Abstract
We report on a hitherto poorly characterized class of genes that are expressed in all tissues, except in one. Often, these genes have been classified as housekeeping genes, based on their nearly ubiquitous expression. However, the specific repression in one tissue defines a special class of “disallowed genes.” In this paper, we used the intersection-union test to screen for such genes in a multi-tissue panel of genome-wide mRNA expression data. We propose that disallowed genes need to be repressed in the specific target tissue to ensure correct tissue function. We provide mechanistic data of repression with two metabolic examples, exercise-induced inappropriate insulin release and interference with ketogenesis in liver. Developmentally, this repression is established during tissue maturation in the early postnatal period involving epigenetic changes in histone methylation. In addition, tissue-specific expression of microRNAs can further diminish these repressed mRNAs. Together, we provide a systematic analysis of tissue-specific repression of housekeeping genes, a phenomenon that has not been studied so far on a genome-wide basis and, when perturbed, can lead to human disease.
Expression of housekeeping genes is generally believed to be required for the maintenance of general tasks that are common to all cells. For example, all living cells continuously need to produce sufficient ATP to stay alive. Most ATP is produced via oxidative phosphorylation in the mitochondrial inner membrane, a process requiring oxygen. When oxygen supply is low, such as during ischemia, cells can survive through ATP production via anaerobic glycolysis, which depends on lactate dehydrogenase (LDHA) (reducing pyruvate to lactate) and members of the monocarboxylic acid transporter (MCT) protein family to transport lactate and pyruvate across the plasma membrane. A remarkable exception is found in pancreatic insulin-producing beta-cells, in which glycolysis is exclusively aerobic (Schuit et al. 1997). Biochemically, beta-cells have vanishingly low expression levels of both the lactate/pyruvate transporter MCT1 (Zhao et al. 2001) and lactate dehydrogenases (Sekine et al. 1994; Schuit et al. 1997). Despite this tissue-specific repression, lactate dehydrogenase A (Ldha), for example, appears in many published lists of housekeeping genes (Warrington et al. 2000; Hsiao et al. 2001; Eisenberg and Levanon 2003; Tu et al. 2006). Inappropriate activation of a tissue-specifically repressed gene was previously demonstrated to be associated with human disease (Otonkoski et al. 2007). Gain-of-function mutation of the Mct1 promoter leads to uptake of lactate and pyruvate by the beta-cells and disruption of correct glucose sensing. When large amounts of lactate and pyruvate are generated by muscles during physical exercise, insulin is secreted, resulting in hypoglycemia and potential brain dysfunction (Otonkoski et al. 2003, 2007).
In this study, we investigated whether tissue-specific repression of genes such as Mct1 (Slc16a1) and Ldha is a typical phenotypic trait of the differentiated beta-cell or whether this phenomenon can be extended to other tissues. Therefore, we assessed gene transcription in various differentiated tissues and integrated bioinformatics and statistical methods to identify these tissue-specifically repressed genes. Furthermore, we investigated how this repression is established and when this occurs during tissue development. In this study, we show for the first time that a developmentally regulated process in pancreas and in liver involves the cell-selective inactivation of genes that are expressed everywhere else, a phenomenon we call “disallowance.”
Results
Identification of repressed genes
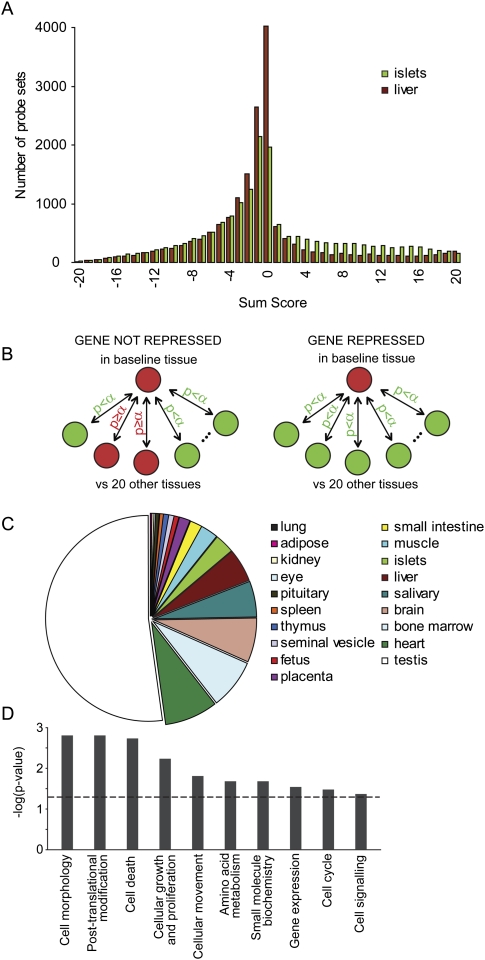
A multi-tissue mRNA expression panel was generated with Affymetrix MOE 430 2.0 microarrays and used to assess gene expression in 21 differentiated mouse tissues. In a first analysis, for each transcript, we compared its mRNA abundance in pancreatic islets versus each of the other tissues in the panel by t-tests. A sum score can then be calculated genewise by addition of the number of pairwise t-tests where expression in islets is significantly lower (−1) or higher (+1) as compared to the other tissues. In addition to the expected existence of islet-specific genes (significantly higher in islets than in each of the other tissues), we found evidence for genes whose expression was significantly lower in islets than in each of the other tissues, although they were fewer in number than the tissue-specific genes (Fig. 1A). This phenomenon was not restricted to islets, but was also observed with other tissues, as illustrated by comparing liver (baseline) to the panel of other tissues (Fig. 1A). We next established a statistical framework to screen systematically for genes that are specifically lower in one tissue as compared to each of the others (Fig. 1B). Both ANOVA and multiple comparison procedures (e.g., Dunnett or Bonferroni) are unsuitable because a significant result means that at least one pair of tissues differs significantly in expression, while we wanted evidence that expression in the baseline tissue is lower than in all other tissues. Therefore, we applied the intersection-union test (Berger 1982; Berger and Hsu 1996; Van Deun et al. 2009) (explained in greater detail in Methods). We did so for each of the 17,334 unique genes that are represented in the mRNA expression analysis and with each of the 21 tissues, in turn, acting as the baseline. This analysis largely confirmed the genes that were found repressed in pancreatic islets and liver in the sum score analysis (Fig. 1A), excluding the possibility that the outcome was an artifact of the filtering strategy. We used information from redundant probe sets (present for more than 7750 genes) to reduce genes found repressed due to a probe set downstream from an alternative polyadenylation site. The distribution of the total number of 1074 repressed genes over the 21 mouse tissues is shown in Figure 1C, and a full list is provided in Supplemental Table S1. Testis accounts for about half of the genes that were selected as being tissue-specifically repressed. We performed a leave-one-out analysis; i.e., we assessed the effect of leaving one tissue out of the data set on the extra number of genes found disallowed in the other tissues. Based on this, we generated a tissue association profile (Supplemental Table S2). The baseline tissues are listed in rows, and the tissues left out of the analysis in columns. Omitting one tissue has the greatest effect on testis, which is not surprising as testis already represents the largest set of disallowed genes. A strong association is found between skeletal muscle and heart, which are functionally closely related as tissues composed mostly of striated muscle fibers and between components of the immune system (spleen and bone marrow). We tested this set of 1074 repressed genes for associations with cellular and molecular functions (Fig. 1D) and found significant enrichment for genes involved in processes determining tissue development and function.
