community from the saliva of our human subjects, in order to assess CRISPR spacer diversity in the broader context of bacterial diversity within samples and subjects. We sequenced the V1-V3 hypervariable regions of the 16S rRNA gene after PCR amplification from samples (Supplemental Table 4), and in general, found a typical picture of bacterial diversity in saliva (Fig. 5). Each subject had a distinct pattern of operational taxonomic unit (OTU) membership in the saliva that differed between time points. As with principal coordinates analysis of CRISPR spacer diversity, the patterns of variation in the bacterial communities reflected a strong contribution from host (Fig. 6).
Figure 5.
Heatmap of bacterial OTU abundance based on analysis of 16S rRNA gene sequences from each of the samples and subjects. OTUs were determined by phylogenetic analysis of 16S rRNA sequence alignments, using a 97% cutoff value. Each row represents a unique OTU sequence based on the cutoff criterion. The intensity scale bar is located to the right. Taxonomic labels are shown along the y-axis, with OTUs from the genus Streptococcus indicated with a blue brace.
Figure 6.
Principal coordinates analysis of OTU composition based on 16S rRNA gene sequence data from the saliva of each subject. Input to the analysis was beta unweighted unifrac distances. (Gray triangles) Subject #1; (gray squares) subject #2; (black circles) subject #3; (black diamonds) subject #4.
We also examined the relative abundance of streptococci in each saliva sample. As a surrogate measure of this community feature, the relative abundance of 16S rRNA reads assigned to the genus Streptococcus as a proportion of the total number of reads was found to be highly variable during the study period in each subject, but especially in subject #1 (Supplemental Fig. 5). In this subject, Streptococcus was the dominant genus present in the oral cavity; however, the relative abundance of Streptococcus varied from ∼11% to 40% of the bacterial population. Streptococcus was predicted to represent no greater than 20% of the population in other subjects (Supplemental Fig. 5).
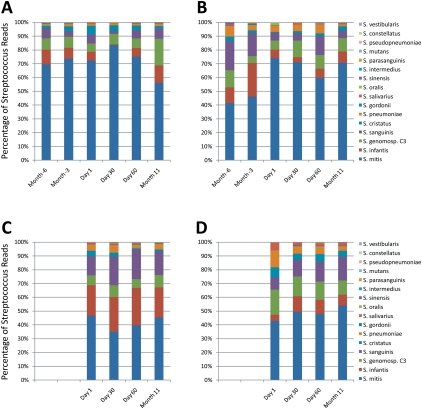
There was even greater variation over time in the relative abundance of certain streptococcal species than there was for the genus overall within subjects (Fig. 7). Subject #1 was dominated by S. mitis, which was relatively stable in its relative abundance; however, the less abundant species, Streptococcus genomo sp. C3, Streptococcus infantis, Streptococcus oralis, and Streptococcus sanguinis were much more variable in their relative abundances. Similar findings were noted for subjects #2 and #4. In contrast, subject #3 had a different streptococcal population structure, with no single dominant streptococcal species and limited variability over time (Fig. 7C). The fact that subject #3 had no dominant Streptococcus species might explain its high CRISPR spacer richness compared to other subjects.
Figure 7.
Streptococcus species in human subject saliva at each time point. Each species is displayed as a percentage of the total number of OTUs identified taxonomically to the Streptococcus genus. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4.
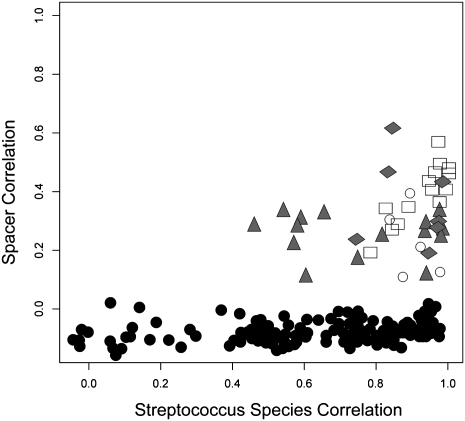
To investigate the relationship between CRISPR spacer diversity and diversity within the streptococcal community, we examined whether the relationship in spacer content between samples predicted the nature of the relationship in streptococcal species content. For intrasubject comparisons, there was a consistently significant correlation between spacer content and streptococcal community composition for all subjects (Fig. 8); however, for intersubject comparisons, there were no significant correlations in spacer composition, and correlations of variable strength in streptococcal community composition (Fig. 8, black circles). Using Fisher z-transformed correlations to assess the predictive power of spacer content on streptococcal community composition, significant P-values were found only for subjects #1 (P < 0.012) and #4 (P < 0.018), while no significance was found for subjects #2 and #3. These data suggest that variation in CRISPR spacer content may predict streptococcal community composition in some subjects.
