Abstract
Despite its importance in cell biology and evolution, the centromere has remained the final frontier in genome assembly and annotation due to its complex repeat structure. However, isolation and characterization of the centromeric repeats from newly sequenced species are necessary for a complete understanding of genome evolution and function. In recent years, various genomes have been sequenced, but the characterization of the corresponding centromeric DNA has lagged behind. Here, we present a computational method (RepeatNet) to systematically identify higher-order repeat structures from unassembled whole-genome shotgun sequence and test whether these sequence elements correspond to functional centromeric sequences. We analyzed genome datasets from six species of mammals representing the diversity of the mammalian lineage, namely, horse, dog, elephant, armadillo, opossum, and platypus. We define candidate monomer satellite repeats and demonstrate centromeric localization for five of the six genomes. Our analysis revealed the greatest diversity of centromeric sequences in horse and dog in contrast to elephant and armadillo, which showed high-centromeric sequence homogeneity. We could not isolate centromeric sequences within the platypus genome, suggesting that centromeres in platypus are not enriched in satellite DNA. Our method can be applied to the characterization of thousands of other vertebrate genomes anticipated for sequencing in the near future, providing an important tool for annotation of centromeres.
Centromeres, physically identified as primary constrictions in chromosomes, carry out important functions in cell biology. They represent the locus where kinetochore fibers bind to the chromatids, thus allowing the correct segregation in daughter cells (Sullivan et al. 2001; Cleveland et al. 2003). Centromeric DNA has been described in different eukaryotes and can be either localized (Saccharomyces cerevisiae) (Pluta et al. 1995) or diffused (Caenorhabditis elegans) (Maddox et al. 2004). Localized centromeres are further classified into two subclasses: point centromeres, whose centromeric function is rigorously specified by a discrete stretch of DNA sequence, and regional centromeres (e.g., Schizosaccharomyces pombe and human), which are composed of much longer and highly homologous tandem repeat arrays, often detectable as satellite bands in CsCl density-gradient centrifugation assays (satellite DNA) (Fowler et al. 1989; Willard et al. 1989; Grady et al. 1992; Vagnarelli et al. 2008). Specific centromere-associated DNA that constitutes the regional centromeres is highly divergent and evolves rapidly during speciation. This suggests that the formation of specialized chromatin structures are more instrumental in centromeric function than specific sequences (Torras-Llort et al. 2009). In human, the centromeres are composed of specific satellite sequences called alphoid DNA. The alpha-satellite is organized in higher-order repeating structures (Willard and Waye 1987b) and is chromosome specific; however, at low-stringency conditions more than one chromosome can show hybridization with the same alphoid sequence. Different organizations of alphoid centromeric satellite were reported in other primates, such as the simple ∼171-bp monomeric structure in orangutan (Haaf and Willard 1998) or the 342-bp dimeric unit structure in New World monkeys (Alves et al. 1994; Cellamare et al. 2009) as lacking any higher-order repeat structure. In a recent study, we characterized the organization and evolution of alpha-satellite DNA in the primate lineage and showed that higher-order repeats evolved more recently in great apes, while the monomeric alpha-satellite in pericentromeric regions is more ancient (Schueler et al. 2005; Alkan et al. 2007).
Satellite DNA constitutes a very unstable part of the genome and is prone to rearrangements. The molecular mechanisms of such rearrangements may include point mutations and amplification of segments of repeated sequences involving one or several copies of a repeat and homogenization, thus forming a pool of related but not identical repetitive sequences (Alexandrov et al. 1988, 2001). Alphoid satellite sequences are also highly variable within species. For example, in humans they represent a source of chromosomal length polymorphism. Unequal crossover between sister chromatids and/or homologous chromosomes may be responsible for this increased variation (Willard and Waye 1987a,b; Waye and Willard 1989; Lee et al. 1997).
Despite their functional significance, the centromeres have largely been omitted from human and other primate genome assemblies (Eichler et al. 2004; Rudd and Willard 2004). In fact, in each chromosome assembly there is no sequence in the existing gap between the p and q arms (Rudd and Willard 2004). However, a few human chromosome assemblies have reached a measurable amount of alpha-satellite (She et al. 2004; Ross et al. 2005), hence providing a valuable resource to understand centromere biology and evolution.
Due to their repetitive and complex nature (millions of highly similar copies of a given repeated sequence), separate efforts need to be carried out to study the centromeric sequences and their organization. In this study, we developed a computational method to predict, identify, and isolate centromeric sequences directly from whole-genome shotgun (WGS) sequence data. We analyzed several representative genomes of the mammalian group, in particular, four placental mammals: horse (Equus caballus [ECA]—Perissodactyla clade), dog (Canis familiaris [CFA]—Carnivora clade), African elephant (Loxodonta africana [LAF]—Afrotheria clade), and armadillo (Dasypus novemcinctus [DNO]—Xenarthra clade); one Methateria, the short-tailed opossum (Monodelphis domestica [MDO]—Marsupialia clade); and one Prototheria, the duck-billed platypus (Ornithorhynchus anatinus [OAN]—Monotremata clade). Polymerase chain reaction (PCR), fluorescence in situ hybridization (FISH), and immunocytohistochemistry (ICHC) were carried out to confirm localization of the extracted sequences to the functional centromere.
This work describes a new genome-wide method to isolate centromeric satellite DNA among various mammalian genomes from WGS sequence data and compare the distribution and organization of these sequences among different mammalian genomes.