Figure 1.
Tissue-specific repression of housekeeping genes. (A) Frequency distribution of the sum score for islets (green) and liver (brown) as baseline. Sum score numbers are calculated genewise and are the addition of the number of pairwise t-tests where expression in baseline is significantly lower (−1) or higher (+1) as compared to the other tissues. Liver- and islet-specific genes reside in the category with the highest possible sum score (right), whereas genes of which expression is specifically repressed in islets and liver are found in the category with the lowest possible sum score (left). (B) Strategy to identify genes that are selectively repressed in one tissue, based on mRNA profiling of 21 mouse tissues. Expression of a gene in a baseline tissue was compared to expression of that gene in all other tissues. When significantly lower compared to all other tissues, this gene was termed repressed in that tissue. This procedure was applied to all genes and repeated for all tissues in the panel. Statistical testing methods are described in Methods. (C) Pie-chart distribution of the number of specifically repressed genes in tissues. (D) Significant associations of specifically repressed genes (in all tissues) with molecular and cellular functions. The Benjamini-Hochberg-corrected P-value threshold of 0.05 is indicated by a dashed line.
Cross-platform validation
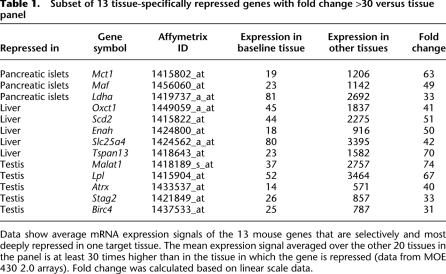
With the Affy MOE430 2.0 array, transcripts with a shorter 3′- untranslated region (UTR) isoform or subject to alternative splicing might be falsely detected as disallowed (example in Supplemental Fig. S1). For an independent confirmation, we performed a new analysis based on a different microarray platform for which we sampled new RNA from a mouse tissue panel. The Affymetrix GeneChip Mouse Gene 1.0 ST Array has each gene represented by a median of 27 probes spread across the full length of the gene, providing a complementary view of gene expression to that generated with 3′-based expression array designs. As a result, this array can more accurately detect the presence of transcripts even when alternative splicing or termination occurs. We examined a set of 13 tissues (each n = 3–5 replicates) on this platform. Since not all 21 tissues from the first analysis were represented, we could only test 900 out of 1074 repressed genes. Out of the 900, 560 (62%) were also confirmed on this platform, and genes passing this additional test were indicated in Supplemental Table S1. We based further studies only on genes that were found repressed on both platforms. As an example, mRNA expression data on both platforms are shown for genes repressed in islets (Supplemental Fig. S2). When adding a second criterion for strong repression in the target tissue (at least 30-fold lower compared to the mean of the tissue panel), we identified 13 deeply repressed genes in three tissues (Table 1).
Table 1.
Subset of 13 tissue-specifically repressed genes with fold change >30 versus tissue panel
Data show average mRNA expression signals of the 13 mouse genes that are selectively and most deeply repressed in one target tissue. The mean expression signal averaged over the other 20 tissues in the panel is at least 30 times higher than in the tissue in which the gene is repressed (data from MOE 430 2.0 arrays). Fold change was calculated based on linear scale data.
Next, we analyzed the Novartis mouse Gene Atlas (which contains data from 91 tissues and cell lines in duplicate) with our method. This resulted in 280 genes that are repressed in one of 28 tissues (Supplemental Table S3). Since the Novartis set has a small number of replications, the standard error on probe-set signal is relatively high as compared to our data set (Supplemental Fig. S3). Also, the selection of tissues affects which genes are being found as disallowed (many samples are different parts of the same tissues or even time series of the same cell type). For the few tissues that can be compared, a good overlap is observed. We find six genes repressed in liver for the Novartis set, of which three (Oxct1, Lass5, and Slc15a36) appear in our set. When using MIN6 cells as a surrogate for islets, we still see three genes (out of seven repressed in the Novartis set) in common (Ldha, Cat, Oat). Interestingly, for testis, 71 out of 97 (73%) genes found repressed in the Novartis set also appear in our analysis.
Disallowance in islets of Langerhans
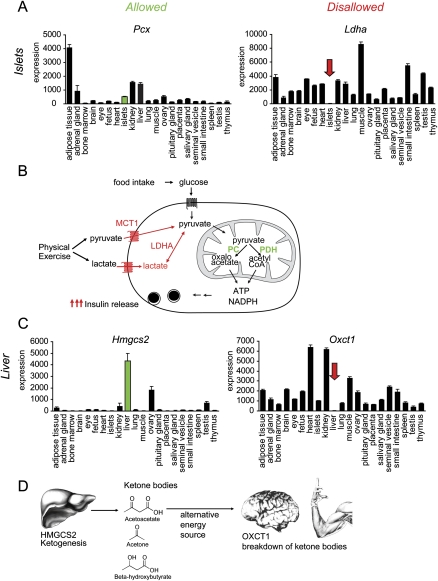
One of the deeply repressed genes in islets of Langerhans is Mct1, encoding the monocarboxylate transporter MCT1, which mediates the transport of lactate and pyruvate across cell membranes and is present in all tissues except adult beta-cells (Otonkoski et al. 2007; Quintens et al. 2008). Inadvertent expression of MCT1 in beta-cells results in hypoglycemia after physical exercise due to inappropriate insulin release (Otonkoski et al. 2003). This example illustrates that loss of metabolic control can be caused by a failure of specific repression of a housekeeping gene in the beta-cell. Therefore, we propose to name these specifically repressed housekeeping genes “disallowed.” Besides Mct1, Ldha was the most strongly repressed gene in islets. LDHA catalyzes the conversion of lactate to pyruvate, and repression in beta-cells is an additional safeguard to ensure insulin release exclusively in response to glucose (Fig. 2B).