Figure 8.
Pearson correlation scores for comparisons between CRISPR spacer content and streptococcal species composition. Intrasubject comparisons: (open squares) subject #1, (gray triangles) subject #2, (open circles) subject #3, (gray diamonds) subject #4; (black circles) intersubject comparisons.
CRISPR spacer homologs
Because CRISPR spacers are believed to contain short sequences from virus genomes, we subjected the spacers from each subject to BLASTN analysis to identify homologs and the possible origins of these spacers. For subject #1, only one of the 823 spacers had homologs to known sequences, while ∼7% (61 of 847) for subject #2, 15% (152 of 1015) for subject #3, and 3% (19 of 559) for subject #4 had known homologs. Most homologs were sequences of streptococcal viruses (Table 2) or sequences of proviruses found in streptococcal genomes. Numerous homologs to Streptococcus phage CP-1 (Podoviridae isolated from S. pneumoniae), Streptococcus phage PH-10 (Siphoviridae isolated from S. oralis), and Streptococcus phage SM-1 (Siphoviridae isolated from S. mitis) were found. Interestingly, the spacer homologs were distributed across the genomes of these viruses, suggesting that these particular virus types were prevalent in the community (Supplemental Fig. 6). A few homologs were non-streptococcal genome sequences, which may reflect the presence of viruses with broad host range, or reflect shared features of viruses that parasitize disparate genera. The lack of identifiable spacer homologs in subject #1 as compared to the other subjects suggested that subject #1 had minimal exposure to these known virus types.
Table 2.
CRISPR spacer homologs
aRepresent Streptococcus viruses.
Human salivary streptococcal isolates
To confirm the presence of Streptococcus species and streptococcal CRISPR sequences in samples from the four subjects, we cultured Streptococcus isolates from samples collected from each subject at Month 11 using Streptococcus-specific media. Each isolate was then subjected to streptococcal CRISPR repeat-based PCR amplification; four to six isolates from each subject were chosen for further analysis. Phylogenetic analysis of the isolates, based on amplification of their 16S rRNA genes, identified most of the isolates as Streptococcus salivarius, and others as S. mitis, S. sanguinis, and Streptococcus anginosus (Supplemental Fig. 7; Supplemental Table 5). Interestingly, isolates of S. mitis and S. sanguinis were found in this study to harbor repeat sequences that differ from those of previously sequenced strains of these species. We analyzed CRISPR spacers from the isolates (Supplemental Table 5) to determine if they had been previously sampled in our direct analysis of the salivary CRISPR population. For subjects #1 and #2, each of the isolates analyzed harbored spacers that had previously been sampled (Supplemental Fig. 8A,B), while many of the strain spacers from subjects #3 and #4 were newly identified (Supplemental Fig. 8C,D).
