Abstract
MicroRNAs (miRNAs) are regulatory noncoding RNAs that affect the production of a significant fraction of human mRNAs via post-transcriptional regulation. Interindividual variation of the miRNA expression levels is likely to influence the expression of miRNA target genes and may therefore contribute to phenotypic differences in humans, including susceptibility to common disorders. The extent to which miRNA levels are genetically controlled is largely unknown. In this report, we assayed the expression levels of miRNAs in primary fibroblasts from 180 European newborns of the GenCord project and performed association analysis to identify eQTLs (expression quantitative traits loci). We detected robust expression for 121 miRNAs out of 365 interrogated. We have identified significant cis- (10%) and trans- (11%) eQTLs. Furthermore, we detected one genomic locus (rs1522653) that influences the expression levels of five miRNAs, thus unraveling a novel mechanism for coregulation of miRNA expression.
The discovery of microRNAs (miRNAs) (19- to 25-nt-long single-stranded RNA molecules) has revealed a new mechanism for the regulation of protein-coding gene expression (Ambros 2004; Bartel 2004; Baek et al. 2008; Selbach et al. 2008). Dosage alterations of miRNA levels are thought to be involved in human disease pathogenesis (Bartel 2004; Kloosterman and Plasterk 2006; Bushati and Cohen 2007; Bartel 2009; Xiao and Rajewsky 2009). One of the least understood aspects of miRNA biogenesis concerns the regulation of its expression levels. Approximately half of the miRNAs identified to date are located in intergenic regions and are therefore likely to possess their own promoter and enhancer elements. The remaining miRNAs map to introns of protein-coding genes and are transcribed from the same strand (Saini et al. 2008). However, it is not yet clear whether these miRNAs are the by-products of protein-coding gene transcription or whether their transcription is controlled by independent regulatory elements. Since miRNA genes are transcribed by RNA polymerase II, it is likely that they share a similar mode of regulation with protein-coding mRNAs.
The goal of this study was to identify genetic variation associated with miRNA levels, as a way to dissect the elements and mechanisms governing miRNA expression.
Recent genetic analyses have demonstrated that transcription levels of protein-coding genes behave as heritable quantitative traits and display significant associations with genetic variants, including single nucleotide polymorphisms (SNPs) and copy number variants (CNVs) (Morley et al. 2004; Cheung et al. 2005; Deutsch et al. 2005; Stranger et al. 2007; Dermitzakis 2008).
In this study, we conducted an association analysis using mature miRNA expression levels as the primary phenotype, with the aim of identifying regulatory polymorphic variants (expression quantitative traits loci [eQTLs]) significantly associated with miRNA expression levels in human primary fibroblasts.
Results
Primary fibroblasts were derived from the umbilical cord of 180 newborns of western European origin recruited for the GenCord project (see Methods). All samples were genotyped using the Illumina Hap550 SNP array. Mature miRNA expression phenotypes were generated using the micro-fluidics-based TaqMan Human MiRNA Array v1.0 (Applied Biosystems). For each sample, the expression levels for 365 known human mature miRNAs were assayed (Supplemental Table S1). We detected expression above the background for 57% (n = 208) of the miRNAs in cultured primary fibroblasts. These were further filtered to include miRNAs with expression above the background in at least 50% of the samples (n = 90). One hundred twenty-one miRNAs were retained for association analysis.
We identified cis-eQTLs, by testing for association between expression levels and SNP genotypes, within 1 Mb 5′ and 3′ of each miRNA. SNPs were considered to be significantly associated with miRNA expression levels (i.e., eQTLs) if they passed the 0.05 permutation level threshold for 10,000 permutations (see Methods).
