Abstract
LSH, a member of the SNF2 family of chromatin remodeling ATPases encoded by the Hells gene, is essential for normal levels of DNA methylation in the mammalian genome. While the role of LSH in the methylation of repetitive DNA sequences is well characterized, its contribution to the regulation of DNA methylation and the expression of protein-coding genes has not been studied in detail. In this report we investigate genome-wide patterns of DNA methylation at gene promoters in Hells−/− mouse embryonic fibroblasts (MEFs). We find that in the absence of LSH, DNA methylation is lost or significantly reduced at ∼20% of all normally methylated promoter sequences. As a consequence, a large number of genes are misexpressed in Hells−/− MEFs. Comparison of Hells−/− MEFs with wild-type MEFs and embryonic stem (ES) cells suggests that LSH is important for de novo DNA methylation events that accompany the establishment and differentiation of embryonic lineage cells. We further show that the generation of normal DNA methylation patterns and stable gene silencing at specific promoters require cooperation between LSH and the G9a/GLP complex of histone methylases. At such loci, G9a recruitment is compromised when LSH is absent or greatly reduced. Taken together, our data suggest a mechanism whereby LSH promotes binding of DNA methyltransferases and the G9a/GLP complex to specific loci and facilitates developmentally programmed DNA methylation and stable gene silencing during lineage commitment and differentiation.
Methylation of cytosine at carbon 5 of the pyrimidine ring (5-meC) in the context of CpG dinucleotides is an abundant modification in vertebrate genomes. In mammals, 70%–80% of all CpGs are methylated with the exception of promoter-associated CpG-rich sequences (CpG islands), and this modification contributes to genome stability, transcriptional silencing of genes and retrotransposons, regulation of genomic imprinting, and X chromosome inactivation in females (Bird 2002; Jaenisch and Bird 2003). Although DNA methylation is relatively stable and heritable in differentiated somatic cells, it undergoes dramatic reprograming during gametogenesis and early embryonic development (Reik et al. 2001). Immediately after fertilization and before DNA replication takes place in the mouse zygote, methyl-cytosine rapidly vanishes from the paternal genome (Oswald et al. 2000). In contrast, the maternal genome loses DNA methylation gradually, and during the late morula stages, the overall genomic content of 5-meC is low (Kafri et al. 1992; Oswald et al. 2000). The very first differentiation event generates a blastocyst with two distinct populations of cells—the pluripotent inner cell mass, later destined to form the embryo proper, and the trophoectoderm, which gives rise to placenta. These differentiation events are also accompanied by reestablishment of DNA methylation (Kafri et al. 1992; Reik et al. 2001; Reik 2007). In the mature blastocyst, the cells of the inner cell mass accumulate more 5-meC than the trophoectoderm, which remains relatively undermethylated (Reik 2007; Ng et al. 2008).
The establishment of de novo DNA methylation in the embryo proper requires DNA methyltransferase enzymes DNMT3A and DNMT3B, while its maintenance is performed mainly by DNMT1 (Okano et al. 1999; Jaenisch and Bird 2003; Goll and Bestor 2005). Embryos null for Dnmt1 or lacking Dnmt3a and Dnmt3b arrest early in gestation and die around day 9.5 (Li et al. 1992; Okano et al. 1999; Jaenisch and Bird 2003). The early lethality of Dnmt knockouts has been attributed to genomic instability resulting from activation of endogenous retroviruses and demethylation of major and minor satellite repeats (Okano et al. 1999; Walsh and Bestor 1999). Nevertheless, Dnmt1−/− as well as Dnmt3a−/−; Dnmt3b−/− embryonic stem (ES) cells are viable and can be maintained in culture (Li et al. 1992; Okano et al. 1999; Jaenisch and Bird 2003). Dnmt1−/− ES cells show 80% loss of DNA methylation and either rapidly die when induced to differentiate or produce trophoectoderm lineage cells (Jackson et al. 2004; Ng et al. 2008).
Although DNMT enzymes play a central role in the establishment and maintenance of DNA methylation, there are additional proteins whose absence affects the levels and patterns of 5-meC in the mouse genome (Dennis et al. 2001; Esteve et al. 2006; Ikegami et al. 2007; Sharif et al. 2007). One such protein is the putative chromatin remodeling ATPase LSH, also known as HELLS and PASG, encoded by the Hells gene. Initially identified as a factor required for proliferation of lymphoid cells, LSH is homologous to Arabidopsis thaliana Deficient in DNA Methylation 1 (DDM1) and closely related to the ISWI family of chromatin remodelers (Geiman and Muegge 2000; Dennis et al. 2001; Meehan et al. 2001). The LSH-null (Hells−/−) mice lose up to 70% of 5-meC from their genome but survive through gestation and die soon after birth with reduced weight (Dennis et al. 2001; Geiman et al. 2001). Interestingly, Hells−/− embryos and cells derived from them display loss of DNA methylation from various repetitive sequences, including major and minor satellite repeats, comparable to Dnmt knockouts (Dennis et al. 2001; Muegge 2005). Few single copy genes affected by LSH deficiency have also been identified (Dennis et al. 2001; Fan et al. 2005; Xi et al. 2007). It has been proposed that LSH remodels chromatin and thereby regulates the accessibility of DNA to de novo DNA methyltransferases DNMT3A and DNMT3B (Meehan et al. 2001; Brzeski and Jerzmanowski 2003). LSH may also serve as a recruitment factor for DNMTs as it interacts with DNMT3A and DNMT3B and, indirectly via DNMT3B, with DNMT1 and histone deacetylases HDAC1 and HDAC2 (Zhu et al. 2006; Myant and Stancheva 2008). Experiments using episomal plasmids and ES cells differentiating in culture have shown that LSH is essential for de novo DNA methylation of exogenous sequences and pluripotency genes but is largely dispensable for maintenance of DNA methylation (Zhu et al. 2006; Xi et al. 2009). However, a knockdown of LSH in primary human fibroblasts leads to a progressive loss of DNA methylation from satellite 3 sequences and other repeats, indicating that apart from facilitating de novo DNA methylation, LSH may have a role in the maintenance of DNA methylation at repetitive sequences (Suzuki et al. 2008).