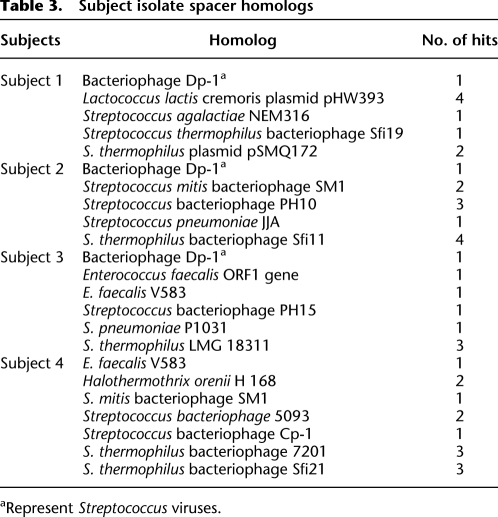
Some (9%) of the spacers derived from the Streptococcus isolates have homologs present in the NCBI non-redundant database (Supplemental Fig. 9). Most of these homologs were to streptococcal viruses or plasmids; however, there were numerous spacers homologous to a plasmid from Lactococcus lactis (Table 3). The presence of spacers in Streptococcus isolates with homology with Enterococcus and Halothermothrix genomes suggests that they are derived from viruses with relatively broad host range. The Halothermothrix spacers were found in both the Streptococcus isolates and the CRISPR spacer population, providing further evidence that Streptococcus was predominantly sampled in the direct PCR-based spacer analysis rather than other genera that had received a Streptococcus-like repeat via lateral gene transfer of the locus.
Table 3.
Subject isolate spacer homologs
aRepresent Streptococcus viruses.
Real-time CRISPR locus evolution
One possible source of newly identified spacers in the CRISPR spacer population is the new viruses or virus variants that are encountered by the host bacterial community. To test this hypothesis, we analyzed the structure of a single CRISPR locus over the course of the study period. We chose a CRISPR locus from subject #2 because a similar locus was identified in a Streptococcus isolate from this subject (2Mut38), and because all of its sampled spacers also were sampled in our analysis of the salivary CRISPR population (Supplemental Fig. 8B). Because in our analysis of the salivary CRISPR population, at each time point we detected numerous CRISPR sequences that began and ended with the same terminal spacer, we developed primers specific for these spacers to verify CRISPR locus structure. We reconstructed each locus at each time point using 100% nucleotide identity over a minimum overlap of 100 nt, and the resulting structure was independently verified by spacer-specific PCR followed by sequencing of the resulting amplicons. Over the 17-mo course of the study, three spacers were added to the locus, while one spacer was lost (Fig. 9). Interestingly, the locus was not detected on Day 60 by either method, but was detected once again at Month 11.
Figure 9.
Structure of a CRISPR locus from subject #2 at different time points. 2Mut38 represents an isolate of S. sanguinis from subject #2 recovered at Month 11.
Discussion
This analysis is unusual in its use of a community-wide sequencing approach and the targeting of a bacterial community over time to provide direct insight into interactions between human indigenous bacteria and their viruses. While the study of the relationships between human bacterial and viral communities remains in its infancy, our data suggest on-going interactions between oral streptococci and their respective viruses, with potential importance for the stability of the human microbiota. The limited degree of shared spacers between human subjects (Supplemental Fig. 4) suggests that either each subject was exposed to different virus populations, which would be supported by a recent finding that fecal viromes are highly subject-specific (Reyes et al. 2010), or that similar virus populations were sampled differently by the streptococcal populations in each of the subjects. The presence of unique CRISPR spacer complements in each subject with shared characteristics across time (Fig. 4) suggests that CRISPR spacer complements may be used to trace individual human subjects; however, further study with a larger group of subjects is needed to verify this potential.
We examined CRISPR loci by means of repeat-based amplification rather than by amplification of an entire locus from flanking sequences, in order to target a broad range of CRISPR loci distributed throughout the genomes of their host bacteria. This strategy allowed us to amplify many loci that could not be detected using primers based on flanking sequences in S. mutans UA159 (data not shown). The disadvantage of a repeat-based amplification strategy was that CRISPR loci had to be assembled from fragments. In fact, there were numerous instances of CRISPR loci with multiple, alternative spacer orders or duplicate spacers that suggested error-prone amplification, perhaps as a result of the repeat-based priming methodology. Spacer-based PCR-priming used for part of this study was not subject to these errors, and based on spacer priming, the definitive arrangement of spacers in CRISPR loci could be defined (Fig. 9). CRISPR locus diversification in this single locus (Fig. 9) over the course of the study suggests real-time virus encounter, genome assimilation, and locus evolution taking place in the salivary environment similar to that seen in acidophilic microbial biofilms (Tyson and Banfield 2008), the oral cavity of a rat (van der Ploeg 2009), and the ocean (Sorokin et al. 2010).