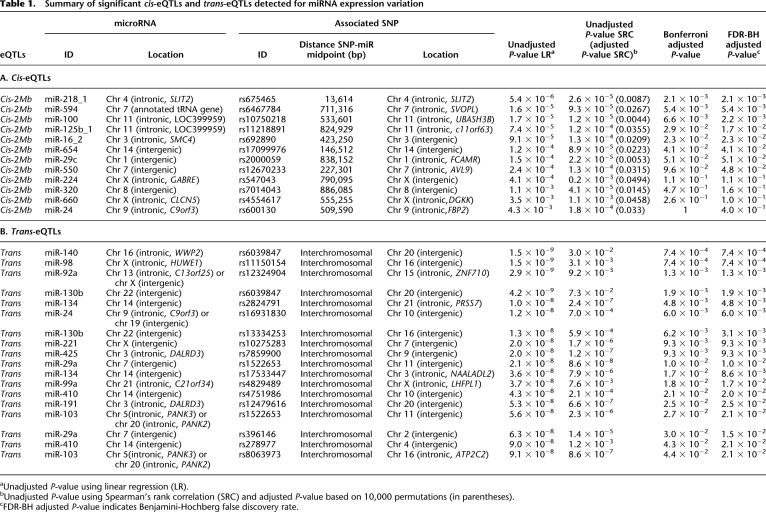
Twelve (i.e., 10%) of the 121 miRNAs tested showed significant evidence for cis-regulatory variation (permutated P-value < 0.05) (Table 1A; Fig. 1; Supplemental Fig. S1). Given that we tested 121 miRNAs and we expect 5% of them to have significant cis- associations by chance at the permutation level 0.05, we estimate that our false-discovery rate (FDR) is about 50% of the 12 miRNA signals.
Table 1.
Summary of significant cis-eQTLs and trans-eQTLs detected for miRNA expression variation
aUnadjusted P-value using linear regression (LR).
bUnadjusted P-value using Spearman's rank correlation (SRC) and adjusted P-value based on 10,000 permutations (in parentheses).
cFDR-BH adjusted P-value indicates Benjamini-Hochberg false discovery rate.
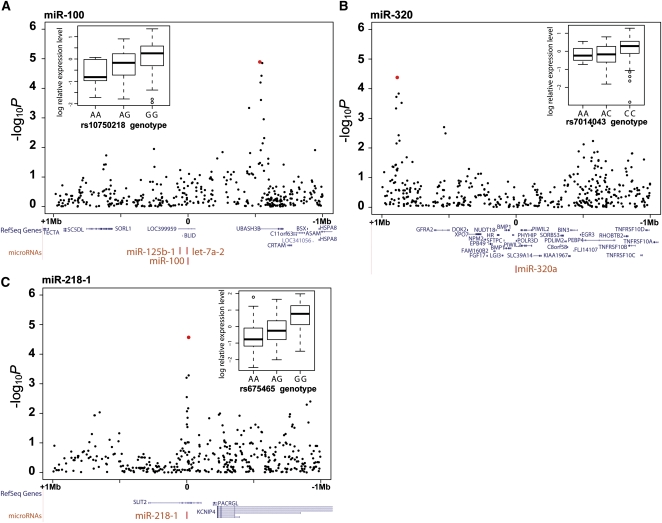
Figure 1.
Examples of cis-eQTLs for miR-100 (A), miR-320 (B), and miR-218-1 (C). The panels show the distribution of −log10 P-values for SNPs across a 1-Mb region surrounding the miRNA (“0” position). The highest significant −log10 P-values are shown as red dots. Also shown are the mapping of RefSeq genes in blue and miRNAs in red. The boxplots depict the relationship between miRNA relative expression levels (log2) and genotypes for the most significant SNPs. Boxplots are divided by median values.