So far, it is unclear to what extent the lack of LSH affects DNA methylation at unique, protein coding sequences in the genome and when during development do the defects in DNA methylation occur in the Hells−/− mice. Does LSH cooperate with other factors, apart from DNMTs, that affect the global levels and patterns of DNA methylation in mammalian cells (Ikegami et al. 2007; Epsztejn-Litman et al. 2008; Tachibana et al. 2008)?
To address these questions, we used methyl-CpG binding domain affinity purification (MAP) of methylated DNA (Illingworth et al. 2008) combined with promoter-specific microarrays to investigate the patterns of DNA methylation in Hells−/− mouse embryonic fibroblasts (MEFs) in comparison with wild-type MEFs and wild-type and EHMT2 (G9a) histone methylase null ES cells. We found that in LSH-deficient cells, DNA methylation is lost or reduced from a large subset of promoters, including genes that normally undergo de novo DNA methylation during the commitment of blastocyst cells to embryonic lineage as well as loci that become methylated during differentiation. As a consequence of LSH deficiency, many genes are inappropriately expressed in Hells−/− MEFs. In addition, we show that the establishment of DNA methylation and stable silencing of pluripotency genes during differentiation of ES cells require cooperation between LSH and G9a. Taken together, our data suggest that LSH promotes de novo DNA methylation and G9a recruitment at specific loci during embryonic lineage commitment and differentiation.
Results
DNA methylation patterns in Hells−/− MEFs
Similar to its plant homolog DDM1, LSH is essential for DNA methylation of repetitive genomic sequences (Geiman et al. 2001; Muegge 2005). Given that less than 30% of 5-meC remains in the genome of LSH-deficient cells (Fig. 1A–C), we decided to investigate in detail the patterns of DNA methylation at unique loci in wild-type and Hells−/− primary MEFs. We purified methylated DNA from both types of cells and hybridized these samples as well as total genomic DNA to microarrays containing approximately 22,036 promoter sequences of mouse protein-coding (RefSeq) genes (Fig. 1D). Each promoter region is represented on the array by 25 probes spanning 2000 bp upstream of and 500 bp downstream from the transcription start site (TSS). As ∼50% of the mouse promoters belong to the class of CpG islands, which do not extend further than 500 bp upstream of the TSS, we subdivided the 2.5-kb regions into “promoters” (+500 to −500 bp relative to TSS) and “upstream regions” (−1500 to −500 from TSS) and analyzed separately the hybridization signal for these two regions on the microarrays. Taking into account that CpG-rich methylated promoters enrich in MAP and MeDIP assays more efficiently than do those with intermediate and low CpG density (Weber et al. 2007; Mohn et al. 2008), we used log2 MAP/input values specific to each of the three classes to identify methylated promoters in wild-type and Hells−/− MEFs (for details, see Supplemental Information). We found 1792 promoters, ∼8% of the total number present on the microarrays, to be methylated in wild-type MEFs (Supplemental Fig. 1). Of those, 376 showed significant (two- to sixfold) reduction of DNA methylation in Hells−/− MEFs compared with the wild-type cells (Fig. 2A, promoters; Supplemental Table 1). Interestingly, a smaller, but detectable, number of promoter sequences (92) showed a gain of DNA methylation in Hells−/− cells (Fig. 2A,E; Supplemental Fig. 4C; Supplemental Table 1). This indicates that in addition to the loss of 5-meC from a subset of gene promoters, LSH deficiency results in rearrangement of DNA methylation patterns at other loci throughout the genome. A similar trend was observed at the sequences upstream of promoters, which may contain essential regulatory elements (Fig. 2A, upstream; Supplemental Table 2). In summary, ∼20% of all methylated promoters, including CpG-rich, intermediate, and CpG poor (Fig. 2B), and ∼17% of all methylated upstream regions show greatly reduced DNA methylation in Hells−/− MEFs.
Figure 1.
DNA methylation profiling of wild-type and Hells−/− MEFs. (A) Western blot on nuclear extracts from wild-type and Hells−/− (LSH-null) MEFs detected with anti-LSH and anti-HDAC1 antibodies. (B) South-Western assay with anti-5meC antibody on genomic DNA purified from wild-type and Hells−/− MEFs. (C) Quantification of the South-Western assay shown in B. Error bars, SD from the mean. (D) MBD affinity purification of methylated DNA from wild-type and Hells−/− MEFs. Input DNA and 5-methylcytosine (5-meC)-enriched MAP samples were labeled with Cy-dyes and cohybridized to a high-density oligonucleotide microarray representing all promoters of mouse RefSeq genes.
Figure 2.
Loss and gain of DNA methylation in Hells−/− MEFs. (A) Scatter plots comparing average DNA methylation values (log2 MAP/input) from replicate microarrays for all 1-kb promoter regions (left) and 1-kb upstream regions (right) in wild-type (y-axis) versus Hells−/− MEFs (x-axis). The red spots indicate sequences showing more than twofold reduction ([log2 MAP/input WT] − [log2 MAP/input Hells−/−] ≥ 1) of DNA methylation in Hells−/− MEFs. The blue spots are sequences showing more than twofold gain ([log2 MAP/input Hells−/−] − [log2 MAP/input WT] ≥ 1) of DNA methylation in Hells−/− MEFs. “n” indicates the number of promoters/upstream regions displaying loss or gain of DNA methylation. “ρ” is the Spearman correlation coefficient. (B) Methylated promoters in wild-type and Hells−/− MEFs divided to low (LCP), intermediate (ICP), and high CpG density (HCP) promoters. The numbers and percentage of promoters in each class are indicated. (C,D) Loss of DNA methylation in Hells−/− MEFs at single genes (C) and clusters of neighboring genes (D). The gray boxes represent tiled 2.5-kb regions. The transcripts originating from these regions are shown above the gray boxes; TSSs are indicated by arrows. The scale represents log2 MAP/input values. The distances between individual promoters are shown in kilobytes. (E) Gain of DNA methylation at Dbt promoter on chromosome 3 in Hells−/− MEFs. (F) A histogram showing the number of LSH-dependent clusters and number of genes per cluster. (G) A heath map comparison of DNA methylation patterns between wild-type ES cells, wild-type MEFs, and Hells−/− MEFs. The three groups of LSH-dependent hypo- and hypermethylated promoters are indicated together with the most significant representative biological functions of the genes in each group (Gene Ontology, P < 0.0001).