Results
Detecting de novo centromeric satellite consensus sequences from whole-genome shotgun (WGS) sequence data
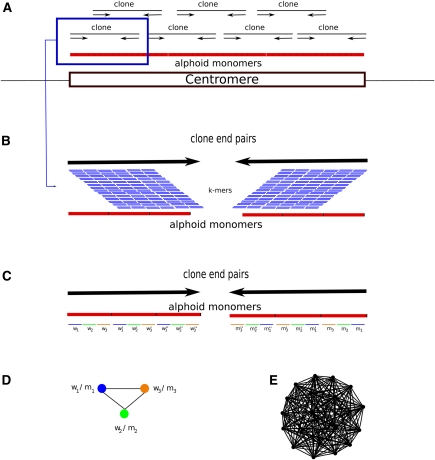
We developed an algorithm, RepeatNet (Fig. 1), that aims to find signatures of long arrays of tandem repeats using paired-end sequencing data generated from long insert clones. In contrast to HORdetect (Alkan et al. 2007), which identifies higher-order repeat structure when the consensus alpha-satellite sequence is known, RepeatNet tries to discover tandem repeats from WGS sequence data with no a priori information about the consensus.
Figure 1.
The RepeatNet algorithm. (A) The layout of the alphoid repeat array in the centromere and the paired-end inserts in the centromeric region is shown. Note that since the centromere is larger than the inserts (fosmids, plasmids, BACs, or short inserts used in next-generation sequencing), both ends of the same insert contain alphoid sequence. (B) Close-up view of a paired-end insert over the alphoid repeat array. We also show all possible k-mers (sliding by 1 bp) that can be generated from the reads. (C) The ideal case for the k-mer structure in the end sequences. When both ends of a paired-end insert contain alphoid sequence, we expect that the k-mers in the forward end will be represented with their reverse-complement counterparts in the reverse end. For simplicity, we show only the nonoverlapping k-mers; however, RepeatNet considers all possible overlapping k-mers. In this figure, w1-m1, w2-m2, w3-m3, w′1-m′1, w′2-m′2, w′3-m′3, w′′1-m′′1, w′′2-m′′2, w′′3-m′′3 are the k-mer pairs that are reverse complements of each other, and the triplet k-mer groups (w1-w′1-w′′1), (w2-w′2-w′′2), (w3-w′3-w′′3) are highly similar k-mers. In the case of exact repeats, these k-mers are identical. (D) Since k-mer pairs w1-m1, w2-m2, and w3-m3 exist in the same read pairs, we put an edge between the nodes that represent such k-mers. (E) The repeat graph for the ideal case of a 31-mer tandem repeat with exact repeat units is shown. This graph includes 20 vertices for 20 k-mer pairs that can be generated from a 31-mer repeat structure, and there exists an edge between all pairs of k-mers. Note that this graph is a clique of size 20. For non-ideal cases, the clique property will be lost; however, the graph will still be very dense in terms of the average degree of the vertices. RepeatNet finds such dense subgraphs of the repeat graph with a heuristic that selects the vertex with the highest degree, and other vertices that share an edge with this selected vertex. Alternatively, a maximum density subgraph algorithm can be used (Fratkin et al. 2006), though this algorithm has a high running time complexity of O[n.m.log(n2m)].
We assume a model where centromeric DNA is organized in tandem array of repetitive DNA. We used the sequences in the WGS databases to first detect “collectively overrepresented k-mers” in both ends of paired-end insert clones (plasmid or fosmid) using our novel method RepeatNet. We then construct consensus satellite sequences by analyzing the read pairs that include these k-mers via phrap (http://www.phrap.org) and Tandem Repeats Finder (TRF) (see Methods). “Collectively overrepresented k-mers” refer to groups of k-mers shared between many independent read pairs that can also be found abundantly in both forward and reverse ends of each pair. As a control, we first tested our algorithm (with k = 12) on the WGS generated from human (Homo sapiens [HSA]), chimpanzee (Pan troglodytes [PTR]), orangutan (Pongo pygmaeus [PPY]), macaque (Macaca mulatta [MMU]), and gibbon (Nomascus leucogenys [NLE]). We could reconstruct the previously published alphoid sequences in these genomes, and furthermore, our algorithms could detect the satellite II and satellite III sequences in the HSA WGS (Supplemental Fig. S1).
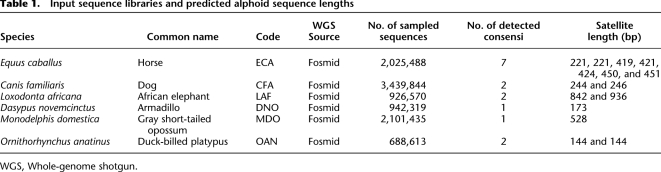
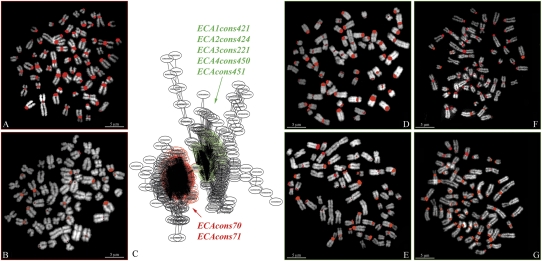
We applied RepeatNet to WGS datasets from six mammalian genomes: horse, dog, elephant, armadillo, gray short-tailed opossum, and duck-billed platypus (NCBI Trace Archive; http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) to discover putative centromeric satellite DNA (Table 1). Sequence reads identified by RepeatNet were then assembled into contigs (phrap), and the basic repeat unit was identified by TRF (Benson 1999). We extracted consensus sequences for each analyzed species, ranging in size from 144 bp (duck-billed platypus) to 936 bp (African elephant). In particular, RepeatNet revealed major clusters of repetitive sequences for all of the analyzed species (see example in Fig. 2 and Supplemental Fig. S2). We extracted seven distinct sequences for ECA (ECAcons70, ECAcons71, ECA1cons421, ECA2cons424, ECA3cons221, ECA4cons450, and ECA5cons451), two each for CFA (CFAcons244 and CFAcons246), LAF (LFAcons842 and LFAcons936) and OAN (OANcons144.1 and OANcons144.2), and only one consensus each for DNO (DNOcons173) and MDO (MDOcons528) (Table 1; Supplemental Fig. S1). In those species where more than one consensus was extracted, we aligned them by BLAST2Sequences in order to extract larger overlapping sequences to use in further experiments (ECAcons421 + 424, CFAcons244 + 246, LAFcons842 + 936, and OANcons144.1 + 144.2). The following analysis was then performed on six ECA consensus sequences and one consensus sequence for all of the other studied species (Supplemental Table S1).