Examples of these cis-eQTLs are shown in Figure 1. The most highly significant cis-eQTL detected was rs10750218, intronic to UBASH3B, which associates with levels of miR-100 533 kb away (Fig. 1). The distance between the cis-eQTLs and their respective miRNA was variable and ranged from 13.6 kb to 886 kb (Table 1A). In one case (miR-218-1), cis-eQTLs mapped within the protein-coding sequences of SLIT2 that also contain the miRNA sequence. This raises the interesting question of whether both the miR-218-1 and the SLIT2 mRNA share regulatory sequences (Table 1). To address this, we investigated whether the specific cis-eQTL for the miRNA was also associated with SLIT2 mRNA levels. Transcription levels of protein-coding genes were assayed using Illumina's WG-6 v3 Expression BeadChip array (Dimas et al. 2009). We found no evidence of shared regulatory variation between mRNA and miRNA, and no correlation between the miR-218-1 and SLIT2 mRNA levels was observed (Pearson correlation = −0.023, n = 55), implying absence of coregulation of these two transcripts in fibroblasts.
We then aimed to identify trans-eQTLs by performing a genome-wide association study (GWAS) for the 121 miRNA expression phenotypes. We observed 18 significant trans-eQTLs for 13 miRNAs (10.7%) after Bonferroni correction for multiple testing at the 95% significance level (Table 1B; Supplemental Fig. S2). Since under the null hypotheses we would expect on average six associations, we can estimate our FDR at about 30% for 18 reported miRNAs.
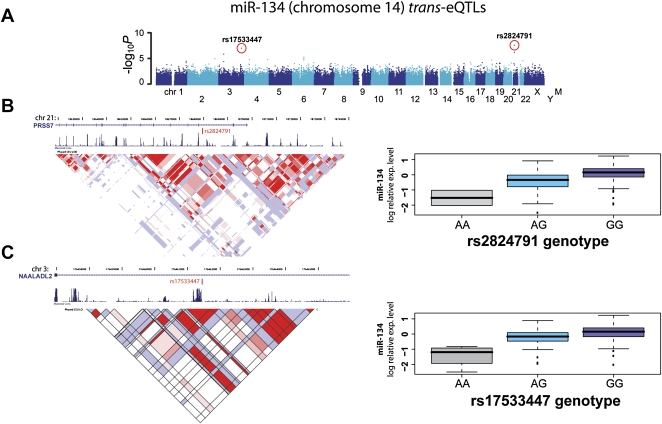
The most significant trans-eQTL was detected for miR-140 (chromosome 16) with SNP rs6039847 located on chromosome 20 (unadjusted P = 1.5 × 10−9). The majority of trans-eQTLs (72%) mapped to intergenic regions. We detected cases where multiple trans-eQTLs, located in different chromosomes, associate with the expression levels of single miRNAs (Table 1B), suggesting that multiple loci may act together to regulate miRNA expression. For example, two significant trans-eQTLs were detected for miR-134, the first on chromosome 21 (rs2824791, unadjusted P = 1 × 10−8) and the second on chromosome 3 (rs17533447, unadjusted P = 3.6 × 10−8) (Fig. 2). Similar observations were made for miR-103, miR-130b, miR-29a, and miR-410 (Table 1B; Supplemental Fig. S2). We also observed two cases in which a single SNP was associated with the expression of multiple, unrelated miRNAs: rs1522653 is significantly associated with the expression of miR-103 and miR-29a; rs6039847, with miR-140 and miR-130b (Table 1B).
Figure 2.
Example of trans- eQTLs for miR-134. (A) Manhattan plot displays −log10 P-values of a GWAS for miR-134 expression variation. Each chromosome is depicted as different shades of blue. Chromosome Y and mitochondrial genotypes have not been included in this study. Red dots indicate SNPs with the highest significant P-values after Bonferroni correction for multiple testing (see Table 1). Those two eQTLs, rs2824791 on chromosome 21 and rs7533447 on chromosome 3, are detailed in panels B and C, respectively. (B,C, left panels) The location of associated SNPs, as well as RefSeq transcripts, conservation, and LD information (LOD scores for CEU population). (B,C, right panels) Boxplots for miRNA expression for different genotypic groups.
These observations prompted us to analyze in-depth for the presence of statistically significant miRNA “master regulators,” defined as trans-eQTLs involved in the regulation of multiple miRNA genes.