When we mapped the position of LSH-dependent hypo- and hypermethylated loci on mouse chromosomes, we observed that they are distributed throughout the genome (Supplemental Fig. 2). Although a proportion of these loci are represented by single genes (e.g., see Fig. 2C), in many cases the hypomethylated promoters in the Hells−/− MEFs tend to cluster together (Fig. 2D). Ninety-seven such clusters can be found on all mouse chromosomes, except Y, and contain from two to 18 promoters (Fig. 2F; Supplemental Table 3). These patterns suggest that LSH activity can spread over large chromosomal domains.
To gain an insight into when during development DNA methylation is normally established at the loci showing LSH dependency, we also compared promoter DNA methylation between wild-type ES cells and wild-type and Hells−/− MEFs (Fig. 2G). These analyses distinguished three groups of LSH-dependent DNA methylation patterns. The first, and largest, group includes promoters that are methylated in wild-type ES cells and MEFs but lack DNA methylation in Hells−/− MEFs. Among these are trophectoderm-specific Rhox and Elf5 loci and genes involved in signal transduction, differentiation, morphogenesis, and cell cycle control (Supplemental Fig. 3). The presence of DNA methylation at these promoters in ES cells suggests that they undergo de novo methylation early, before, or during establishment of pluripotent embryonic lineage cells. The second group of promoters is methylated in MEFs but not in ES cells and Hells−/− MEFs, indicating that these de novo DNA methylation events have taken place during differentiation of embryonic cells. This group includes pluripotency markers, imprinted loci, and genes involved in neurogenesis and ion homeostasis. The third, and smallest, group of promoters shows gain of DNA methylation in Hells−/− MEFs and represents genes involved in metabolic processes, cell differentiation, and proliferation. As promoters in this group lack methylation in ES cells and wild-type MEFs, we cannot unambiguously establish the timing and origin of these methylation events. Altogether, our analyses indicate that LSH facilitates de novo DNA methylation at specific loci during the establishment and differentiation of embryonic lineage cells.
LSH is required for DNA methylation of the Rhox gene cluster
The largest region affected by LSH deficiency is the Rhox cluster of reproductive homeobox genes located on the X chromosome (Fig. 3A), in this case the sole X, as both the wild-type and the Hells−/− MEFs are male. The majority of Rhox genes are expressed specifically in the trophectoderm and placenta, as well as in male and female reproductive tissues (testis and ovaries) (Maclean et al. 2005). In ES and most somatic cells, these genes are transcriptionally silenced and heavily methylated at their promoter sequences (Maclean et al. 2005; Oda et al. 2006). Previous studies have shown that the entire Rhox cluster undergoes DNMT3A/3B-dependent de novo DNA methylation and transcriptional silencing in the inner cell mass of the developing blastocyst and the embryo proper, but remains unmethylated and expressed in trophectoderm (Oda et al. 2006). In our experiments, we observed partial loss of DNA methylation from the promoter of the Gm9 gene, located immediately upstream of the Rhox cluster, and complete loss of DNA methylation from the promoters of 15 out of 22 Rhox genes represented on the arrays (Supplemental Tables 1, 3). The promoters of the remaining seven Rhox genes are extremely CpG-poor, and these sequences may not enrich sufficiently well by MAP. The promoter of cullin 4B, a gene located downstream from the Rhox cluster, is methylated in wild-type MEFs as well as in Hells−/− cells. Altogether, the domain affected by LSH deficiency spans ∼1 Mbp of the X chromosome (Fig. 3A).
Figure 3.
DNA methylation of Rhox gene cluster is LSH-dependent. (A) Schematic representation of the Rhox gene cluster and neighboring genes on mouse chromosome X A.33. The light-colored genes are provisional or predicted RefSeq. The GC content of the region is shown as a histogram. (B) DNA methylation at the promoters of the Rhox genes and genes neighboring the Rhox cluster was examined by PCR following digestion with methylation-sensitive HpaII (H) or methylation insensitive MspI (M) restriction enzymes of genomic DNA from wild-type and Dnmt3a/3b−/− ES cells as well as wild-type and Hells−/− MEFs. “−” represents undigested DNA. “Mr” is a DNA size marker. (C) Bisulfite DNA sequencing confirms lack of DNA methylation at Rhox2a and Rhox9 promoters, but not at the Culin4b promoter in Hells−/− compared with the wild-type MEFs. Methylated CpGs are shown as black circles. The average percentage of methylated CpGs relative to the total number of CpGs investigated by bisulfite DNA sequencing for any given sequence is shown in parentheses.
We used a methylation-sensitive PCR assay (see Methods) as well as bisulfite DNA sequencing to validate our microarray data. We investigated in detail the DNA methylation at the promoters of several Rhox genes and genes located either upstream of or downstream from the Rhox cluster in wild-type and Hells−/− MEFs. As the Rhox loci are hypomethylated in DNMT-deficient cells (Oda et al. 2006), we used wild-type and Dnmt3a/3b−/− ES cells as controls. A methylation-sensitive PCR assay showed that the promoters of Gm9, Rhox2, Rhox5, Rhox6, Rhox9, Rhox11, and Rhox13 were methylated in wild-type ES cells and MEFs, but not in Dnmt3a/3b−/− and Hells−/− cells (Fig. 3B). The Cullin 4b promoter was the only exception, as it was methylated in all cell lines with the exception of DNMT3A/3B-deficient cells. In agreement with the microarray data and methylation-sensitive PCR, bisulfite DNA sequencing confirmed that DNA methylation was either absent or significantly reduced at the promoters of Gm9, Rhox2a, Rhox6, Rhox9, and other loci in the Hells−/− MEFs compared with wild-type cells (Fig. 3C; Supplemental Fig. 4A,B). In contrast, we detected only minor differences in the number of methylated CpGs at the Culin4b promoter (Fig. 3C). Interestingly, the hypomethylation at the Rhox cluster in the Hells−/− MEFs was not restricted to gene promoters, as sequences located several kilobases upstream of and downstream from the Rhox6 promoter also displayed significantly reduced methylation when examined by bisulfite DNA sequencing (Supplemental Fig. 4A). This largely resembles the patterns observed at the Rhox loci in Dnmt3a/b −/− embryos and ES cells (Oda et al. 2006). Notably, a stable knockdown of LSH in the ES cells did not lead to hypomethylation of Rhox2a or other Rhox promoters (Supplemental Fig. 5A–C; data not shown), indicating that LSH is largely dispensable for maintenance of DNA methylation at these loci. Taken together, our data suggest that LSH, similar to DNMT3A/3B, is required for the developmental stage- and lineage-specific establishment of DNA methylation at the Rhox gene cluster and other loci.