Figure 2.
Allowed and disallowed genes. (A) Microarray expression profiles of pyruvate carboxylase (Pcx) versus lactate dehydrogenase A (Ldha), which are, respectively, highly expressed and deeply repressed in adult mouse pancreatic beta-cells. Mean mRNA expression signals ±SEM of three to five biological replicate microarray experiments. (B) Glucose levels in the blood are measured by the beta-cell through conversion of glucose to pyruvate, which is further metabolized in the mitochondria for ATP generation and anaplerosis. Downstream signals lead to insulin secretion. In many other cells, pyruvate does not necessarily come from glucose uptake. Extracellular pyruvate can be directly taken up by monocarboxylate transporter 1 (Mct1), or lactate can be converted to pyruvate by Ldha. However, when this happens in beta-cells, the glucose-sensing mechanism is disrupted, leading to inappropriate insulin release (Otonkoski et al. 2007). (C) Microarray expression profiles of 3-hydroxy 3-methylglutaryl-CoA synthase 2 (Hmgcs2) versus 2-oxoacid CoA transferase (Oxct1), which are, respectively, highly expressed and deeply repressed in adult liver. Mean mRNA expression signals ±SEM of three to five biological replicate microarray experiments. (D) In the fasted state, ketone bodies are generated in the liver. The first and rate-limiting enzyme for ketogenesis is Hmgcs2. Ketone bodies are transported in the blood and can be used by many organs as an alternative energy source. Oxct1 plays a central role in extrahepatic ketone body catabolism by catalyzing the reversible transfer of coenzyme A from succinyl-CoA to acetoacetate.
Cellular heterogeneity affects detection of disallowed genes
Because pancreatic islets are composed of several endocrine and non-endocrine cell types, the possibility exists that the data obtained from whole islets even underestimate the degree of repression in the beta-cell (Schuit et al. 1999). This was further investigated by comparing mRNA abundance of Mct1 in whole islets versus acinar cells and FACS-purified beta-cells (>99% pure beta-cells) (Supplemental Fig. S4A,B). Expression of Mct1 was determined by microarray (Supplemental Fig. S4A) and qPCR (Supplemental Fig. S4B). An even lower expression can be observed in purified beta-cells versus whole islets. This indicates that for a number of repressed genes, a low-level expression may be due to contaminating cell types in which the gene is not repressed. This contamination is on the order of 1%–5%, consistent with our observation that the expression of the acinar transcription factor Ptf1a in isolated islets is at 2% of expression in isolated acini (Supplemental Fig. S4C).
Disallowance in liver
We further examined repression of 2-oxoacid CoA transferase mRNA (Oxct1) in mouse liver. The encoded protein is an enzyme that plays a key role in ketone body degradation, which provides an alternative energy source to many tissues during fasting (Fig. 2D). Disallowance in liver seems appropriate as this tissue is specialized in ketone production in the fasted state to support metabolism of other tissues (Cahill and Veech 2003). In fact, the specific repression of Oxct1 in the mouse adult liver mirrors the expression of 3-hydroxy 3-methylglutaryl-CoA synthase 2 (Hmgcs2), which is required for ketogenesis and is highly expressed in adult liver, even in the fed state (Fig. 2C). Other enzymes necessary for ketogenesis (such as Acat1-3 and Hmgcl) were also highly expressed in adult liver (data not shown).
Disallowance is established during tissue maturation
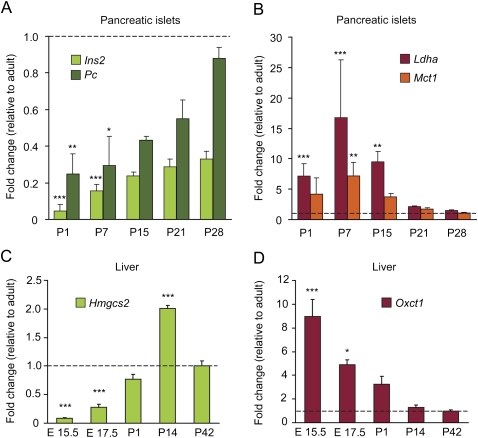
Next, we analyzed whether the tissue-specific disallowance of Mct1, Ldha, and Oxct1 was linked to the development of islets and liver. As is shown in Figure 3A, neonatal rat islets progressively induce expression of typical beta-cell genes during the first month after birth. For instance, the expression of insulin (Ins2) as well as pyruvate carboxylase (Pc) rises fivefold to 10-fold. In contrast, during neonatal beta-cell maturation the mRNA expression of both Mct1 and Ldha decreases progressively (Fig. 3B). Also the third deeply repressed gene in pancreatic islets, c-Maf (also known as Maf), displays a similar neonatal time-dependent down-regulation (Supplemental Fig. S5A). The same time course for establishing the tissue-specific repression is also observed in liver. In mouse embryonic and neonatal liver, we observed a parallel pronounced up-regulation of Hmgcs2 and progressive loss of expression of Oxct1 (Fig. 3C,D), again suggesting that during differentiation cell-specific genes are turned on while repressed housekeeping genes are turned off. The reduction of Hmgcs2 mRNA in rat liver after weaning (Fig. 3C) has been reported before in rodents (Serra et al. 1996).
Figure 3.