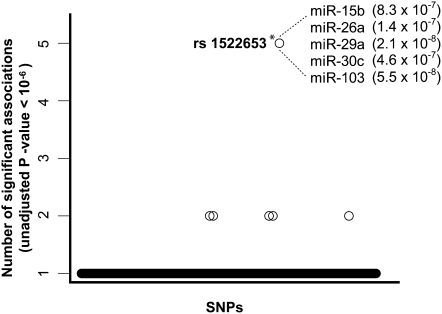
To this end, we ascertained for each SNP the number of miRNA associations detected using a reduced stringency (unadjusted P-value < 10−6) (Supplemental Table S2). This analysis identified one trans-eQTL, rs1522653 on chromosome 11 that was associated with the expression of five miRNAs (miR-15b, miR-26a, miR-29a, miR-30c, and miR-103) (Fig. 3). To determine the significance of this finding, we permuted 1000 times the expression levels of all miRNAs (preserving the miRNA expression matrix per individual) and performed GWAS for each permuted data set. From this, we estimated the empirical significance of our master regulator to be equal to 0.005 (Fig. 3; see Methods).
Figure 3.
Master miRNA trans-eQTLs. Plot shows SNPs associated with the expression variation of multiple miRNAs (using a threshold of an unadjusted P-value < 10−6 per association) (see Supplemental Table S2). Each circle represents a single SNP. Only SNPs with at least one association below the P < 10−6 threshold are shown. One SNP (rs1522653) is significantly associated with the expression of five miRNAs (*, permutated P-value of 0.005). The identities and unadjusted P-values for these miRNAs are shown.
Remarkably, rs1522653 is an intergenic SNP, located in a large gene desert (3.29 Mb with no annotated protein-coding or noncoding RNAs); the nearest gene, FAM181B, maps 1.59 Mb away (Supplemental Fig. S2). The identification of regulatory variants associated with the expression levels of multiple miRNAs may point to potential “master regulatory” properties and suggests that the expression levels of groups of miRNAs may be coordinated through the use of common regulatory elements. This hypothesis predicts that the five miRNAs associated with rs1522653 should display related expression profiles. To test this hypothesis, we compared the average of the correlation values of the five miRNAs associated with rs1522653 to 10,000 sets of five randomly selected miRNAs. We found that the observed average correlation of 0.44 is higher than that expected by chance (permutated P-value of 0.0012) (Supplemental Fig. S3). We also examined whether the predicted target transcripts of the five miRNAs associated with a master regulator share molecular functions. We investigated Gene Ontology (GO) terms from computational target predictions of the five coregulated miRNAs (miRanda [John et al. 2004] from the miRBase-Targets database [Griffiths-Jones et al. 2008]). This analysis revealed that the mRNA targets for these five miRNAs are significantly enriched for “protein-binding process” (P = 4.4 × 10−8, Fisher's exact test), “transcription regulator activity” (P = 7.8 × 10−8), and “transcription factor activity” (P = 1.2 × 10−6) (Supplemental Table S3).
We therefore propose a model in which certain eQTLs act as master regulators by comodulating the expression of multiple miRNAs, thus revealing a novel mechanism for coregulation of miRNA expression.
Discussion
This study provides an initial assessment of the expression level variation of mature human miRNAs and explores how these levels are regulated by common genetic variants in fibroblasts from European individuals. Since we only studied one cell type, the eQTLs identified here are likely to represent a small subset of regulatory variation affecting miRNA levels. Indeed, many miRNAs are expressed in a tissue-restricted manner (Landgraf et al. 2007) and are thus likely to have tissue-specific regulators, as reported recently for protein coding genes (Dimas et al. 2009).