While we cannot exclude the possibility that our analysis of streptococcal CRISPRs included non-streptococcal loci, our data strongly suggest that the salivary CRISPR loci were largely Streptococcus-specific. Most of the identified homologs were sequences from known streptococcal virus isolates or proviruses within Streptococcus genomes. Homologs from non-streptococcal database sequences also matched sequences found in Streptococcus isolates cultivated from each subject. The isolates of S. salivarius, S. sanguinis, S. anginosus, and S. mitis characterized in this study contained repeat sequences that amplified with the streptococcal repeat primers, expanding the spectrum of streptococci known to harbor these repeats.
An examination of CRISPR spacer populations in a complex environment of the sort illustrated in this study can only be as complete as the sampling of each individual specimen at each time point. Good's coverage and rarefaction analysis demonstrated that there was reasonably deep sampling of our subjects (Fig. 1; Supplemental Fig. 3). In this context, we believe that the variable number of unique spacers in each population over time cannot be explained by sampling bias alone. This observation could indicate diversification of CRISPR loci over time or differential representation of streptococcal strains at each time point (due to virus predation or other factors). The possibility of changes in streptococcal composition over time is supported by the finding of heterogeneity in repeat sequence representation at certain time points (Supplemental Fig. 2).
We suggest the presence of two separate phenomena in the CRISPR population, both of which may have important implications for understanding bacteria–virus interactions in the human oral cavity. The first is the maintenance of a core of shared spacers over time. This could reflect selective pressure to maintain certain spacers from repeated exposure to the same virus types or inheritance of spacers along strain lineages. We observed numerous spacer homologs spread out over virus genomes (Supplemental Fig. 6), indicating that the host bacteria may have been repeatedly sampling these virus types. The second observation is the rapid change in spacer complements across the time periods sampled. A large proportion of novel spacers were not identified at subsequent time points (Fig. 2). Given that the species composition of the Streptococcus population in each subject was relatively stable (Fig. 7), it is less likely that these newly identified spacers were the result of new species entering the community; however, we cannot rule out Streptococcus strain variation over time, of a type that 16S rRNA gene sequence analysis might fail to resolve. It is well-known from other studies that CRISPR spacers vary at the strain level (Horvath et al. 2008; McShan et al. 2008; Salzberg et al. 2008; Heidelberg et al. 2009; Diez-Villasenor et al. 2010), which reinforces that some of the CRISPR spacer variation found in the present study may result from the presence of new and diverse streptococcal strains.
As we continue to explore the diversity and temporal dynamic within the microbial communities of human ecosystems, a wealth of bacteria–virus interactions are likely to be uncovered. Our analysis of salivary streptococcal CRISPR populations provides only a glimpse into the potential complexities of these interactions. The choice of a single streptococcal repeat sequence for our experimental approach in this study underscores this point, as there are many other known CRISPR repeat sequences in streptococci and other organisms that might provide a similar but distinct picture of diversity. Despite the limitations of our approach, there are numerous benefits, such as the ability to identify virus types to which the community has been exposed without isolating the individual viruses, and the ability to identify the portions of virus genomes targeted by host bacteria. With the ever-increasing depth of virus genome databases, CRISPR spacer community-wide sequencing constitutes a powerful tool for understanding host–virus dynamics in complex ecosystems.
Methods
Human subjects
All subjects were enrolled and donated saliva samples over a 17-mo period from February 2008 to July 2009. Subject recruitment and enrollment were approved by the Stanford University Administrative Panel on Human Subjects in Medical Research. All subjects completed a questionnaire demonstrating their willingness to participate in the study. Four subjects were enrolled under the criteria that no antibiotics were to be given either during the study or had been given for 3 mo prior to beginning the study, and that they had no preexisting medical conditions associated with significant immunosuppression. All subjects self-reported their health status. Each subject was subjected to a full baseline periodontal examination consisting of measurements of probing depths, clinical attachment loss, Gingival Index, Plaque Index, and gingival irritation (Loe 1967). Each subject was found to be overall periodontally healthy (overall mean clinical attachment loss of <1 mm) with a diagnosis of slight localized gingivitis, and were free of nonrestored carious lesions. A minimum of 3 mL of saliva was collected at each time point, and saliva was stored at −20°C until further analysis.