Table 1.
Input sequence libraries and predicted alphoid sequence lengths
WGS, Whole-genome shotgun.
Figure 2.
Example of FISH results on ECA metaphase spreads using horse PCR products obtained with primers designed on ECA consensus sequences (Table 1). Partial RepeatNet graph is reported in C showing two different clusters colored in red and green, respectively. ECAcons70 and ECAcons71 were extracted from the red cluster, while ECA1cons421, ECA2cons424, ECA3cons221, ECA4cons450, and ECA5cons451 were obtained from the green cluster. (A) FISH with PCR product of ECAcons70. (B) FISH with PCR product of ECAcons71. (D) FISH with PCR product of ECAcons421 + 424. (E) FISH with PCR product of ECA3cons221. (F) FISH with PCR product of ECA4cons450. (G) FISH with PCR product of ECA5cons451.
To test whether our sequences were previously classified as satellite DNA elements, we searched for repetitive elements in the consensus sequences using RepeatMasker (http://www.repeatmasker.org/) and BLAST against the “nt” database. In three species the sequence was not previously identified as satellite DNA (RepeatMasker), and in two cases only a portion of the consensus sequence was annotated as satellite DNA. Only LAF and DNO were not previously described as satellite repetitive elements, while MDO sequence was recognized as an LTR/ERV1 element (Supplemental Table S2).
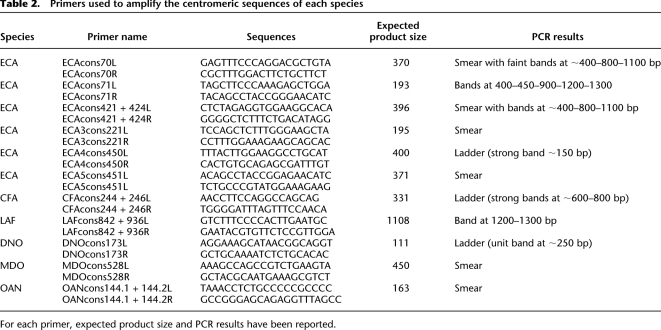
We used each sequence to design species-specific oligo primers (L, left; R, right) covering the entire sequence (Table 2). The PCR products for all of the species were consistent with highly repetitive DNA but showed different amplification patterns: smear, smear with more representative bands and clear ladder patterns (Supplemental Fig. S3). All three patterns were observed in ECA when we used different pairs of primers corresponding to the different consensus sequences. The ladders we obtained using the primers ECAcons71, ECAcons421 + 424, and ECA4cons450 showed different monomeric units: roughly 400, 420, and 150 bp, respectively. This, in addition to the smeared patterns observed using other consensus primers, shows a great diversity and variability in structure and organization of centromeric sequences in ECA. Amplification smears were observed in MDO and OAN showing homogeneity of centromeric sequences in these species. Consensus primers in these cases would anneal to multiple sites, thus resulting in a variety of amplification products not detectable as discrete bands. Loxodonta africana showed the most interesting results in post-amplification as compared with the high-complexity patterns observed in the other species. We observed a single band, sized roughly at 1800 bp, which could represent a unique centromeric sequence without any higher-order structure, or which can correspond to the smaller monomeric unit in this species. In the latter, the ladder pattern cannot be detected, mostly due to the limitation of the technique. DNO, on the other hand, most resembles human centromeric structure, showing a perfect ladder in gel electrophoresis, whose unit size is roughly 130 bp (Table 2).
Table 2.
Primers used to amplify the centromeric sequences of each species
For each primer, expected product size and PCR results have been reported.
Next, we used the PCR products as probes in species-specific FISH experiments. ECA PCR amplification products revealed different hybridization patterns: ECAcons70 hybridized to all centromeres except ECA11; ECAcons421 + 424, ECA3cons221, ECA4cons450, and ECA5cons451 hybridized to all centromeres except ECA7 and ECA11, while ECAcons71 showed signals on 12 out of 32 homologous chromosomes (Fig. 2; Yang et al. 2004). LAF and DNO PCR products hybridized to all of the centromeres; MDO-specific PCR products hybridized on centromeres of four homologous chromosomes (Rens et al. 2001); and OAN PCR products showed strong signals to heterochromatic pericentromeric DAPI-positive regions of the chromosomes 1, 2, 3, 6, 11, 12, and X3 (McMillan et al. 2007) (Supplemental Fig. S4; Supplemental Table S1).
The CFA amplification product did not show any signals in FISH experiments, so further analysis was performed in this case. We modified our approach to discover CFA centromeric sequences. While centromeric DNA spans 3–5 Mb, the typical higher-order repeat unit (343–1197 bp) is sufficiently small enough that it can be traversed by a plasmid. RepeatNet takes this into account during its search for WGS clone mapping within a centromeric region by flagging those that have both forward and reverse ends containing centromeric satellite sequences. We reasoned that one possible explanation for why we may not have recovered centromeric repeats was that the CFA was larger than the average insert size of the plasmids used for WGS (insert size ∼ 2000 bp). Thus, we repeated the analysis for CFA using the end-sequence data set generated from a larger insert clone library (40 kbp), and obtained four collectively overrepresented k-mers. Next, we selected the fosmid clones that include these k-mers in both forward and reverse end sequences. Finally, we randomly selected eight CFA clones (two for each cluster) to test by FISH on dog metaphases. Four out of eight clones showed signals on all dog centromeres except for CFA1; one clone detected only the centromere of CFA37; two showed signals on chromosome 36, 37, and 38 in centromeric position and one out of eight clones showed strong signals on the heterochromatic block of CFA1 (Supplemental Fig. S2; Supplemental Table S3; number of chromosomes according to Breen et al. 1999).