Gene expression patterns in Hells−/− MEFs
Previous studies have shown that the failure to establish DNA methylation at the Rhox loci results in the misexpression of these genes in Dnmt3a/3b−/− embryonic day (E) 9.5 embryos, while they are normally methylated and stably silenced in wild-type littermates (Oda et al. 2006). In agreement with this, RT-PCR experiments showed that Gm9 and a proportion of the Rhox genes (Rhox2a, Rhox6, and Rhox9) were expressed in Hells−/− cells, but not in wild-type MEFs (Fig. 4A). Despite being hypomethylated, Rhox11 and Rhox13 were not expressed in Hells−/− MEFs, indicating that they can be silenced by other means in the absence of promoter DNA methylation. In Hells−/− MEFs, but not in wild-type cells, we could also detect hypomethylation and the expression of imprinted genes Mkrn3, Ndn, and Peg12 located on chromosome 7 (Fig. 4B; Supplemental Fig. 6A,B); pluripotency genes Dppa2 and Dppa4 (Fig. 4C); and, in agreement with previous studies (Xi et al. 2007, 2009), homeobox genes Hoxa5, Hoxa6, and Hoxa7 (Fig. 4D). The expression of Hox loci was somehow surprising given that the promoters of all the genes in the Hoxa cluster as well as other Hox loci were heavily methylated in the wild-type and Hells−/− MEFs according to the microarray data (Supplemental Fig. 6C; Supplemental Table 3).
Figure 4.
Changes in gene expression in Hells−/− MEFs. (A) RT-PCRs detect expression of the normally silenced Gm9 and Rhox genes in Hells−/− MEFs. (B) Expression of the imprinted genes Mkrn3, Ndn, and Peg12 of the mouse PWS imprinted locus on chromosome 7 in Hells−/− and wild-type MEFs. Ube3a is a neighboring imprinted gene. (C) Pluripotency markers Dppa2 and Dppa4, but not Dppa3 and Gdf3, are expressed in Hells−/− MEFs. (D) As previously reported (Xi et al. 2007), some Hox genes, including Hoxa5, Hoxa6, and Hoxa7, are aberrantly expressed in Hells−/− MEFs. In A–D, the triangles represent an increasing concentration of cDNA. “Mr” is a DNA size marker. (E) Global changes in gene expression in Hells−/− MEFs compared with wild-type controls as detected by expression microarrays. “n” indicates the number of up-regulated fourfold or more (log2 Hells−/−/wild type ≥ 2) and down-regulated fourfold or more (log2 wild type/Hells−/− ≥ 2) transcripts. Log2 values of normalized intensities for Hells−/− MEFs (y-axis) versus wild-type MEFs (x-axis) are shown.
A global investigation of the gene expression in Hells−/− cells compared with wild-type MEFs identified a large number of misexpressed transcripts. Seven hundred forty-four of these were up-regulated and 502 were down-regulated by fourfold or more (Fig. 4E), including one-third of significantly hypo- or hypermethylated loci in Hells−/− MEFs. Taken together, the number of misexpressed transcripts accounts for 4.8% of all protein coding genes. Although only a proportion (∼10%) of these expression changes can be directly attributed to significant (twofold or more) loss or gain of DNA methylation from the promoters or upstream regulatory sequences, it is clear that many genes are inappropriately expressed in LSH-deficient MEFs.
Methylation of Rhox loci requires G9a/GLP complex of histone methylases in addition to LSH
Studies from several laboratories have implicated histone methylase G9a (EHMT2) in the establishment and maintenance of DNA methylation patterns in the mouse genome via interactions with DNMT1 and DNMT3A/3B (Esteve et al. 2006; Epsztejn-Litman et al. 2008). G9a forms a heterodimer with a similar enzyme, GLP (also known as EHMT1), and the stable G9a/GLP complex is responsible for most of the dimethylation of histone H3 at lysine 9 in chromatin as well as DNA methylation at repetitive and unique loci, including Oct4 (also known as Pou5f1), in differentiating ES cells (Tachibana et al. 2002, 2005; Epsztejn-Litman et al. 2008). Interestingly, the DNA methylation facilitated by the G9a/GLP complex is independent of the histone H3K9 methylation activity of these enzymes, as DNA methylation at specific loci can be restored to a large extent in Ehmt2−/− (G9a-null) ES cells expressing a transgene with inactivating point mutations (Y1120V; Y1207F) in the G9a catalytic domain (Dong et al. 2008; Tachibana et al. 2008).
We noticed that DNA methylation was absent from the promoter of the G9a/GLP target gene Wfdc15 (Tachibana et al. 2008) in Hells−/− MEFs, despite the normal levels of G9a protein and histone H3K9 mono-, di-, and trimethylation in these cells (Fig. 5A,B). This led us to examine the overall levels and specific patterns of DNA methylation in Ehmt2−/− ES cells and cells expressing the catalytically inactive Ehmt2 (G9a) transgene (Tg) on a Ehmt2−/− background. Despite the normal levels of LSH (Fig. 5A) and DNMT enzymes (Tachibana et al. 2008), the 5-meC was reduced by 50% in the genome of Ehmt2−/− ES cells compared with the wild type (Fig. 5C). A proportion of 5-meC (18%) was restored in Ehmt2−/− cells expressing the catalytically inactive G9a protein (Fig. 5C, Ehmt2−/− Tg). Investigation of the Rhox and Elf5 promoters, which also display LSH-dependent DNA methylation (group I), by methylation-sensitive PCR and/or bisulfite DNA sequencing showed that all these loci were methylated in wild-type ES cells but were significantly hypomethylated in Ehmt2−/− ES cells (Fig. 5D,E). DNA methylation was partially restored in ES cells expressing the Ehmt2 transgene (Fig. 5D,E). However, we did not observe a complete loss of DNA methylation from the Rhox2 and Elf5 promoters in Ehmt2−/− ES cells. This suggests that DNMT3A and DNMT3B can access and methylate promoter sequences, although inefficiently, in the absence of G9a. In agreement with single gene analyses, global investigation of DNA methylation in Ehmt2−/− ES cells by MAP coupled with microarrays identified 153 hypomethylated promoters; 56 (36.6%) of these were shared with promoters hypomethylated in Hells−/− MEFs compared with wild-type ES and MEFs (Fig. 5F). Taken together, these data suggest that LSH and the G9a/GLP complex act independently of each other at some loci and cooperatively at a subset of specific promoters.