Amplification of CRISPR spacers
From each subject, genomic DNA was prepared from 180 μL of saliva using the QIAGEN QIAamp DNA MINI kit (QIAGEN). Primers SMRPF-1 (5′-GAAACAACACAGCTCTAAAAC-3′) and SMRPR-1 (5′-TGTTTCGAATGGTTCCAAAAC-3′) were designed based on their specificity for the CRISPR repeat sequences present in S. mutans UA159, S. thermophilus LMD-9, S. pyogenes MGAS 10270, and S. agalactiae A909, and were used to amplify CRISPRs from salivary DNA by PCR. Reaction conditions included 5 μL of 10× PCR buffer (Applied Biosystems), 3 μL of MgCl2 (25 mM), 1 μL of each of the forward and reverse primers (20 pmol each), 0.5 μL of AmpliTaq DNA polymerase (Applied Biosystems), 5 μL of salivary DNA template, and 34.5 μL of H2O. The following were used as PCR cycling parameters: 3 min initial denaturation at 95°C, followed by 30 cycles of denaturation (60 sec at 95°C), annealing (60 sec at 45°C), and extension (5 min at 72°C), followed by a final extension (10 min at 72°C). CRISPR amplicons were purified using the QIAGEN QIAquick PCR Purification kit (QIAGEN), and purified amplicon mixtures were cloned into the pCR4 vector using the Invitrogen TOPO TA Cloning Kit for Sequencing. For each sample, 384 clones were picked and subjected to Sanger sequencing using standard M13 primers.
Analysis of repeats and spacers
CRISPR sequences were analyzed using Sequencher 4.9 (Gene Codes Corporation). Primer sequences were removed, and only those sequences with a length of ≥100 nt and a Sequencher quality score >80% were chosen for further analysis. CRISPR repeats were identified based on an algorithm that searches for the first 5 nt of the CRISPR repeat sequence (GTTTT) followed by the last 5 nt (AAAAC) of the sequence, with allowances for a single nucleotide polymorphism in the repeat at any nucleotide position. The repeats were defined as any set of nucleotides ∼36 nt long that begins and ends with the aforementioned nucleotides. In addition, for all samples the sequences were manually examined to ensure no repeat motifs went undetected and that no errors occurred in the classification of repeat motifs. Spacers were defined as any sequence (length ≥ 20) flanked by repeat motifs. Only clone sequences containing at least two repeat motifs flanking a single spacer were retained; all others were removed from the analysis. Contigs were created for each subject at each time point using 100% identity over a minimum overlap of 100 nt to prevent the creation of quasi-CRISPR loci. Spacers were grouped according to three rules: (1) spacers that were identical; (2) spacers that were identical, with the exception of a single nucleotide polymorphism; and (3) spacers that differed in length, but were identical over the length of the shorter spacer. For each sample, a database of spacers and repeat motifs was generated and was used to create heatmaps using Java TreeView (Saldanha 2004) and to determine shared spacers and repeats. Heatmap input data were normalized by the total number of spacers for each time point, and then multiplied by 100 so that the heatmap color intensity was represented as percentages of the total number of spacers. Good's coverage was determined as the estimation of the number of singletons in the population (n), compared to the total number of sequences (N), using the equation [1 − (n/N)] × 100 (Good 1953). Rarefaction analysis was performed based on species richness estimates of 10,000 iterations using EcoSim (Lee et al. 2005). Beta diversity was determined using Sorensen's similarity, which also was used as input for principal coordinates analysis. Correlations in CRISPR spacer content and streptococcal species composition were performed using Pearson's correlation in the R Statistical Package (http://www.R-project.org). Regression analysis was performed on Fisher z-transformed correlations to determine significant P-values. Spacers from each subject were subjected to BLASTN analysis based on the NCBI non-redundant database. Hits were considered significant if they had bit scores of ≥50, which roughly correlates to 2-nt differences over the 30-nt average length of the spacers.