Tissue-specific gene expression and repression of certain housekeeping genes during maturation of rat islets (A,B) and mouse liver (C,D). (A) mRNA expression level in extracts from rat pancreatic islets from age 1 d (P1) to 4 wk (P28) after birth measured by real-time qPCR; data were normalized for signal in islets of adult animals (10 wk; dashed line). Maturation-dependent up-regulation of genes that are important for beta-cells, such as insulin 2 (Ins2) and pyruvate carboxylase (Pc), was observed during this period. (B) In parallel we observed maturation-dependent repression of monocarboxylate transporter 1 (Mct1) and lactate dehydrogenase (Ldha), genes that are profoundly repressed in adult mouse islets (Table 1). (C,D) mRNA expression level in extracts from mouse fetal and neonatal liver between embryonic day E15.5 and 6 wk after birth. A progressive up-regulation (linear increasing trend P < 0.001) of Hmgcs2 (C) mirrors a progressive loss (linear decreasing trend P < 0.001) of expression of Oxct1 (D). Data in panels A–D are means ± SEM of three to four individual experiments. Statistical significance of the difference between postnatal and adult expression signal was calculated by one-way ANOVA with a Dunnett post-hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
Mechanisms for disallowance
Epigenetic mechanisms have been described to establish tissue-specific gene expression through chromatin modification and remodeling (Li 2002; Missaghian et al. 2009), and histone modifications are well known to be stably introduced during cellular differentiation (Li 2002; van Arensbergen et al. 2010) and to profoundly alter the transcriptional activity of genes. Therefore, we tested whether the differentiation-dependent repression of certain housekeeping genes is similarly dependent on changes in chromatin structure. Histone H3 trimethylation at lysine 27 (H3K27me3) is associated with dense packaging of the nucleosome and low transcriptional activity, while acetylation at lysine 9 (H3K9ac) is associated with loosely packaged DNA and enhanced transcriptional activity. To examine a relationship between nucleosome structure and tissue-specific repression, we tested H3K9ac and H3K27me3 of the Mct1 and Ldha promoter regions in chromatin from adult mouse islets and liver. As is shown in Figure 4, A and B, the H3K27me3 content of chromatin associated with the Mct1 and Ldha genes was higher in islets than in liver. For the Mct1 gene in islets, we found a gradient of H3K27me3 along the length of the gene, with the highest level around the promoter (data not shown). In contrast, the signal for H3K9ac was significantly higher in liver than in islets for both genes. Therefore, the low Mct1 and Ldha mRNA levels in mature islets correlate to epigenetic silencing that is expected to be established when beta-cells mature postnatally. Also, c-Maf was associated with dense packaging (H3K27me3) in pancreatic islets, in contrast to liver, where an open nucleosome structure (H3K9ac) was predominant (Supplemental Fig. S5B). To gain insight into the temporal pattern of epigenetic modification, we examined histone modifications of immature pancreatic progenitors and embryonic stem (ES) cells versus mature islets, the MIN6 beta-cell line, and other mature tissues (Supplemental Fig. S6). In immature pancreatic progenitors, no enrichment of H3K27me3 was detected for these genes. On the contrary, in whole islets, beta-cells, and MIN6 cells, a clear H3K27me3 mark was detected. Moreover, in ES cells, either the inactivation mark was not present (Mct1 and Ldha) or a bivalent mark was observed (Maf), which serves to poise key developmental genes for lineage-specific activation or repression (Mikkelsen et al. 2007). This indicates a striking overlap between the acquired epigenetic inactivation in primary beta-cells and our observation of disallowance in adult islets.
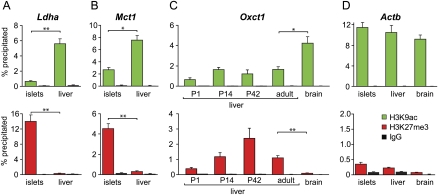
Figure 4.
Epigenetic mechanism for islet- and liver-specific silencing of repressed genes. Histone H3 lysine9-acetylation (green bars) and lysine27-trimethylation (red bars) of chromatin around promoter region of repressed genes were compared in adult mouse tissues with and without observed repression. (A,B) For Ldha and Mct1, a high level of H3 lysine27-trimethylation is seen in islets but not in liver, whereas H3 lysine9-acetylation is more extensive in the liver. This difference parallels differences between liver and islets for mRNA expression (see also Fig. 2). (C) Histone modifications during postnatal liver maturation (age 1, 14, and 42 d and adult). Histone H3 trimethylation in the Oxct1 promoter increases during liver development and is significantly higher in adult liver than in adult brain, while acetylation at the lysine9 residue is lower for all liver time points versus brain. (D) Nucleosomes of the beta actin (Actb) promoter, which is active in all tissues, display a high level of histone H3 lysine9 acetylation and a low level of histone 3 lysine 27 trimethylation in islets, brain, and liver. (Black bars) IgG immunoprecipitation control. Data are means ± SEM of three to four QPCRs.
We next looked at the Oxct1 gene, which is repressed in the liver and highly expressed in many other tissues such as heart, kidney, muscles, and brain (Fig. 2C). In line with the results found in islets, nucleosomal chromatin around the promoter of Oxct1 was more trimethylated and less acetylated in liver than in brain (Fig. 4C). In isolated livers from mice of age 1, 14, and 42 d, we found that H3K27me3 histone modification of the Oxct1 promoter increases during the neonatal period, whereas H3K9ac initially follows the rise, reminiscent of bivalent genes, but then decreases again at day 42. These data further support the idea that histone modifications correlate with gene repression in the postnatal period when tissues mature. Together, these data show that epigenetic changes in chromatin structure are associated with the selective disallowance of housekeeping genes in specialized cell types.