To further verify the centromeric location of PCR probes, we performed in situ immunocytohistochemistry (ICHC). First, we tested specific antibodies (see Methods) against CENPB + CENPC centromeric proteins on the studied species (Supplemental Table S4). Previous data reported the interaction between these two proteins in assembling an active centromere on alphoid DNA (Suzuki et al. 2004). Immunoassay failed on CFA, LAF, and DNO, and gave only two signals on OAN metaphases, likely due to high divergence between our antibodies and the centromeric protein in these species or the reduced amount of protein (under optical resolution) in the target region. We then combined antibodies and PCR-specific products to study mutual localization between DNA and centromeric proteins. Perfect colocalization of anti-CENP antibodies and PCR probes was only observed in ECA and MDO (data not shown).
Next, we performed CENPA chromatin immunoprecipitation (ChIP) on genomic DNA of all of the species in order to localize the active portion of the centromeres (Gopalakrishnan et al. 2009; Trazzi et al. 2009). The ChIP experiments were successful in horse, opossum, and elephant cells; however, they failed in dog, armadillo, and platypus. We then carried out FISH experiments using the immunoprecipitated DNA on horse, opossum, and elephant metaphases in cohybridization with our species-specific probes. In horse and opossum, the signal patterns agreed perfectly with the results of our previous experiments, where most signals colocalized (yellow signals in Supplemental Fig. S5 and Supplemental Table 1). Conversely, while ICHC experiments using the mixture of antibodies (anti-CENPB and anti-CENPC) failed in elephant, the additional ChIP experiment using CENPA was successful and helped us to unequivocally prove the centromeric localization of our probes in this species (Supplemental Fig. S5). As an attempt to detect colocalization between centromeric protein and DNA in dog, armadillo, and platypus, we performed cohybridization and immunoFISH experiments using antibodies for CENPA obtained from multiple sources (CENPA [A-15], sc-11277 CENPA [C-17] sc-11278 [Santa Cruz Biotechnology], and rabbit anti-CENPA monoclonal antibody, unconjugated, clone EP800Y [Abcam]) and the centromeric probes we isolated in this work. In all cases, even the high-quality antibodies failed on the metaphases, preventing us from further investigating the centromeres of these species. We can speculate that these negative results might be due to the extremely low reactivity of the anti-CENPA in dog, opossum, and platypus.
CENPB box is known to be a DNA-binding domain for the centromeric protein CENPB, is present in all mammalian centromeres from human to marsupials, and is highly conserved (Earnshaw and Tomkiel 1992; Bulazel et al. 2006). Thus, to support the robustness of our strategy in detecting centromeric satellite DNA, we searched any putative CENPB-like box (CTTCGTTGGAAACGGGA) (Muro et al. 1992; Yoda et al. 1992) in the extracted sequences using ClustalW (Larkin et al. 2007), focusing on the most evolutionarily conserved domain (ECD) in the box (nTTCGnnnAnnCGGGn) (Stitou et al. 1999). We found a strong conservation of CENPB box motifs in all of the mammalian consensus sequences: they showed 10–12 out of 17 conserved bases. OANcons144.1 + 144.2 showed the lowest similarity (8/17) with five out of nine bases in the ECD compared with the human CENPB box (Fig. 3). The results in platypus support the pericentromeric instead of the centromeric locations found using the amplification product OANcons144.1 + 144.2 for FISH experiments (Supplemental Table S1). Furthermore, this comparison showed that the African elephant, armadillo, and short-tailed opossum shared exactly the same CENPB-like box element, suggesting high conservation of this DNA domain across the phylogeny.
Figure 3.
CENPB box-like motifs extracted from consensus sequences. Conserved bases in the evolutionarily conserved domain (ECD) have been reported in red, and conserved bases compared with human (HSA) other than the ECD domain are reported in blue. The number of total conserved bases is reported in last column. At left, a phylogenetic tree according to Prasad et al. (2008).
Discussion
The centromere is the most characteristic landmark on monocentric eukaryotic chromosomes, appearing as a structural constriction on condensed metaphase chromosomes. Despite its importance, centromeric sequences are typically triaged during genome sequence and assembly, in part due to their molecular complexity. So far, several genomes have been completed, including Drosophila melanogaster, human, mouse, rice, Arabidopsis thaliana, several primates, and mammals, but relatively few centromeres have been fully sequenced as largely separate efforts (Dong et al. 1998; Cheng et al. 2002; Schueler et al. 2005; Kawabe et al. 2006; Roizes 2006; Bulazel et al. 2007; Morris and Moazed 2007; Eckardt 2008; Cellamare et al. 2009). A critical step in such work is identifying centromeric DNA sequence motifs and distinguishing them from other repetitive sequences within genomes.