Analysis of bacterial 16S rRNA sequences
We amplified the V1-V2-V3 region of the bacterial 16S rRNA gene sequence from salivary DNA from each sample using primers that were optimized for pyrosequencing (Liu et al. 2007). The forward primer consisted of a 10:1:1 ratio of the following primers (8FM-B, 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGAGTTTGATCMTGGCTCAG-3′; 8FT-B, 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGAGTTTGATTCTGGCTCAG-3′; and 8Fbif-B, 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGGGTTCGATTCTGGCTCAG-3′). This primer incorporated the 454 Life Sciences (Roche) primer B sequence and a two-base linker sequence “CA,” and modifications of the broad range 16S rRNA primer 8F. The reverse primer (515R-A, 5′-CATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNGGTACCGCGGCKGCTGGCAC-3′) incorporated the 454 Life Sciences (Roche) primer A sequence, a unique 10-nt barcode for each subject sample (represented in the above sequence by N), the broad range bacterial 16S rRNA primer 515R, and a two-base linker sequence “CA.” PCRs were performed in 50-μL reaction volumes using the Roche FastStart HiFi polymerase kit (Roche Applied Science). Each reaction consisted of 39.8 μL of H2O, 5 μL of HiFi buffer with MgCl2, 1 μL of dNTPs, 1.2 μL of forward primer, 1 μL of reverse primer, 1 μL of HiFi polymerase, and 1 μL of salivary DNA template. The following were used as cycling parameters: 3 min of initial denaturation at 95°C, followed by 25 cycles of denaturation (30 sec at 95°C), annealing (45 sec at 51°C), and extension (5 min at 72°C), followed by a final extension (10 min at 72°C). Products were ∼550 bp and were gel-purified using a QIAGEN QIAquick Gel Extraction kit (QIAGEN), and further purified using Ampure bead purification (Beckman Coulter Genomics). Purified amplicons were quantified using PicoGreen (Invitrogen) and were pooled in equimolar ratios. Pyrosequencing was performed using primer A on a 454 Life Sciences (Roche) Genome Sequencer FLX Titanium instrument.
Sequences were processed in a manner similar to that previously described (Hamady et al. 2008). Sequences were removed from the analysis if they were <200 nt or >800 nt, had an uncorrectable barcode, contained any ambiguous characters, or contained more than 10 homopolymers. These sequences were deposited in the NCBI Sequence Read Archive database under accession number SRA024393.1. Sequences were assigned to their respective samples based on their 10-nt barcode sequence, and similar sequences were clustered into OTUs using a minimum identity of 97% using CD-Hit (Li and Godzik 2006). To limit overestimation of the microbial diversity present, pyrosequencing noise was reduced using Pyronoise (Quince et al. 2009). Representative sequences from each OTU were chosen and aligned using NAST (DeSantis et al. 2006b) based on the Greengenes database (DeSantis et al. 2006a). Phylogenetic trees were constructed using FastTree based on Kimura's two-parameter distances, and taxonomy was assigned to each OTU using the RDP classifier with a minimum support threshold of 60% (Wang et al. 2007; Price et al. 2009). Shared OTUs were compared between each subject at each time point to generate heatmaps using Java TreeView (Saldanha 2004). Heatmap input data were normalized by the total number of sequences for each time point and then multiplied by 100 so that the heatmap color intensity was represented as percentages of the total number of sequences. Principal coordinates analysis was performed based on beta diversity using weighted Unifrac distances. The presence of streptococcal species was determined by identifying sequences assigned to the genus Streptococcus from each subject at each time point, and analyzing each sequence with RDP Seqmatch (Cole et al. 2009). Each of the taxonomic assignments that were processed had a threshold value ≥0.85 at the species level; therefore, each sequence was assigned at the species level. Results of RDP Seqmatch were confirmed independently for representative OTUs by phylogenetic analysis using RDP Tree Builder (Cole et al. 2009).
Isolation of Streptococcus
On Month 11, fresh saliva was collected from each of the four subjects. Saliva was stored at room temperature for <2 h prior to culturing. Samples were diluted in sterile normal saline at 1:1000, 1:10,000, 1:100,000, and 1:1,000,000; and 100 μL of each was plated on Mitis-Salivarius agar (Remel Inc.). Plates were incubated overnight at 37°C in a 5% CO2 environment, and 100 colonies from each were picked and placed into 1 mL of Brain-Heart Infusion medium (BD Diagnostics). Each isolate was incubated overnight at 37°C with shaking, and 500 μL of each suspension was used for genomic DNA extraction using the Invitrogen PureLink 96-Well Genomic DNA Purification kit. The protocol was modified to include the use of lysozyme for lysis of Gram-positive organisms.