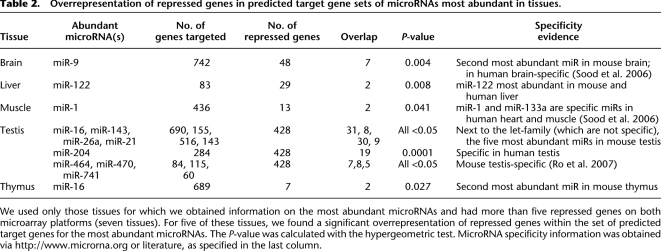
Do other levels of control, besides epigenetic suppression, exist to induce disallowance? A possible mechanism could occur via microRNAs, short noncoding RNAs with widespread influence on mRNA stability and mRNA translation (Baek et al. 2008; Selbach et al. 2008). In line with this idea, the 3′ UTR of mouse Oxct1 mRNA is a predicted target for miR-122, which is specifically expressed in mouse liver and which is the most abundant liver microRNA species (Supplemental Fig. S7; Lagos-Quintana et al. 2002; Landgraf et al. 2007). During the course of mouse embryonic liver differentiation, miR-122 expression increased 12-fold between embryonic day 15.5 (E15.5) and birth (Fig. 5A). In parallel, Oxct1 expression gradually decreased from E15.5 until 2 wk after birth (Fig. 3D). In order to assess whether the expression of miR-122 is responsible for repression of Oxct1 mRNA, we performed experiments with a bipotential mouse embryonic liver (BMEL) cell line, which is derived from E14.5 hepatoblasts and can be differentiated to hepatocyte-like cells (Strick-Marchand et al. 2004). During differentiation of cultured BMEL cells, miR-122 expression increased ninefold. Expression of miR-122 could be inhibited by the addition of miR-122 antagomir (Fig. 5B), which was specific since a mutated antagomir abolished the inhibition (Fig. 5B) and a non-target microRNA (miR-20) was not affected by either antagomir (data not shown). In differentiated BMEL cells, Oxct1 expression increased 1.6-fold by adding miR-122 antagomir, while the mutated control antagomir did not alter Oxct1 expression (Fig. 5B). To verify if miR-122 represses OXCT1 expression in vivo, we compared RNA and protein levels in wild-type and transgenic mouse livers at E15.5. The transgenic embryos overexpressed miR-122 specifically in the liver under control of alpha-fetoprotein and albumin regulatory sequences (data not shown). A 1.7 ± 0.07-fold overexpression of miR-122 (mean ± SEM) was associated with repression of Oxct1 RNA and protein, as determined by qRT-PCR and Western blotting (Fig. 5C). miR-124 increases during differentiation of beta-cells (Baroukh et al. 2007) and is highly expressed in MIN6 cells (Supplemental Fig. S8A) and mature islets (Supplemental Fig. S8B). This microRNA has multiple predicted target sites on the 3′ UTR of Mct1 that are conserved among human, rat, mouse, dog, and chicken. Down-regulation of Mct1 by miR-124a has been experimentally validated by overexpression experiments (Lim et al. 2005; Wang and Wang 2006). In addition, other microRNAs abundant in islets (e.g., miR-27b, miR-29a) are also predicted to recognize the Mct1 3′ UTR. This also indicates that for Mct1 an additional layer of post-transcriptional control is present to ensure an appropriate repression in islets. More generally, if repressed genes are down-regulated by microRNAs, we expect to find binding sites for microRNAs in the 3′ UTRs of these genes. We would then expect these binding sites to correspond to microRNAs that are abundant in the specific tissue. Therefore, we checked for overrepresentation of tissue-specific microRNA binding sites in the 3′ UTRs of the repressed genes for the particular tissue (where such information was available). For several tissues, we found such strong overrepresentation (Table 2), indicating that at least a subset of the repressed genes can be down-regulated by tissue-specific and abundant microRNAs.
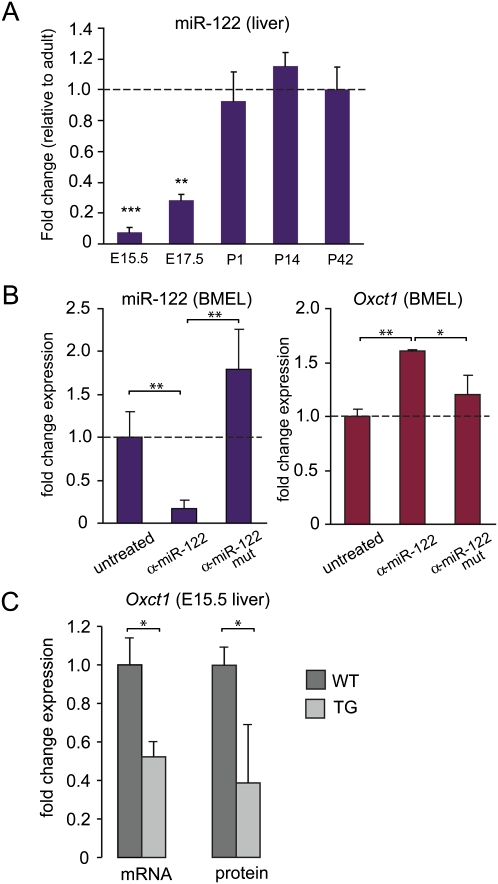
Figure 5.
MicroRNA 122 (miR-122) contributes to liver-specific silencing of Oxct1 expression. (A) Time course of miR-122 expression in vivo during liver maturation as measured by qPCR. miR-122 increases with a linear trend, P = 0.01. Statistical significance of the difference between P42 and other time points was calculated by one-way ANOVA with a Dunnett post-hoc test. (B) Treatment of in vitro differentiated BMEL cells with antagomiR-122 (α-miR-122) or a mutated antagomiR (α-miR-122 mut) control. Changes in miR-122 expression (left) and Oxct1 expression (right) were measured by Q-PCR. MiR-122 expression decreases sixfold by treatment with antagomiR-122 (α-miR-122). A mutated antagomiR (α-miR-122 mut) control did not significantly influence miR-122 expression. Oxct1 expression is increased by miR-122 knockdown with antagomiR-122, indicating that miR-122 inhibits Oxct1 expression. A mutated antagomiR did not significantly alter Oxct1 expression. Data are means ± SEM of three individual experiments. (C) Oxct1 RNA and protein expression in livers at E15.5 of wild-type versus liver-specific miR-122 overexpressing transgenic mice. A 1.7 ± 0.07-fold overexpression of miR-122 (mean ± SEM; n = 4 [wild-type] and n = 6 [transgenic]) was associated with a significant (t-test, P < 0.05) repression of Oxct1 RNA and protein, as determined by Q-RT-PCR (n = 4 or 6 for wild-type or transgenic, respectively) and Western blotting (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
Table 2.
Overrepresentation of repressed genes in predicted target gene sets of microRNAs most abundant in tissues.
We used only those tissues for which we obtained information on the most abundant microRNAs and had more than five repressed genes on both microarray platforms (seven tissues). For five of these tissues, we found a significant overrepresentation of repressed genes within the set of predicted target genes for the most abundant microRNAs. The P-value was calculated with the hypergeometric test. MicroRNA specificity information was obtained via http://www.microrna.org or literature, as specified in the last column.