Figure 5.
The Rhox and other LSH-dependent loci are hypomethylated in Ehmt2−/− ES cells. (A) Western blots show G9a, LSH, and HDAC1 in nuclear extracts from wild-type and mutant ES cells and MEFs. (B) Western blots for H3K9me1, H3K9me2, and H3K9me3 in wild-type and mutant ES cells and MEFs. “Ehmt2−/− Tg” indicates Ehmt2−/− (G9a-null) cells expressing catalytically inactive Ehmt2 (G9a) transgene. (C) ELISA assays with anti-5meC antibody on genomic DNA purified from wild-type and mutant ES cells and MEFs. Note than Hells−/− MEFs and Ehmt2−/− ES cells have comparable amounts of 5-meC. Catalytically inactive Ehmt2 transgene partially rescues DNA methylation defects in Ehmt2−/− ES cells. (D,E) Methylation-sensitive PCR (D) and bisulfite sequencing (E) detect loss of DNA methylation at Rhox and other LSH-dependent promoters in Ehmt2−/− ES cells. DNA methylation is partly restored in Ehmt2−/− cells expressing catalytically inactive Ehmt2 transgene (Tg). MspI and HpaII digested genomic DNA is indicated by “M” and “H,” respectively. “Mr” is DNA size marker. In F, methylated CpGs are shown as black circles. The percentage of methylated CpGs relative to the total number of CpGs investigated by bisulfite sequencing for any given sequence is shown in parentheses. (F) A Venn diagram showing the overlap between hypomethylated (relative to ES cells and MEFs, group I) promoters in Hells−/− MEFs and hypomethylated loci in Ehmt2−/− ES cells.
G9a/GLP binding and H3K9 dimethylation are compromised in Hells−/− MEFs at specific loci
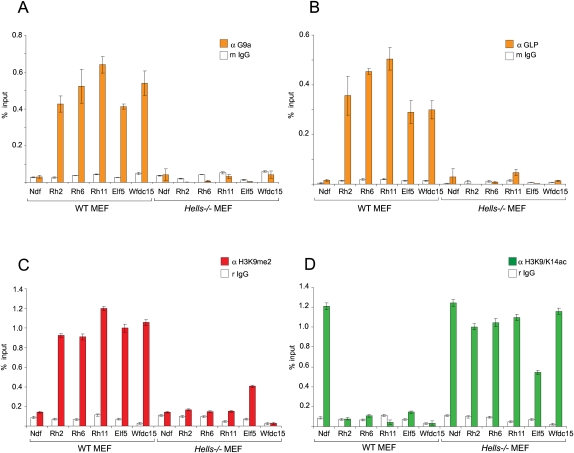
Given that the DNA methylation at Rhox, Elf5, Wfdc15, and other promoters cannot be correctly established in the absence of LSH and/or G9a, we asked if G9a/GLP complex binds to these loci in Hells−/− MEFs to methylate histone H3 at lysine 9. We performed chromatin immunoprecipitation (ChIP) experiments with antibodies against G9a, GLP, H3K9me2, and acetylated H3 followed by Q-PCR. These experiments showed that in wild-type MEFs the promoters of Rhox2a, Rhox6/9, Rhox11, Elf5, and Wfdc15 were hypoacetylated, enriched in dimethylated H3K9, and bound by G9a/GLP compared with the control promoter of the Ndufa1 gene located upstream of the Rhox gene cluster (Fig. 6A–D). In contrast, we could not detect G9a and GLP binding at these promoters in Hells−/− MEFs (Fig. 6A,B). H3K9me2 was also significantly reduced at these sequences, and the levels of H3 acetylation at the Rhox2a, Rhox6/9, Rhox11, Elf5, and Wfdc15 promoters in Hells−/− MEFs were comparable to H3 acetylation at the promoter of the expressed Ndufa1 gene (Fig. 6C,D). Notably, H3 acetylation at the Rhox11 and Elf5 promoters was independent of transcription, as these two genes are not expressed in Hells−/− MEFs (Fig. 4A; Supplemental Table 3). These data indicate that binding of G9a/GLP complex and establishment of silenced chromatin at the Rhox and other loci are compromised in the absence of LSH.
Figure 6.
H3K9me2 and G9a/GLP are absent from Rhox and other promoters in Hells−/− MEFs. (A,B) ChIP experiments detect loss of G9a and GLP binding (gray bars) from the promoters of Rhox2, Rhox6/9, Rhox11, Elf5 and Wfdc15 in Hells−/− compared with wild-type (WT) MEFs. The promoter of the expressed Ndufa1 gene was used as a control. Total mouse IgG served as a nonspecific control antibody (white bars). (C,D) ChIP assays detect loss of H3K9me2 (C) and gain of H3 K9/K14 acetylation at Rhox, Elf5, and Wfdc15 promoters in Hells−/− MAFs. All graphs show the average enrichment ± SD from two independent chromatin preparations and ChIP experiments performed and analyzed in triplicate.
LSH promotes recruitment of G9a to pluripotency genes during differentiation
To examine in more detail the cooperation of LSH with G9a, we followed the silencing of pluripotency genes during differentiation to embryoid bodies (EBs) of control ES cells expressing scrambled shRNA (shScr), ES cells with stable 80% shRNA knockdown of LSH (shLsh), wild-type, and Ehmt2−/− (G9a-null) ES cells. As previously reported (Xi et al. 2009), the levels of LSH decreased steadily during differentiation of ES cells (Fig. 7A). A decline of LSH levels was also observed in shLsh EBs, and LSH became virtually undetectable at day 6 of differentiation (Fig. 7A; Supplemental Fig. 5D). In contrast, G9a and HDAC1 remained constant throughout differentiation, while DNMT3B (and DNMT3A; data not shown) was present at steady amounts in ES cells and EBs at days 2 and 4 but was lost at day 6 (Fig. 7A). Quantitative investigation of mRNA levels of Oct4, Dppa2, Dppa3, and Dppa4, and Gdf3 relative to Gapdh showed that the pluripotency genes were expressed at similar levels in shScr, shLsh, wild-type, and Ehmt2−/− ES cells but were silenced 2.5-fold to sevenfold less efficiently in day 8 EBs when either G9a was absent or LSH was greatly reduced (Fig. 7B–D). Consistent with the mRNA levels, ChIP experiments detected reduced H3K9me2 and G9a binding and increased H3 acetylation at the Oct4; Dppa2, Dppa3, and Dppa4; and Gdf3 promoters at day 8 of differentiation in shLsh EBs compared with the controls (Fig. 7E). In Ehmt2−/− EBs at day 8, we also detected a lack of H9K9me2 and a modest increase of H3 acetylation (Fig. 7F).