Characterization of Streptococcus isolates
Genomic DNA from each isolate was subjected to PCR amplification of CRISPR spacers using primers SMRPF-1 and SMRPR-2 as specified, and four to six isolates from each subject were chosen for further analysis. Amplicons were cloned into pCR4 (Invitrogen), and 24 clones from each were subjected to Sanger sequencing using standard M13 primers. The database of spacer sequences amplified directly from salivary DNA from each subject was compared with the database of spacer sequences from isolates to determine shared spacers.
The 16S rRNA gene sequence was amplified from each strain using broad-range bacterial primers 8F and 1391R (Lane et al. 1985; Edwards et al. 1989). Reaction conditions included 5μL of 10× PCR buffer (Applied Biosystems), 3 μL of MgCl2 (25 mM), 1 μL of of each the forward and reverse primers (20 pmol each), 0.5 μL of AmpliTaq DNA polymerase (Applied Biosystems), 1 μL of strain genomic DNA, and 38.5 μL of H2O. The following were used as cycling parameters: 3 min of initial denaturation at 95°C, followed by 25 cycles of denaturation (60 sec at 95°C), annealing (60 sec at 45°C), and extension (2 min at 72°C), followed by a final extension (10 min at 72°C). Amplicons were purified using the QIAGEN QIAquick Gel Extraction kit (QIAGEN) and subjected to Sanger DNA sequencing using primers 8F and 1391R. Species assignment was performed using RDP Seqmatch and RDP Tree Builder to determine phylogenetic relationships among closely related Streptococcus species (Cole et al. 2009).
Analysis of CRISPR locus structure
CRISPR locus structure was analyzed by examining assemblies created from both strain databases and salivary DNA databases from each sample. A single locus was chosen for further analysis, as it was present in both the isolate 2Mut38 and in the subject #2 spacer database. Primers (1190-1F, 5′-CGACGCTAGCCATGCCAG-3′; 1137-1F, 5′-GTCAAAAGATAAGTCCAG-3′; 1194-1F, 5′-TCAATCAAAGTGTAGTAG-3′; 1376-1R, 5′-TTCCTTAAAACTCATGGC-3′; and 978-1R, 5′-CGGGGTGTTTGTCAAAGG-3′) were developed that were specific for spacers in this CRISPR locus, and were used in various combinations to amplify the CRISPR locus from the genomic DNA of the isolates and the subject #2 salivary DNA. The following were used as cycling parameters: 3 min of initial denaturation at 95°C, followed by 25 cycles of denaturation (60 sec at 95°C), annealing (60 sec at various different annealing temperatures based on the primer pair used), and extension (1 min at 72°C), followed by a final extension (10 min at 72°C). The resulting amplicons were purified using the QIAGEN QIAquick PCR Purification kit (QIAGEN) and subjected to Sanger DNA sequencing. Resulting sequences were examined using Sequencher 4.9 (Gene Codes Corporation), and the resulting locus structure was displayed as it varied over time.
Acknowledgments
This work was supported by the Robert Wood Johnson Foundation, the UNCF-Merck Science Initiative, and the Burroughs Wellcome Fund to D.T.P.; and the National Institutes of Health Director's Pioneer Award DP1OD000964 to D.A.R. D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University. We thank Les Dethlefsen for his design of the 16S rRNA V1-V3 amplification scheme, and Elies Bik for helpful suggestions.
Author contributions: D.T.P., C.S., J.B., and D.A.R. conceived and designed experiments; D.T.P. and N.R. performed the experiments; D.T.P., C.S., J.S., J.B., and D.A.R. analyzed the data; P.L. and G.C.A. contributed reagents and performed examinations; and D.T.P. and D.A.R. wrote the manuscript.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA024393.1.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111732.110.