Earlier studies have shown that common genetic variants contribute significantly to the individual differences in protein-coding gene expression variation (Cheung et al. 2003, 2005; Morley et al. 2004; Deutsch et al. 2005; Stranger et al. 2005, 2007; Spielman et al. 2007; Storey et al. 2007) and transcript isoform variation (Hull et al. 2007; Kwan et al. 2007, 2008; Zhang et al. 2009). Our study adds a level of complexity to cellular gene expression regulation by revealing that cis- and trans-eQTLs can affect the expression of miRNAs that are themselves regulatory molecules. eQTLs identified in this study are potential candidates for the involvement in human phenotypes. Differences in the quantity of mature miRNAs have a clear impact on the level of targeted proteins and result in phenotypic differences (Sethupathy et al. 2007; Baek et al. 2008; Selbach et al. 2008; Bartel 2009). The subsequent identification of the functional variation related to each eQTL type may provide important genomic targets for dissecting the molecular basis of susceptibility to genetic disorders.
Methods
Cell culture and RNA preparation
We obtained primary fibroblasts from 180 individuals of the GenCord project. This collection was established from umbilical cords of newborns of western European origin (following appropriate informed consent and approval by the Geneva University Hospital's ethics committee). All cell lines were grown in DMEM with Glutamax I (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) and 1% penicillin/streptomycin/fungizone mix (Amimed, BioConcept) at 37°C and 5% CO2. Confluent cell lines were trypsinized and diluted at a density of 7 × 105 cells/mL (40% of confluence) and harvested the following day. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. RNA quality was assessed using RNA 6000 NanoChips with the Agilent 2100 Bioanalyzer (Agilent), and RNA was quantified with a NanoDrop spectrophotometer (NanoDrop Technologies).
miRNA expression measurement and data normalization
Expression of 365 known human miRNAs was analyzed using the TaqMan Human MiRNA Array v1.0 early access (Applied Biosystems), according to the manufacturer's instructions. Briefly, 800 ng of total RNA samples was used as template for eight multiplex reverse transcriptions containing up to 48 specific primers, using the Multiplex RT for TaqMan miRNA Assays Kit (Applied Biosystems) under conditions defined by the supplier. Each cDNA generated was amplified by quantitative PCR using 365 sequence-specific primers from the TaqMan miRNA Assays Human Panel on an Applied Biosystems 7900 Fast Real Time PCR system. Absolute threshold cycle values (Ct) were determined with the SDS 2.2 software (Applied Biosystems). A threshold value was determined for each miRNA and used for all the 180 samples. All signals with a Ct value of ≥34 (background threshold) were manually set to undetermined. Indeed, we considered miRNA with a Ct value of <34 as an “expressed miRNA.” Values were normalized across individuals using median normalization and were reported as an expression relative to the population mean for each miRNA as described (Deutsch et al. 2005; Prandini et al. 2007). Log2 values were used for the association analysis. TaqMan miRNA data sets have been submitted to the NCBI Gene Expression Omnibus (GEO) database under accession number GSE24610.
Genotyping
Genotyping was performed using the Illumina Hap550 or Hap550-duo arrays. Genotype calling was performed using the BeadStudio 3.1 software. SNPs were filtered in a stepwise fashion using the following criteria: (1) a SNP call frequency of at least 99%, (2) cluster separation greater than 0.3, (3) SNPs with Het Excess values between [−1.0 to −0.1] and [0.1 to 1.0] were removed, (4) SNPs that violate Hardy-Weinberg equilibrium (HWE = P < 0.05) were removed, (5) SNPs with a minimum allele frequency (MAF) < 0.02 were removed (at least seven heterozygous in our sample). After filtering, 479,314 SNPs were retained for statistical analyses. Genotyping data sets have been submitted to the European Genome-phenome Archive (EGA) database under accession number EGAS00000000056.