Evolutionary conservation
We verified the expression of the most strongly repressed mouse genes in human tissue microarrays. A data set was used encompassing 17 human tissues equivalent to the murine tissues. Of the genes we find deeply repressed in liver and testis (islets are not present in the human set), six (out of 10) had the lowest expression in the same human tissue as the murine tissue in which it was found to be repressed (including Oxct1). Additionally, detection of protein by immunohistochemistry confirmed the absence of OXCT1 in human liver, and MCT1 and LDHA in human pancreatic islets (Human Protein Atlas-MCT1 2010, http://www.proteinatlas.org/normal_unit.php?antibody_id=3324&mainannotation_id=506755; Human Protein Atlas-LDHA 2010, http://www.proteinatlas.org/normal_unit.php?antibody_id=15336&mainannotation_id=1473283; Human Protein Atlas-OXCT1 2010, http://www.proteinatlas.org/normal_unit.php?antibody_id=12047&mainannotation_id=1169580). In the NCBI Gene Expression Omnibus (GEO), we did not find data from adult islets or pancreas in comparison to other tissues for species other than human and rodents. As for liver, data in pig and zebrafish (Supplemental Fig. S9) illustrate that Oxct1 is also disallowed in the liver of these species. Taken together with a similar repression of Mct1 and Ldha in rat islets (Fig. 3B), these results suggest that tissue-specific repression of many of the genes identified in this study is evolutionarily conserved.
Discussion
Unlike what is sometimes claimed, expression of housekeeping genes can differ widely among different tissues (Thorrez et al. 2008). In this study, we propose that profound repression of certain housekeeping genes in specialized tissues is possible and necessary and that this repression is the opposite of expression of cell-specific genes. Traditionally, tissue function has mostly been investigated by studying genes with high tissue-specific expression; however, we propose that the class of disallowed genes described in this paper provides additional important insights in tissue function. Tissue-specific gene repression is known to occur during development and requires the spatial and temporal organization of an interacting network of transcriptional repressors and activators. However, specific absence of one gene in a particular tissue has not been systematically studied so far.
By applying a custom statistical intersection-union test (Berger 1982; Berger and Hsu 1996; Van Deun et al. 2009) on a set of mouse tissue microarrays, we identified genes with significantly lower expression in one tissue versus all other (20) tissues. To avoid false-positives due to (3′-biased) microarray design, we used another microarray platform to confirm gene repression. Many of these genes are associated with tissue development and function. We show that this type of repression is conserved in several mammalian species such as mice, rats, and human. A limitation of our screen is based on mRNA extracts from whole tissues that are composed of a number of different cell types. This mixed population may result in the situation that genes that are specifically repressed in a certain cell type are not detected in the screen because other cell types in the same tissue do express the gene(s) and thus mask the repression. If this phenomenon were re-examined at the level of primary differentiated cell types, a more extensive degree of repression may be found, as was previously shown for hexokinase I in rat beta cells (Schuit et al. 1999). The fact that we have underestimated the repression of some of the islet genes is illustrated by the analysis of FACS-purified mouse beta-cells, which shows that the residual expression signal of disallowed genes in pancreatic islets derives from non-beta-cells.
To demonstrate the functional relevance of the specific repression, we further focused on a few examples of disallowed genes related to metabolism in two different tissues. In pancreatic islets, lactate dehydrogenase (Ldha) and the lactate/pyruvate transporter Mct1 are disallowed since the inadvertent uptake of lactate or pyruvate would interfere with the glucose sensing by the beta-cells. In liver, the key enzyme for ketone body utilization (Oxct1) is strongly repressed since the liver needs to synthesize and export ketone bodies, which serve as an alternative energy source during starvation. A known example of a human disease illustrates that ignoring this disallowance can destroy specific tissue function (Otonkoski et al. 2007). However, association of disallowed genes with (human) disease is challenging since redundant mechanisms may be in use to protect the cell against inadvertent disallowed gene expression and because a clinical phenotype may only be observed in certain stress conditions. Indeed, exercise-induced hypoglycemia caused by the Mct1 promoter mutation only occurs during physical exercise, and it is conceivable that Oxct1 mutations may only manifest in starvation conditions.
Other examples of disallowed genes outside the realm of metabolism likely exist. For instance, in spleen, a lymphoid organ, ring finger protein 128 (Rnf128), also known as “gene related to anergy in lymphocytes” (GRAIL), is deeply repressed. Rnf128 encodes a ubiquitin ligase and is associated with T-cell suppression (Kostianovsky et al. 2007; MacKenzie et al. 2007). Disallowance of Rnf128 in spleen and expression in other tissues may help to shape the balance in central and peripheral T-cell reactivity. In testis, Lpl (lipoprotein lipase) is repressed, and a lack of LPL protein in Sertoli and Leydig cells has been described (Nielsen et al. 2010), which might be due to interference with a distinct function of LIPG, another (endothelial) lipase. Another example in testis is Stag2, which is a component of cohesin complexes and is described to be mutually exclusive with other STAG proteins (STAG1 and STAG3) (Uniprot 2010, http://www.uniprot.org/uniprot/Q8N3U4). STAG3, a subunit of the meiotic cohesin complex, is expressed specifically in testis (Pezzi et al. 2000); therefore, it is not surprising to find Stag2 repressed in testis. Summarized, disallowed genes display the observed expression profile because the presence of the gene product would interfere with correct tissue function. The physiological relevance of disallowance of individual tissue-specifically repressed genes might be explored further by transgenic mouse models.
In the pancreas, the beta-cells expand rapidly in the first month of life (Bonner-Weir 2000) and only become truly functional in their response to glucose about weaning, when insulin release becomes controlled by oral food intake. Therefore, we might expect that establishment of pathways that mediate tissue-specific processes occurs at this time. Indeed, by means of qRT-PCR time series, we demonstrate that tissue-specific genes (such as insulin in the pancreatic islets and HMG CoA synthase 2 in liver) become highly up-regulated in the late fetal/early postnatal period and reach mature expression levels at 1–1.5 mo. In parallel, expression of the disallowed genes is shut down during this phase of tissue maturation, and repression is subsequently maintained through adulthood.