In this work we developed a computational approach to discover centromeric satellite sequences from unassembled whole-genome shotgun sequences and characterized them experimentally. All eukaryotic DNA sequences in regional centromeres reported until now are arranged in arrays of tandem repetitive motifs variable in size (e.g., 155-bp CentO in rice and to 171-bp alpha-satellite in human). Due to the highly repetitive structure, these sequences have been isolated as separate DNA bands from the bulk of genomic DNA in a CsCl density-gradient centrifugation, and for this reason they are named as satellite DNA. Even though several observations reveal that neither specific DNA sequences (e.g., alphoid satellite) nor the DNA-binding proteins CENPB are essential or enough to dictate the assembly of a functional centromere, it is clear that both of them are common features of centromeres. Taking into consideration the high conservation of organization in eukaryotes, we performed a computational analysis on fully sequenced genomes of six mammals, looking for satellite sequences located at centromeres and containing CENPB box-like domains. We analyzed four representatives of the Eutherian class: two in the superorder of the Laurasiantheria horse in Peryssodactyla and dog in the Carnivora; one representative of the Afrotherian superorder, the African elephant; and one representative of the superorder of the Xenarthra, the armadillo. In addition, the short-tailed opossum and the platypus were considered as members of the other two classes of mammals, the Methateria and Prototheria, respectively (Fig. 3; Prasad et al. 2008).
Using this approach, we defined consensus satellite sequences for the six studied species and localized them in pericentromeric or centromeric regions. Searches for repetitive elements by RepeatMasker and BLAST in ECA, CFA, and OAN sequences showed previously reported species-specific satellite DNA. On the other hand, MDO-isolated sequence recognized ERV elements, while no previously reported repetitive elements resulted for LAF and DNO.
Recently, a high density of endogenous retroviruses (ERVs) and LINE1s (L1s) has been reported within centromeres and evolutionary breakpoints of the tammar wallaby (a marsupial in the Methateria group) and centromeric and/or telomeric regions of most Monodelphis chromosomes, displaying a common feature of this group of mammals (Gentles et al. 2007; Mikkelsen et al. 2007; Longo et al. 2009). The finding of these repetitive sequences in the isolated sequences from the armadillo supports their centromeric function and localization, suggesting that this use of common repeats as the primary source of centromeric satellites may represent a mammalian ancestral state.
Centromeric sequences were isolated for most of the analyzed mammals and showed different levels of complexity in the studied genomes. Horse and dog showed the greatest variability and complexity in centromeric organization. In these species, we found the greatest number of different consensus sequences, which varied in length and FISH hybridization patterns. In horse, for example, we found six centromeric sequences derived from two different clusters (Fig. 2), with ECAcons71 showing the more specific hybridization pattern on ECA2, ECA4, ECA8, ECA14, ECA15, ECA17, ECA18, ECA19, ECA22, ECA24, ECA27, and ECA28. We conclude that the centromere DNA in these chromosomes is a patchwork of different satellite sequences since they showed signals with all of the consensus sequences, further supported by the great diversity in the amplification pattern observed for this species (Supplemental Fig. S3). In contrast, ECAcons70 is the main centromeric sequence in ECA chromosomes since it was detected on all chromosomes except ECA11. Similarly, dog centromeric sequences showed the same complexity with four different clusters and three different hybridization patterns. CFA1 was not detected by any fosmid probes, three dog fosmids (e.g., G630P89020G11_BI1-3524M21) gave signals on all chromosomes except for CFA1, and two fosmids (e.g., G630P88303G10_BI1-3749N20) showed signals on CFA36, CFA37, and CFA38 and one (G630P88580D11_BI1-3897G21) only on CFA37 (Supplemental Fig. S2; Supplemental Table S3). These findings strongly support the hypothesis of a complex patchwork organization in the dog centromeres similar to what we observed in horse.
FISH analysis of the amplification products and ICHC (when available) showed a pancentromeric distribution of isolated centromeric sequences in the African elephant and armadillo. This is characteristically different from human and great ape species, where higher-order structures and chromosome-specific patterns have been reported (Alkan et al. 2007; Ventura et al. 2007; Cellamare et al. 2009); instead, elephant and armadillo resemble the macaque centromere organization (Ventura et al. 2007). While this could represent an example of convergent evolution, it is more likely that the archetype for Eutherian centromere organization was simple tandem arrays and lacked the higher-order structure prevalent among human and African great ape chromosomes. Furthermore, we observed a single 1800-bp band in the elephant and could not detect any ladder pattern. This leaves us with the uncertainty of the centromeric structure in this species. The 1800-bp fragment could represent a single centromeric unit or a monomeric unit for which high-order organization cannot be detected in PCR, mostly due to the limitation of the technique in amplifying fragments larger than 3000 bp. Future work could focus on the Loxodonta africana genome to clarify the organization of centromeres in this species.
We note that without colocalization with CENPB, CENPC, and CENPA for CFA and DNO, we cannot exclude that these sequences could be pericentromeric in nature.
A limitation of our method is that not all centromeres were detected (negative centromeres). However, this is not surprising since other studies previously reported similar patterns in gibbon and New World monkeys that show the existence of chromosome-specific centromeric sequences (Cellamare et al. 2009). Our approach is biased in selecting regions organized in a highly repetitive manner; thus, it avoids finding centromeric sequences with a more degenerate structure. Negative centromeres might have a sequence organization with no satellite-like signatures or, in contrast, the centromeric satellite sequences might not be represented in the analyzed WGS libraries due to cloning bias (e.g., ECA11 and CFA1 that never showed FISH signals) (Alkan et al. 2007).
We searched for CENPB box-like elements in the consensus sequences two different ways: (1) by looking for conservation in the ECD and (2) by searching for similarity with the human CENPB box. We showed that the pancentromeric satellite, ECAcons70, has a perfect conservation, while CFAcons244 + 246 and OANcons144.1 + 144.2 showed the lowest conservation of the ECD. The comparison with the full-length human CENPB box further supports the higher divergence of the platypus centromeric sequence from the rest of the mammals we analyzed. This finding agrees with the location of the OANcons144.1 + 144.2 in the heterochromatic pericentromeres in this species. According to our data, the centromeres in platypus are not defined by satellite DNA; instead, they are embedded in a satellite territory, where it is possible to detect the highly divergent CENPB box-like domains. We theorize that in platypus, the real binding domain CENPB box exists, but it is not located in satellite DNA. Functional studies need to be carried out to address this question.