Figure 7.
LSH is required for recruitment of G9a to promoters of pluripotency genes during differentiation. (A) Western blots display G9a, LSH, HDAC1, and DNMT3B protein levels in shScr and shLsh ES cells during differentiation to embryoid bodies (EB) at days 2, 4, and 6. (B) Fold down-regulation of pluripotency markers mRNA as detected by qRT-PCR in wild-type and mutant EBs at day 8 relative to ES cells. (C) mRNA expression ratios relative to GAPDH of pluripotency genes in shScr versus shLsh ES cells and EBs at day 8 of differentiation indicate that these genes have equal expression in shScr and shLsh ES cells, but higher expression in shLsh EB8 relative to shScr EB8. (D) mRNA expression ratios relative to GAPDH of pluripotency genes in wild-type versus Ehmt2−/− (G9a-null) ES cells and EB at day 8. (E) ChIP for acetylated H3, H3K9me2, and G9a at promoters of pluripotency genes in shScr and shLsh EBs at day 8 of differentiation. Ndf is a control promoter of constitutively expressed Ndufa1 gene. (F) ChIP for acetylated H3, H3K9me2, and G9a at promoters of pluripotency genes in wild-type and Ehmt2−/− EBs at day 8 of differentiation. In E and F, the scale on the left applies to H3ac and H3K9me2; the scale on the right applies to G9a. (G,H) ChIP enrichment profiles of acetylated H3, H3K9me2, LSH, and G9a at the promoter of the Dppa4 gene during differentiation of shScr and shLsh ES cells. The scale on the left applies to H3ac and H3K9me2; the scale on the right applies to LSH, G9a, and the nonspecific IgG.
We also followed the dynamics of binding of LSH and G9a as well as the levels H3K9me2 and H3 acetylation at Dppa4 promoter by ChIP throughout the differentiation process. We detected a peak of LSH binding at day 2, which decreased at days 4, 6, and 8 (Fig. 7G). The peak of LSH binding at day 2 also coincided with a sharp drop in H3 acetylation (Fig. 7G). In contrast, we observed a gradual increase in G9a binding and H3K9me2 from day 4 to day 8 (Fig. 7G). Compared with shScr, these events were compromised during differentiation of shLsh ES cells (Fig. 7H). H3 acetylation declined less steeply at days 2 and 4 and remained higher at day 8 of differentiation, while G9a binding and H3K9 dimethylation were dramatically reduced (Fig. 7H). We observed similar patterns at the Dppa3 promoter during the differentiation of shScr and shLsh cells (data not shown). In summary, these experiments demonstrate that LSH binding precedes G9a and promotes G9a recruitment and H3K9 dimethylation at promoters of pluripotency genes during differentiation.
Discussion
DNA methylation is essential for normal embryonic development in all vertebrate species studied so far (Li et al. 1992; Okano et al. 1999; Stancheva and Meehan 2000; Rai et al. 2006). Although DNMT enzymes are indispensable for the establishment and maintenance of the 5-meC patterns in the genome, other proteins such as LSH, the G9a/GLP complex, NUP95, ZFP57, DPPA3 (Stella), and LSD1 contribute as well (Dennis et al. 2001; Ikegami et al. 2007; Nakamura et al. 2007; Sharif et al. 2007; Wang et al. 2009). In this study, we examined in detail the genome-wide patterns of gene expression and DNA methylation at the gene promoters arising as a consequence of LSH loss during mouse development. In addition to the previously reported hypomethylation of repetitive sequences (Dennis et al. 2001), we found that Hells−/− MEFs exhibit loss and rearrangement of DNA methylation patterns at multiple unique genomic loci. This is accompanied by the misexpression of a large number of genes. Notably, many of the hypomethylated genes (53) are transcription factors. Therefore, some of the effects of LSH deficiency on gene expression might be indirect, as only a proportion of the expression changes can be directly attributed to significant loss or gain of DNA methylation.
Notably, LSH-dependent methylated loci include the trophectoderm-expressed Rhox cluster and Elf5 transcription factor, which are targeted by DNMT3A and DNMT3B in a lineage-specific manner early in mouse development (Oda et al. 2006; Ng et al. 2008). Given that some of these genes are inappropriately expressed in Hells−/− MEFs, we hypothesize that the function of LSH in promoting the action of DNMT enzymes is essential to reinforce the commitment to embryonic lineage fate by stable silencing of trophectoderm-specific genes. However, not all de novo DNA methylation events are LSH-dependent. Many promoters that are methylated in wild-type MEFs have normal levels of DNA methylation in Hells−/− MEFs. This may explain why DNMT-deficient embryos die during gestation while Hells−/− mice develop to birth.
We also found that LSH and G9a act cooperatively to promote DNA methylation at a subset of gene promoters, including the Rhox loci. A detailed investigation of methylated CpGs, G9a/GLP binding, and H3K9me2 at Rhox and other loci suggest that LSH may function “upstream” of the DNM3A/3B and G9a/GLP complex. Thus in LSH-deficient embryos and cells, DNMT3A/3B (Dennis et al. 2001) and G9a enzymes are intact, but they are unable to methylate DNA or chromatin at the LSH-dependent loci. In contrast, when G9a is absent, the levels of LSH and DNMTs are normal (Fig. 5; Tachibana et al. 2008), and DNA methylation occurs at these sequences, but with reduced efficiency. Our observations are consistent with previous findings that the Mage-a2 and Wfdc15a promoters, which are hypomethylated in Ehmt2−/− ES cells, display normal patterns of DNA methylation in Ehmt2−/− E9.5 embryos (Tachibana et al. 2008). Therefore G9a deficiency may lead to a delay rather than an absence of de novo DNA methylation during mouse embryogenesis.