Immature cells that are the direct precursors of the studied differentiated cells have not fully exercised the level of disallowance; therefore, specific elements that are induced during the developmental program are responsible for repression in the differentiated cell. Chromatin condensation and the resulting gene silencing play a key role in differentiation. A hallmark for transcriptional silencing is trimethylation of lysine27 on histone H3 (H3K27me3), a process that is catalyzed by the methyl transferase activity of the Polycomb complex PCR2 (Vire et al. 2006; Shen et al. 2008) and antagonized by the demethylases UTX and JMJD3 (Agger et al. 2007; De Santa et al. 2007; Lan et al. 2007; Lee et al. 2007). The group of tissue-specifically repressed genes we present in this paper is expressed in embryonic stem cells and is distinct from the genes associated with Polycomb complexes in embryonic stem (ES) cells (Boyer et al. 2006). Cell-specific combinations of transcription factors might activate the PCR2 complex specifically to repress expression of disallowed genes during tissue differentiation. We checked histone modifications at the promoter regions of Mct1, Ldha, and Oxct1 and found them to be associated with H3K27me3 in the tissues where they are disallowed. Additionally, acetylation of histone H3 at lysine9 (H3K9ac), a mark of transcriptional activation, was found to be significantly lower around the promoter of these genes in the tissue of disallowance. Certain promoters in ES cells carry a bivalent mark, with large regions of H3K27me3 harboring smaller regions of H3 lysine 4 methylation (Bernstein et al. 2006). This mark serves to poise key developmental genes for lineage-specific activation or repression (Mikkelsen et al. 2007). Most promoters of the repressed genes in this paper were found to be negative for H3K27me3 in ES cells (Mikkelsen et al. 2007). These data fit with our observations that the expression of the disallowed genes is turned off only during late fetal liver maturation and early postnatal islet maturation. Indeed, in immature pancreatic progenitors, no enrichment of H3K27me3 was detected for the genes we found deeply repressed in pancreatic islets. In addition, we observed an increase of H3K27me3 in the promoter of Oxct1 during postnatal liver development.
In a tissue in which transcription is low and mRNA is scarce, further repression by miRNAs could add additional control of disallowance. The most abundant microRNA in liver, miR-122, (Lagos-Quintana et al. 2002; Landgraf et al. 2007) is predicted to target Oxct1. We showed by Q-PCR that during the same time when Oxct1 is down-regulated, miR-122 is up-regulated. When miR-122 was inhibited by miR-122 antagomir, Oxct1 expression increased 1.6-fold. In a transgenic mouse model where miR-122 is overexpressed in liver, a similar decrease of Oxct1 mRNA and protein is observed at E15.5. Although this is only a modest effect, it is exactly what can be expected for the impact of a single microRNA (Baek et al. 2008; Selbach et al. 2008) and confirms that reduced protein output is mainly caused by destabilization of target mRNAs (Guo et al. 2010). This modest degree of repression may function as a secondary control level to further inhibit translation of mRNA that is strongly repressed by a first layer of epigenetic repression. When we generalized this analysis to other tissues, we found strong overrepresentation of tissue-specific microRNA binding sites in the 3′ UTRs of the genes repressed in that tissue, which indicates that tissue-specific microRNAs provide an additional level of control. We also investigated whether certain (tissue-specific) transcription factors might be responsible for down-regulation of a subset of the repressed genes by network analysis and detection of common regulatory promoter elements, but could not find any evidence for such a mechanism in any of the tissues.
Two studies previously reported on disallowance in islets (Quintens et al. 2008; Pullen et al. 2010), but this study is the first to analyze the phenomenon in a broad set of tissues and provide mechanistic insights. In summary, we propose that tissue-specific disallowance of housekeeping genes is required for the specialized function of differentiated tissues. Profound repression is maintained through a multi-tier mechanism comprising chromatin remodeling and microRNAs. We propose that this phenomenon has widespread consequences in cell biology and can lead to disease when disrupted.
Methods
Preparation of tissues and purified cells
All experiments based on laboratory animals were approved by committees for animal welfare at the Katholieke Universiteit Leuven and Harvard University. The following tissues were hand-dissected from 10- to 12-wk-old C57Bl6 mice: liver, gastrocnemius muscle, brain, heart, adrenal gland, eye, small intestine, thymus, epidydimal adipose tissue, pituitary gland, kidney, parotid gland, spleen, lung, bone marrow, testis and seminal vesicles (males), and ovary and placenta (females). Fetal tissue was isolated at day 16. Tissues were rinsed in phosphate-buffered saline, frozen in liquid nitrogen, and stored at −80°C. Islets of Langerhans were isolated from the male and female adult (12 wk) mouse pancreata after injecting collagenase solution into the pancreatic duct. For adult rats, islets from one pancreas were used as a sample. For liver time-series experiments, livers were isolated from CD1 mice, stored overnight at 4°C in RNAlater (Ambion), and then stored at −80°C. Mouse pure beta-cells were purified from islets of RIPYY mice expressing EYFP under the rat insulin promoter, i.e., specifically in beta-cells (Quoix et al. 2007). Additional details on tissue preparation and RNA extraction are provided in the Supplemental Material.
mRNA expression analysis via microarray
For the Affymetrix mouse MOE 430 2.0 arrays, 2 μg of total RNA was used to prepare biotinylated cRNA with an IVT labeling kit (Affymetrix) according to the Genechip Expression Analysis Technical Manual 701025 Rev. 5, except for islets, adrenal gland, and pituitary gland, where 1 μg of total RNA was used. For the Affymetrix mouse Gene Arrays 1.0 ST, 100 ng of total RNA was used according to the manufacturer's manual 701880Rev4 (Affymetrix). The concentration of labeled cRNA was measured using the NanoDrop ND-1000 spectrophotometer, and quality was analyzed using the Agilent bioanalyzer 2100. Fragmented cRNA was hybridized to the arrays during 16 h at 45°C. The arrays were washed and stained in a fluidics station (Affymetrix) and scanned using the Affymetrix 3000 GeneScanner.
The MOE 430 2.0 (69 arrays) and Gene Array (40 arrays) sets consist of 21 and 13 different murine tissues, respectively, with three to five replicates for each tissue. Quality controls of the arrays were according to manufacturer's criteria. All CEL files were analyzed using GCOS (Affymetrix GeneChip Operating Software) and the affy library (Gautier et al. 2004) of the BioConductor project (Gentleman et al. 2004). Data were processed with the robust multichip average (RMA) algorithm with default parameters (RMA background correction, probe-level quantile normalization, and average difference summarization). We independently applied the MAS5 algorithm using global scaling to 150. All data were log2-transformed for normalization, and all further data analysis was performed on log2-transformed data. The data files are accessible through the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE24207 (430 2.0 arrays) and GSE24940 (1.0 ST gene arrays). For the human data, we selected 69 arrays from GSE7307, representing 17 human tissues (3–5 arrays/tissue) equivalent to the mouse tissues. Other GEO accession numbers of data sets used are GSE10246 (Novartis data set), GSE10898 (pig), and GSE11107 (zebrafish).