Despite the high divergence we found in platypus, we detected a highly conserved CENPB box, both in the ECDs and compared with human, in the African elephant, armadillo, and opossum. These species share exactly the same CENPB box domains, greatly supporting the importance of this element and the robustness of our method in detecting functional centromeric sequences when they show satellite properties. All of the mammals we studied have repeated DNA satellites at their centromeres except platypus. This further supports the hypothesis that centromeres are composed of repeated arrays evolved from simple monomeric structures, and this structure is strongly linked to their functions (Warburton et al. 1996; Harrington et al. 1997; Schueler et al. 2001, 2005).
Our work represents the first study to systematically detect, analyze, and isolate centromeric satellite sequences from the bulk of whole-genome shotgun sequence data, identifying for the first time centromeric satellite sequences in species such as elephant. The data provide an important baseline for further studies to address questions of centromere biology and evolution. More importantly, the methods we developed should be directly applicable to next-generation sequence datasets from genomic libraries. Such approaches may complement efforts to characterize and assemble genomes in the future.
Methods
Prediction of alphoid sequences
We designed a computational method to detect the consensus centromeric sequences from paired-end whole-genome shotgun sequence libraries. Our pipeline starts with a novel algorithm (RepeatNet) used to locate the read pairs likely to include the alphoid sequences, and such read pairs are further processed with the readily available tools phrap and TRF (Benson 1999). RepeatNet makes use of the fact that the alpha-satellite repeat array is larger than the cloning vectors (fosmid, plasmid, or BAC) used in sequencing, and both ends of a vector lie within the repeat array (Fig. 1). Therefore, we expect that the sequence content of both ends are identical and include a highly similar tandem repeat structure. However, calculating all pairwise comparisons of the read pairs in a WGS library would be computationally infeasible; therefore, RepeatNet builds “collectively overrepresented k-mer graphs” to look for signatures of such high-sequence identity among end sequences of the same clone as well as end sequences from different clones. Collectively, overrepresented k-mers are defined as k-mers that are shared between end sequences and frequently occur within the entire WGS sequence library (Fig. 1). Note that this method is independent from the read length. In our experiments, we set k = 12 and require perfect matches between k-mers to achieve high sensitivity while keeping memory requirements low.
RepeatNet algorithm
We load all reads from a WGS sequence library (or a random subsample) into memory and parse the mate-pair information from sequence names. We then create a counter array and a location array of size 4k (for k = 12, 4k = 16,777,216) for all possible k-mers to store both the frequency information and the “source clone” for each k-mer. Next, we process the reads clone-by-clone; if a k-mer occurs on the forward end and its reverse complement occurs in the reverse end, we increase its counter by one and add the corresponding clone name to that k-mer's entry in the location array. After all end-sequences are processed, we merge the counters and location lists of all pairs of k-mers that are a reverse complement of each other. This is necessary because the source strands of the WGS are unknown and the merged k-mers in this step are equivalent. We discard k-mers with counter value (frequency) less than 100 (arbitrary cutoff to remove nonsignificant k-mers and reduce computational cost) and then pairwise compare the location lists of all remaining k-mers. If the location lists of two k-mers overlap by at least 100 clones (arbitrary cutoff), we create an edge between the two vertices corresponding to the two k-mers (Fig. 1C,D) to indicate their collectively overrepresentation relationship. We select the vertex in the graph with the highest degree (maximum number of edges) and its “neighbors” (i.e., vertices that share an edge with the selected node). Next, we retrieve the clone list and corresponding sequences of all k-mers in the selected subgraph. Finally, we assemble the selected sequences using phrap and build the consensus tandem repeat sequences from the assembled contigs using TRF. The current implementation of the RepeatNet algorithm is available at http://eichlerlab.gs.washington.edu/software/repeatnet/.
PCR
Genomic DNA from different species were obtained from fibroblastoid cell lines by standard methods. Primer pairs (Table 2) designed on the consensus sequences of each species were used to amplify DNA by PCR.
The PCR cycling parameters used were as follows: 2 min initial denaturation at 94°C, followed by 30 cycles of: 94°C for 20 sec, 60°C for 1 min, and 72°C for 2 min. Final extension was at 72°C for 10 min (and then at 12°C hold).
Reaction mixture consisted of 5 μL of dNTPs (10×), 0.5 μL of each primer (10 μM), 0.3 μL of Platinum Taq DNA polymerase (5 U/μL), 1.5 μL MgCl2 (50 mM), 5 μL of reaction buffer (Invitrogen) (10×), 3 μL of DNA template (50 ng/μL), and water up to 50 μL.
PCR products were analyzed by 1% agarose gel electrophoresis.
Cell line
Metaphase preparations were obtained from the fibroblastoid cell line of Canis familiaris (CFA), Equus caballus (ECA), Loxodonta africana (LAF), Dasypus novemcintus (DNO), Monodelphis domestica (MDO), and Ornithorhynchus anatinus (OAN) following standard procedures. Cell lines from LAF, DNO, MDO, and OAN were obtained by Professor O'Brien's repository (Laboratory of Genomic Diversity, NCI-Frederick, Frederick, MD); CFA and ECA chromosomes were obtained from common dog and horse blood samples, following the standard procedure (Carbone et al. 2006; Cardone et al. 2006).
FISH
FISH experiments were essentially performed as previously described (Ventura et al. 2003). Briefly, DNA probes were directly labeled with Cy3-dUTP (Perkin-Elmer) or fluorescein-dCTP (Fermentas) by PCR labeling for each PCR product and by nick-translation for specific dog fosmid clones. The use of PCR labeling avoids the possible contamination from genomic DNA by nick-translation labeling of PCR products.