References
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 [DOI] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, et al. 2006. The marine viromes of four oceanic regions. PLoS Biol 4: e368 doi: 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77: 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA 2010. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F 2002. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci 99: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, et al. 2008. Viral diversity and dynamics in an infant gut. Res Microbiol 159: 367–373 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6: 417–424 [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL 2006a. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL 2006b. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villasenor C, Almendros C, Garcia-Martinez J, Mojica FJ 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156: 1351–1361 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17: 7843–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci 104: 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE 2006. Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IJ 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264 [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP 2009. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D 2009. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS ONE 4: e4169 doi: 10.1371/journal.pone.0004169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitch G, Pratten J, Taylor PW 2004. Isolation of bacteriophages from the oral cavity. Lett Appl Microbiol 39: 215–219 [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML 2007. Microbial population structures in the deep marine biosphere. Science 318: 97–100 [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4: e1000255 doi: 10.1371/journal.pgen.1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ 2005. Oral microbial communities in sickness and in health. Trends Microbiol 13: 589–595 [DOI] [PubMed] [Google Scholar]
- Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F, et al. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res 18: 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci 82: 6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, Francois P 2009. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods 79: 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SG, Kim CM, Hwang KS 2005. Development of a software tool for in silico simulation of Escherichia coli using a visual programming environment. J Biotechnol 119: 87–92 [DOI] [PubMed] [Google Scholar]
- Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA 2004. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci 101: 6176–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI 2006. Microbial ecology: Human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A 2006. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35: e120 doi: 10.1093/nar/gkm541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe H 1967. The Gingival Index, the Plaque Index and the Retention Index Systems. J Periodontol 38: S610–S616 [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, et al. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J Bacteriol 190: 7773–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, Goto N, Takahashi K, Yasunaga T, Ikuta K, et al. 2009. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS ONE 4: e4219 doi: 10.1371/journal.pone.0004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M 2009a. Global diversity in the human salivary microbiome. Genome Res 19: 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Li J, Li M, Tang K, Stoneking M 2009b. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal Biochem 391: 64–68 [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO 2007. Development of the human infant intestinal microbiota. PLoS Biol 5: e177 doi: 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151: 653–663 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP 2009. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Schoenfeld T 2008. Genome signature analysis of thermal virus metagenomes reveals Archaea and thermophilic signatures. BMC Genomics 9: 420 doi: 10.1186/1471-2164-9-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6: 639–641 [DOI] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. 2010. Microbes and Health Sackler Colloquium: Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci doi: 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI 2010. Viruses in the faecal microbiota of monozytotic twins and their mothers. Nature 466: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J 4: 739–751 [DOI] [PubMed] [Google Scholar]
- Rohwer F, Thurber RV 2009. Viruses manipulate the marine environment. Nature 459: 207–212 [DOI] [PubMed] [Google Scholar]
- Roucourt B, Lecoutere E, Chibeu A, Hertveldt K, Volckaert G, Lavigne R 2009. A procedure for systematic identification of bacteriophage–host interactions of P. aeruginosa phages. Virology 387: 50–58 [DOI] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9: 204 doi: 10.1186/1471-2164-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova II, Severinov K 2009. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Lett 296: 110–116 [DOI] [PubMed] [Google Scholar]
- Sorokin VA, Gelfand MS, Artamonova II 2010. Evolutionary dynamics of clustered irregularly interspaced short palindromic repeat systems in the ocean metagenome. Appl Environ Microbiol 76: 2136–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10: 200–207 [DOI] [PubMed] [Google Scholar]
- van der Ploeg JR 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155: 1966–1976 [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Li Y, Gorge O, Cui Y, Song Y, Zhou D, Grissa I, Dentovskaya SV, Platonov ME, Rakin A, et al. 2007. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv Exp Med Biol 603: 327–338 [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG, Rassoulzadegan F 2004. Are viruses driving microbial diversification and diversity? Environ Microbiol 6: 1–11 [DOI] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 4: e7370 doi: 10.1371/journal.pone.0007370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Andremont A, Ruimy R, Sola C 2009. Mycobacterium tuberculosis complex CRISPR genotyping: Improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J Med Microbiol 59: 285–294 [DOI] [PubMed] [Google Scholar]