Our experiments with ES cells differentiating in culture demonstrate further that LSH and G9a are required for the stable silencing of pluripotency genes and that LSH promotes recruitment of G9a to specific promoters. Given that we did not convincingly detect an interaction between LSH and the G9a/GLP complex in coimmunoprecipitation experiments (data not shown) and that LSH and G9a bind to the Dppa4 promoter at different times during differentiation, we hypothesize that removal of acetyl groups from H3 tails by LSH-associated histone deacetylases (Myant and Stancheva 2008; Zhou et al. 2009) may generate a suitable substrate for mono- and dimethylation of H3K9 and thus facilitate binding of the G9a/GLP complex to specific loci (Supplemental Fig. 7). It is important to acknowledge that in the Hells−/− MEFs some pluripotency genes remain expressed (Dppa2 and Dppa4) and others remain hypomethylated (Oct4, Dppa3, and Gdf3) but silenced. Therefore, additional, LSH-independent mechanisms for silencing of pluripotency genes must exist.
It is yet unclear whether LSH functions as a chromatin remodeler, which destabilizes nucleosomes to facilitate the access of DNMT3A/3B to DNA. LSH interacts with all three DNA methyltransferases (Zhu et al. 2006; Myant and Stancheva 2008; Xi et al. 2009) and may promote local accumulation of these proteins without inducing significant conformational changes in chromatin. It will be important to establish whether DNA methylation can take place in embryos or cells expressing a mutant LSH protein entirely lacking ATPase activity.
Our data also indicate that in addition to single promoters dispersed throughout the genome, LSH is required for the DNA methylation of large chromosomal domains often containing multiple genes. Given that chromatin remodeling proteins can cause rearrangement of positioned nucleosomes over extended genomic regions (Becker and Horz 2002; Whitehouse et al. 2007), it is likely that LSH functions as an active ATPase that affects chromatin structure in vivo. How LSH is recruited to specific loci in the genome and what determines the boundaries of LSH-dependent methylated domains is currently unclear. In most cases, these domains are flanked by either strong CpG islands or promoters of actively transcribed genes, as it is in the case of the Rhox cluster. Therefore, chromatin modifications associated with CpG islands and active promoters, such as histone H3K4 di- and trimethylation, may serve as a boundary that counteracts the function of LSH.
In summary, our investigation of LSH-deficient MEFs and ES cells demonstrates that LSH is essential for developmentally programmed de novo DNA methylation at the promoters of protein coding genes and recruitment of G9a/GLP complex to a subset of genomic loci.
Methods
Biological samples
Wild-type and Hells−/− MEFs (Zhu et al. 2006) were cultured in DMEM supplemented with 10% FCS and penicillin, streptomycin, and L-glutamine. Wild-type TT2 (c57BL/6xCBA), Ehmt2−/−, Ehmt2−/− Tg (G7), and Ehmt1−/− ES cells (Tachibana et al. 2008); wild-type (129S4/SvJae) and Dnmt3a/3b−/− ES cells (Okano et al. 1999; Oda et al. 2006); and ES cells with stably integrated pSEC-neo vectors expressing either scrambled or LSH shRNA (Zhu et al. 2006; Athanasiadou et al. 2010) were cultured in MEM supplemented with 10% FCS, nonessential amino acids, Na-pyruvate, β-mercaptoethanol, and LIF in T175 flasks coated with gelatin. shScr and shLsh ES cells were differentiated as described (Athanasiadou et al. 2010).
DNA methylation profiling by methyl-CpG binding domain (MBD) affinity purification (MAP)
MAP purification of methylated DNA was performed essentially as described by Cross et al. (1994). Genomic DNA from wild-type and Hells−/− MEFs and from wild-type and Ehmt2−/− ES cells was sonicated to an average size of 300 bp, and 30 μg was loaded on a 1-mL chromatography column (Tricorn, GE Healthcare) containing 50 mg of His-tagged MBD domain of MeCP2 protein bound to nickel-charged Chelating Sepharose Fast Flow (GE Healthcare). The column was washed with 10 volumes of buffer A (20 mM HEPES at pH 7.5, 100 mM NaCl, 0.1% Tween, 10% glycerol, 0.5 mM PMSF) followed by buffer A with increasing concentration of NaCl (0.1–0.7 M). Methylated DNA was eluded with buffer A with 1 M NaCl, concentrated, and subjected to whole-genome amplification (WGA kit, Sigma Aldrich) in parallel with the total sonicated genomic DNA. Amplified samples were labeled with Cy3 and Cy5, respectively, and cohybridized to the MM8 mouse RefSeq promoter microarrays (Roche NimbleGen). Final methylation log2 ratios of bound over input signals represent the average of two or three (for ES cells) independent experiments performed with independent biological replicas. The normalization of microarray data and analyses of promoter and upstream regions were carried out with the custom designed software “Prometheus” (I de Las Heras, YMA Sundaram, S Schatlowski, and I Stancheva, in prep.). Details concerning data analyses can be found in the Supplemental Materials and Methods. The data from promoter microarray experiments can be downloaded from ArrayExpress, accession nos. E-MEXP-2383 and E-MEXP-2872.
Gene expression analyses
Total RNA was purified using TRIzol reagent (Invitrogen). cDNA was synthesized from 100 μg of RNA using poly-dT primer and SuperscriptII reverse transcriptase (Invitrogen) and was hybridized in quadruplicate to MM8 4x72K expression microarrays (Roche NimbleGen). The raw data were quantile normalized using the limma package from the BioConductor project and were analyzed in R environment, using limma and standard R/BioConductor tools. The RT-PCRs and qRT-PCRs were performed on 120-, 60-, and 30-fold dilutions of cDNA using the primers listed in the Supplemental Methods. The expression data from the microarray experiments can be found on ArrayExpress, accession no. E-MEXP-2385.
Quantification of total DNA methylation
Total methylation in genomic DNA purified from wild-type or mutant cells was quantified using an Imptint Kit (Sigma Aldrich) according to the manufacturer's instructions. For South-Western analyses, sonicated genomic DNA was slot blotted to a Z-probe nitrocellulose membrane (Bio-Rad) and detected with 1 μg/mL anti-5meC antibody (Eurogentec) in 4% milk, 1× TBS buffer followed by a secondary anti-mouse IR800 antibody (LiCor Biosciences). DNA was stained with 0.002% of methylene blue dye. DNA and 5-meC signals were detected with Odyssey Imager (LiCor Biosciences) and quantified by Odyssey V.3.0 software.