Genome-wide and cis-association analysis
eQTLs were detected using linear regression as implemented in the PLINK package (Purcell et al. 2007). For the cis-analysis, the association of genotype with expression levels was calculated for each miRNA within a 2-Mb window around its transcription start site (1 Mb either side). Association was also calculated using Spearman's rank correlation and was compared to the extreme P-value distribution of similar associations calculated for 10,000 permutations of the expression phenotype for each miRNA (permutation threshold) as previously reported (Stranger et al. 2007; Dimas et al. 2009). We applied a permutation threshold of 0.05 per gene, and we subsequently estimated the FDR on our number of discoveries based on the fact that we expected 5% of the miRNA genes to have a significant signal under the null. This design, which we have extensively applied in the past (Stranger et al. 2005, 2007; Bartel 2009; Dimas et al. 2009; Montgomery et al. 2010), allows for simultaneous assessment of the multiple testing effect of all markers tested within a 2-Mb window as well as across all phenotypes tested. For visualization and graphical displays, we used WGAviewer (Ge et al. 2008).
Gene Ontology annotation analysis
Analysis were conducted using Bioconductor GO stats version 2.8.0 and annotation Ms.eg.db version 2.2.6 packages (FDR adjusted P-value < 0.05) (Falcon and Gentleman 2007).
Expression clustering analysis
Hierarchical clustering was performed using Pearson correlation as a similarity measure and average linkage as an agglomerative hierarchical clustering algorithm.
Statistical analysis for master regulator identification
We tested for each SNP how many miRs were associated using an unadjusted P-value < 10−6. To estimate the significance for our findings, we permuted 1000 times the miR expression phenotypes (preserving the miR expression matrix per individual) and performed GWAS for each permuted data set.
Acknowledgments
We thank P. Descombes and the members of the genomics platform of the University of Geneva for their assistance, A.J. Sharp for comments on the manuscript, and Vital-IT for computational support. This study was funded by the Swiss National Science Foundation, the National Center for Competence in Research (NCCR) “Frontiers in Genetics,” the European Union FP6 “AnEUploidy” integrated project, and the Infectigen, ChildCare, and J. Lejeune foundations.
Footnotes
[Supplemental material is available online at http://www.genome.org. The miRNA expression data from this study has been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE24610. The genotyping data from this study have been submitted to the EMBL-EBI European Genome-phenome Archive (http://www.ebi.ac.uk/ega/) under accession no. EGAS00000000056.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109371.110.
References
- Ambros V 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33: 422–425 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET 2008. From gene expression to disease risk. Nat Genet 40: 492–493 [DOI] [PubMed] [Google Scholar]
- Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, Parand L, Gagnebin M, Rougemont J, Jongeneel CV, et al. 2005. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum Mol Genet 14: 3741–3749 [DOI] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325: 1246–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB 2008. WGAViewer: Software for genomic annotation of whole genome association studies. Genome Res 18: 640–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, et al. 2007. Identification of common genetic variation that modulates alternative splicing. PLoS Genet 3: e99 doi: 10.1371/journal.pgen.0030099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human microRNA targets. PLoS Biol 2: e363 doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH 2006. The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Serre D, Zuzan H, Clark TA, Schweitzer A, Staples MK, Wang H, et al. 2007. Heritability of alternative splicing in the human genome. Genome Res 17: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J 2008. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40: 225–231 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini P, Deutsch S, Lyle R, Gagnebin M, Delucinge Vivier C, Delorenzi M, Gehrig C, Descombes P, Sherman S, Dagna Bricarelli F, et al. 2007. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet 81: 252–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Enright AJ, Griffiths-Jones S 2008. Annotation of mammalian primary microRNAs. BMC Genomics 9: 564 doi: 10.1186/1471-2164-9-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE 2007. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet 81: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG 2007. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39: 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM 2007. Gene-expression variation within and among human populations. Am J Hum Genet 80: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, et al. 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet 1: e78 doi: 10.1371/journal.pgen.0010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K 2009. MicroRNA control in the immune system: Basic principles. Cell 136: 26–36 [DOI] [PubMed] [Google Scholar]
- Zhang W, Duan S, Bleibel WK, Wisel SA, Huang RS, Wu X, He L, Clark TA, Chen TX, Schweitzer AC, et al. 2009. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum Genet 125: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]