PCR labeling was carried out in a final volume of 50 μL, which contained 100 ng of PCR product, 5 μL of reaction buffer 10×, 4 μL of 50 mM MgCl2, 1 μL of each 10 μM primer, 1 μL of 2 mM dACG, 2.5 μL of 1 mM Cy5-dUTP, 5 μL of BSA 1%, and 0.6 μL of 5 U/μL Taq polymerase.
DNA extraction from fosmids was performed as already reported (Ventura et al. 2001).
Two hundred nanograms of labeled probe were used for the FISH experiments. Hybridization was performed at 37°C in 2× SSC (sodium chloride and sodium citrate), 50% (v/v) formamide, 10% (w/v) dextran sulfate, and 5 mg of sonicated salmon sperm DNA in a volume of 10 μL. Post-hybridization washing was at 60°C in 0.1× SSC (sodium chloride and sodium citrate) (three times, high stringency).
Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), fluorescein (green), and DAPI (blue) fluorescence signals, detected with specific filters and recorded separately as grayscale images. Pseudocoloring and merging of images was performed using Adobe PhotoShop software.
Immunolocalization
Immunofluorescence using a mix of CENPB + CENPC antibody was performed as suggested by Earnshaw and Tomkiel (1992) with some modifications. Fibroblasts from different species were grown and treated by standard procedure to obtain metaphases. As soon as the surface was dry, each slide was rehydrated by immersion in a 1× PBS-Azide (10 mM NaPO4 at pH 7.4, 0.15 M NaCl, 1 mM EGTA, 0.01% NaN3) for 5–15 min. The chromosomes were then swollen by washing the slides three times (2 min each) with 1× TEEN (1 mM treithanolamine-HCl at pH 8.5, 0.2 mM NaEDTA, 25 mM NaCl) + 0.5% Triton X-100 + 0.1% BSA. The primary antibody was diluted 1:100 in the same solution and then added (200 μL) on the surface of the slide. Each slide was incubated for 1.5–2 h at 37°C. Unlabeled primary antibody was removed by washing the slides three times with 1× potassium buffer (KB) (10 mM Tris-HCl at pH 7.7, 0.15 M NaCl, 0.1% BSA) for 2 min, 5 min, 3 min at room temperature. Secondary antibody conjugated with Cy3 was diluted 1:200 in the same solution and 200 μL were then added to the slide, avoiding air dry, and incubated, or 30–60 min at 37°C in a dark chamber. After detection the slide was washed once with 1× KB for 2 min at RT, stained with DAPI (200 ng/mL in 2× SSC) for 5 min, and mounted with antifade (0.233 ng of DABCO [1,4-diazabicyclo-(2.2.2)octane, Sigma], 800 μL of H2O, 200 μL of 1 M Tris-HCl, 9 mL of glycerol). For immunoFISH after the incubation with the secondary antibody, the slide was washed once with 1× KB for 2 min, prefixed with 4% paraformaldeide in 1× KB for 45 min, washed with distilled H2O by immersion for 10 min at RT, and fixed with methanol and acetic acid (3:1) for 15 min. After that, the standard procedure was followed for FISH.
ChIP analysis
Native chromatin immunoprecipitation (N-ChIP) analysis was performed as previously described (Umlauf et al. 2004). Briefly, fibroblastoid cells from ECA, LAF, CFO, DNO, MDO, and OAN were processed, and the native chromatin was prepared by micrococcal nuclease (New England BioLabs) digestion of cell nuclei. A portion of digested DNA was used as INPUT DNA. Then, immunoprecipitation was performed using a polyclonal antibody against the human centromeric protein CENPA (Trazzi et al. 2009). Both purified DNA samples were amplified using the Whole Genome Amplification kit (Sigma-Aldrich).
Acknowledgments
We thank G. Aksay for her help in implementing RepeatNet and T. Brown for proofreading the manuscript. This work is partly supported by a HG002385 grant to E.E.E. and a PRIN 2007 grant to M.V. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at http://www.genome.org. The RepeatNet algorithm is freely available at http://eichlerlab.gs.washington.edu/software/repeatnet/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111278.110.