Methylation-sensitive PCR
Five micrograms of genomic DNA was digested with either MspI or HpaII restriction enzymes and PCR amplified with primers flanking HpaII restriction sites in gene promoters. The list of primers can be found in the Supplemental Methods. Each amplicon contains two to four HpaII sites.
Bisulfite DNA sequencing
Bisulfite treatment of genomic DNA was carried out as described (Feil et al. 1994) and prepared for sequencing as outlined by Suzuki et al. (2007). Sequences of bisulfite-specific primers were as described (Oda et al. 2006) or were designed both manually and with the aid of the MethPrimer software and are listed in the Supplemental Materials and Methods.
Western blots
Nuclear extracts were prepared from wild-type and mutant cells by resuspending 1 × 107 cells in 1 mL of hypotonic NE buffer (100 mM HEPES at pH 7.5, 10 mM KCl, 1 mM MgCl2, 1% Triton) followed by Dounce homogenization. The nuclei were spun down at 3000 rpm for 5 min, resuspended in 0.5 mL of NE containing 300 units of Benzonase endonuclease (Merck), and incubated on ice for 30 min. NaCl was added to a 400 mM final concentration followed by rotation for 1 h at 4°C. Extracted nuclei were spun down for 15 min at 13,000 rpm, 4°C, to separate the insoluble fraction from the nuclear extract. Fifty micrograms of each nuclear extract was resolved on 8.5% or 15% SDS-PAGE, transferred to a nitrocellulose membrane (Bio-Rad), and detected by anti-LSH (Santa Cruz, sc-46665), anti-G9a (Perseus Proteomics), anti-HDAC1 (Santa Cruz, sc-7872), anti-DNMT3B (Peirce, PA1-884-C100), anti-monomethyl H9K9, anti-dimethyl H3K9, and anti-trimethyl-H3K9 (Millipore) antibodies followed by secondary anti-mouse IR800 and anti-rabbit IR680 (LiCor Biosciences). The Western blots were imaged on the Odyssey Imager (LiCor Biosciences) with the aid of Odyssey V3.0 software.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out as described elsewhere. Briefly, the cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by adding 2.5 M glycine to the final concentration of 125 mM. Chromatin was sonicated to achieve DNA fragments of an average size of 300–400 bp and immunoprecipitated with antibodies against G9a and GLP (Perseus Proteomics), H3K9me2 (Millipore, 07-212), acetylated H3 (Millipore, 06-599), LSH (Bethyl Laboratories, A300-226A), or total mouse and rabbit IgG (Sigma, I5381; I5006) immobilized on Protein G Dynabeads (Invitrogen). The immunoprecipitated chromatin was de–cross-linked overnight at 65°C; the DNA was purified by PureLink kit (Invitrogen) and analyzed by real-time PCR using SYBR Green master mix and 480 Light Cycler (Roche). The primers can be found in the Supplemental Materials and Methods.
Acknowledgments
We thank Kathrin Muegge (National Cancer Institute, Frederick, MD), Yoichi Shinkai (Institute for Virus Research, Kyoto University, Japan), En Li (Novartis Institute for Biomedical Research, Cambridge, MA) and Adrian Bird for kindly providing cell lines; Masaki Okano (Center for Developmental Biology, RIKEN, Kobe, Japan) for primer sequences; and Wendy Bickmore, Ken Sawin, Adrian Bird, and members of the Stancheva laboratory for helpful comments during the manuscript preparation. This work was supported by Cancer Research UK grants C7215/A4399 and C7215/A8983. J.B. is a BBSRC PhD student supported by a CASE studentship with CellCentric Ltd.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. E-MEXP-2383, E-MEXP-2385, and E-MEXP-2872.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108498.110.
References
- Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A 2010. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS ONE 5: e9937 doi: 10.1371/journal.pone.0009937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W 2002. ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A 2003. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem 278: 823–828 [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP 1994. Purification of CpG islands using a methylated DNA binding column. Nat Genet 6: 236–244 [DOI] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 15: 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K 2005. Lsh controls silencing of the imprinted Cdkn1c gene. Development 132: 635–644 [DOI] [PubMed] [Google Scholar]
- Feil R, Charlton J, Bird AP, Walter J, Reik W 1994. Methylation analysis on individual chromosomes: Improved protocol for bisulphite genomic sequencing. Nucleic Acids Res 22: 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Muegge K 2000. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci 97: 4772–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K 2001. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta 1526: 211–220 [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Ikegami K, Iwatani M, Suzuki M, Tachibana M, Shinkai Y, Tanaka S, Greally JM, Yagi S, Hattori N, Shiota K 2007. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6: e22 doi: 10.1371/journal.pbio.0060022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 24: 8862–8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev 6: 705–714 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Maclean JA II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF 2005. Rhox: A new homeobox gene cluster. Cell 120: 369–382 [DOI] [PubMed] [Google Scholar]
- Meehan RR, Pennings S, Stancheva I 2001. Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes Dev 15: 3231–3236 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Muegge K 2005. Lsh, a guardian of heterochromatin at repeat elements. Biochem Cell Biol 83: 548–554 [DOI] [PubMed] [Google Scholar]
- Myant K, Stancheva I 2008. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol 28: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M 2008. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 10: 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M 2006. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev 20: 3382–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA 2006. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol 26: 7077–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev 14: 313–327 [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Suzuki MM, Kerr AR, De Sousa D, Bird A 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res 17: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Farrar JE, Yegnasubramanian S, Zahed M, Suzuki N, Arceci RJ 2008. Stable knockdown of PASG enhances DNA demethylation but does not accelerate cellular senescence in TIG-7 human fibroblasts. Epigenetics 3: 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH 1999. Cytosine methylation and mammalian development. Genes Dev 13: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. 2009. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41: 125–129 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K 2007. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci 104: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S, Geiman TM, Briones V, Tao YG, Xu H, Muegge K 2009. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells 27: 2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Han L, Li G, Tong T 2009. Senescence delay and repression of p16INK4a by Lsh via recruitment of histone deacetylases in human diploid fibroblasts. Nucleic Acids Res 37: 5183–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K 2006. Lsh is involved in de novo methylation of DNA. EMBO J 25: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]