References
- Alexandrov IA, Mitkevich SP, Yurov YB 1988. The phylogeny of human chromosome specific alpha satellites. Chromosoma 96: 443–453 [DOI] [PubMed] [Google Scholar]
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y 2001. Alpha-satellite DNA of primates: Old and new families. Chromosoma 110: 253–266 [DOI] [PubMed] [Google Scholar]
- Alkan C, Ventura M, Archidiacono N, Rocchi M, Sahinalp SC, Eichler E.E 2007. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol 3: 1807–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Seuanez HN, Fanning T 1994. Alpha satellite DNA in neotropical primates (Platyrrhini). Chromosoma 103: 262–267 [DOI] [PubMed] [Google Scholar]
- Benson G 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF 1999. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics 61: 145–155 [DOI] [PubMed] [Google Scholar]
- Bulazel K, Metcalfe C, Ferreri GC, Yu J, Eldridge MD, O'Neill RJ 2006. Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics 172: 1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulazel KV, Ferreri GC, Eldridge MD, O'Neill RJ 2007. Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol 8: R170 doi: 10.1186/gb-2007-8-8-r170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P et al. 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87: 777–782 [DOI] [PubMed] [Google Scholar]
- Cardone MF, Alonso A, Pazienza M, Ventura M, Montemurro G, Carbone L, de Jong PJ, Stanyon R, D'Addabbo P, Archidiacono N, et al. 2006. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol 7: R91 doi: 10.1186/gb-2006-7-10-r91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellamare A, Catacchio CR, Alkan C, Giannuzzi G, Antonacci F, Cardone MF, Della Valle G, Malig M, Rocchi M, Eichler EE, et al. 2009. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol Biol Evol 26: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF 2003. Centromeres and kinetochores. From epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J 1998. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci 95: 8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Tomkiel JE 1992. Centromere and kinetochore structure. Curr Opin Cell Biol 4: 86–93 [DOI] [PubMed] [Google Scholar]
- Eckardt NA 2008. Defining a functional centromere. Plant Cell 20: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X 2004. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat Rev Genet 5: 345–354 [DOI] [PubMed] [Google Scholar]
- Fowler JC, Skinner JD, Burgoyne LA, Drinkwater RD 1989. Satellite DNA and higher-primate phylogeny. Mol Biol Evol 6: 553–557 [DOI] [PubMed] [Google Scholar]
- Fratkin E, Naughton BT, Brutlag DL, Batzoglou S 2006. MotifCut: Regulatory motifs finding with maximum density subgraphs. Bioinformatics 22: e150–e157 [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J 2007. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res 17: 992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD 2009. DNMT3B interacts with constitutive centromere protein CENPC to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18: 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady DL, Ratliff RL, Robinson DL, McCanlies EC, Meyne J, Moyzis RK 1992. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci 89: 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Willard HF 1998. Orangutan alpha-satellite monomers are closely related to the human consensus sequence. Mamm Genome 9: 440–447 [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355 [DOI] [PubMed] [Google Scholar]
- Kawabe A, Hansson B, Hagenblad J, Forrest A, Charlesworth D 2006. Centromere locations and associated chromosome rearrangements in Arabidopsis lyrata and A. thaliana. Genetics 173: 1613–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC 1997. Human centromeric DNAs. Hum Genet 100: 291–304 [DOI] [PubMed] [Google Scholar]
- Longo MS, Carone DM, Green ED, O'Neill MJ, O'Neill RJ 2009. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics 10: 334 doi: 10.1186/1471-2164-10-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Oegema K, Desai A, Cheeseman IM 2004. “Holo”er than thou: Chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12: 641–653 [DOI] [PubMed] [Google Scholar]
- McMillan D, Miethke P, Alsop AE, Rens W, O'Brien P, Trifonov V, Veyrunes F, Schatzkamer K, Kremitzki CL, Graves T, et al. 2007. Characterizing the chromosomes of the platypus (Ornithorhynchus anatinus). Chromosome Res 15: 961–974 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177 [DOI] [PubMed] [Google Scholar]
- Morris CA, Moazed D 2007. Centromere assembly and propagation. Cell 128: 647–650 [DOI] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T 1992. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 116: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC 1995. Centromere: Hub of chromosomal activities. Science 270: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Prasad AB, Allard MW, Green ED 2008. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol 25: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, O'Brien PC, Yang F, Solanky N, Perelman P, Graphodatsky AS, Ferguson MW, Svartman M, De Leo AA, Graves JA, et al. 2001. Karyotype relationships between distantly related marsupials from South America and Australia. Chromosome Res 9: 301–308 [DOI] [PubMed] [Google Scholar]
- Roizes G 2006. Human centromeric alphoid domains are periodically homogenized so that they vary substantially between homologues. Mechanism and implications for centromere functioning. Nucleic Acids Res 34: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Willard HF 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet 20: 529–533 [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115 [DOI] [PubMed] [Google Scholar]
- Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L, Rocchi M, Willard HF, Green ED 2005. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci 102: 10563–10568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, et al. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430: 857–864 [DOI] [PubMed] [Google Scholar]
- Stitou S, Diaz de la Guardia R, Jimenez R, Burgos M 1999. Isolation of a species-specific satellite DNA with a novel CENP-B-like box from the North African rodent Lemniscomys barbarus. Exp Cell Res 250: 381–386 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH 2001. Determining centromere identity: Cyclical stories and forking paths. Nat Rev Genet 2: 584–596 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Nakano M, Nozaki N, Egashira S, Okazaki T, Masumoto H 2004. CENP-B interacts with CENP-CCENPC domains containing Mif2 regions responsible for centromere localization. J Biol Chem 279: 5934–5946 [DOI] [PubMed] [Google Scholar]
- Torras-Llort M, Moreno-Moreno O, Azorin F 2009. Focus on the centre: The role of chromatin on the regulation of centromere identity and function. EMBO J 28: 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S, Perini G, Bernardoni R, Zoli M, Reese JC, Musacchio A, Della Valle G 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE 4: e5832 doi: 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296– 1300 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro SA, Earnshaw WC 2008. Centromeres: Old tales and new tools. FEBS Lett 582: 1950–1959 [DOI] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M 2001. Centromere emergence in evolution. Genome Res 11: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res 13: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246 [DOI] [PubMed] [Google Scholar]
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF 1996. Characterization of a chromosome specific chimpanzee alpha satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics 33: 220–228 [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF 1989. Concerted evolution of alpha satellite DNA: Evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma 98: 273–279 [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS 1987a. Chromosome-specific subsets of human alpha satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 25: 207–214 [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS 1987b. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet 3: 192–198 [Google Scholar]
- Willard HF, Wevrick R, Warburton PE 1989. Human centromere structure: Organization and potential role of alpha satellite DNA. Prog Clin Biol Res 318: 9–18 [PubMed] [Google Scholar]
- Yang F, Fu B, O'Brien PCM, Nie W, Ryder OA, Ferguson-Smith MA 2004. Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: Insight into the occasional fertility of mules. Chromosome Res 12: 65–76 [DOI] [PubMed] [Google Scholar]
- Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T 1992. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol 119: 1413–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]