Statistical analysis
To reduce multiple testing, we defined a core of 17,334 probe sets, each corresponding to a unique gene symbol, where only the probe set with the highest average expression was selected when multiple probe sets were targeting the gene. To find those genes that are significantly repressed in a particular baseline tissue compared to each of the 20 other tissues, we initially used a sum score, which is the summation of the number of times the gene is more (+1), less (−1), or equally (0) expressed in the baseline tissue versus the 20 other members in the tissue panel, using unpaired Student's t-tests and P < 0.001 as a threshold for significance. To control the type I error, we improved this test statistic by applying the testing procedure described by Berger (1982), also known as the intersection-union test (Berger and Hsu 1996). This tests the null hypothesis that at least one other tissue scores lower than or equal to the baseline against the alternative that expression in each of the other tissues is larger than in the baseline tissue (see Supplemental material). When available, we used redundant probe sets as an additional control by also requiring the redundant probe sets to have the lowest expression in baseline tissue when not in background range. For significance testing in time-course experiments, we used one-way ANOVA with Dunnett post-hoc testing. *P < 0.05; **P < 0.01; ***P < 0.001. We also used contrasts to test for increasing or decreasing trends. The remaining comparisons were based on t-tests with the standard error from ANOVA. All statistical analysis was performed in SAS 9.2.
Bioinformatics
Benjamini-Hochberg-corrected P-values for associations with molecular and cellular functions were calculated with Ingenuity Pathway Analysis. Prediction of microRNA targets in 3′ UTRs was based on context scoring (Grimson et al. 2007) performed with TargetScanMouse 5.0. We identified the most abundant microRNAs in various mouse tissues via a public microRNA expression database (Betel et al. 2008) or literature (Sood et al. 2006; Ro et al. 2007). Overrepresentation of repressed genes in the target list was assessed with the hypergeometric test.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to a modified protocol of Upstate (Millipore) EZ-ChIP. The detailed method is provided in the Supplemental material. Precleared chromatin was incubated overnight at 4°C using rabbit anti-acetyl-histone H3 (Lys9), rabbit anti-trimethyl-histone H3 (Lys27), or normal rabbit IgG (Upstate). Antibody–chromatin complexes were collected by adding blocked protein A-TSK beads for 1 h at 4°C and washed. Immune complexes were eluted in 1% SDS, 0.1 M NaHCO3 at room temperature. Samples were treated with RNase A and RNase T1 (Fermentas) for 30 min at 37°C and 0.6 U of proteinase K (Fermentas) for 1.5 h at 45°C. DNA was purified using spin columns (Geneclean Turbo; Qbiogene) and eluted in 60 μL of DNase-free water. ChIP samples were analyzed with probe-based quantitative PCR (Rotor-Gene 3000; Corbett). PCRs were carried out in a 25-μL reaction volume containing ABsolute QPCR mix (Abgene), 1.5 μL of DNA, 300 nM or 900 nM forward and reverse primer, and 50 nM dual-labeled fluorogenic probe. Primers and probes were obtained from Sigma-Aldrich. The amplification conditions were 15 min at 95°C and 45 cycles of 20 sec at 95°C and 45 sec at 60°C. Enrichment of DNA was calculated using the equation: 2(CtInput − CtAb), where Ct is the mean threshold cycle of PCR done in triplicate. Input is 1% input sample, and Ab is the specific antibody or IgG sample.
Cell culture and antagomir treatment
BMEL cells were cultured as previously described (Plumb-Rudewiez et al. 2004), plus glutamine and hepatocyte-like differentiation was obtained by inducing the formation of floating aggregates of BMEL cells (Strick-Marchand et al. 2004). For miR-122 inhibition, we used antagomirs (Eurogentec). Antagomir sequences are in the Supplemental material. BMEL cells cultured in aggregates were exposed to 10 μg/mL antagomirs, and RNA was extracted after 3 d.
Western blot
Proteins were extracted from embryonic livers at E15.5 with TriPure reagent (Roche) according to the manufacturer's protocol. For Western blot analysis, protein extracts were subjected to electrophoresis on 7.5% polyacrylamide gels (SDS-PAGE) and transferred onto a nitrocellulose Hybond-C membrane (Amersham). The membrane was revealed with anti-OXCT1 antibody (ProteiTech Group; 1:1000 dilution in TBS/0.1% Tween, 5% BSA), or anti-beta actin (Santa Cruz; 1:1000) overnight at 4°C. The membrane was incubated with HRP-anti-rabbit (Santa Cruz; 1:5000) or HRP-anti-goat (Santa Cruz; 1:10,000) antibodies for 2 h at room temperature. Signals were visualized by chemiluminescence. The OXCT1 signals were quantified using Scion Image software and normalized for the beta-actin signals.
Reverse transcription and qRT-PCR
Rat islets
Reverse transcription was done in 25 μL containing 500 ng of total RNA, 5 μL of superscript buffer, 0.1 M dithiothreitol (DTT), 50 ng of random hexanucleotide primers, 10 mM dNTP, 200 units of RNasin, and 200 U of Superscript II reverse transcriptase (Invitrogen); incubated for 10 min at 25°C, 50 min at 42°C, and 15 min at 72°C. cDNA was diluted to 3 ng/μL in nuclease-free water and stored at −20°C. qRT-PCR was performed using the ABI7000 (Applied Biosystems). Primers were synthesized by MWG-Biotech, and specificity was confirmed by dissociation curve analysis. The reaction mix contained 10 μL of SYBR green master mix (Applied Biosystems), 1 μL of each 5′ and 3′ oligonucleotide (10 pmol/μL), and 1 μL of cDNA. The reaction conditions for PCR were 15 sec at 95°C, 30 sec at 55°C–58°C (depending on the primers), and 30 sec at 72°C for 40 cycles. After normalization of the gene of interest to a control gene (S25), the comparative CT (threshold cycle) method was used to calculate relative gene expression levels for the selected genes.
Mouse islets
Quantitative RT-PCR was performed as earlier described (Otonkoski et al. 2007). mRNA expression levels were normalized using polymerase II alpha (RNA Pol II).
Mouse liver
Reverse transcription on total RNA was performed in 10–25 μL containing 1× First Strand Buffer (Invitrogen), 10 mM DTT, 200 U of M-MLV reverse transcriptase (Invitrogen), 20–40 U of RnaseOUT (Invitrogen), 4 mM dNTPs (Amersham Pharmacia Biotech), and 200–400 ng of random hexamers (Roche) for 10 min at room temperature and 2 h at 37°C (mRNA) or 1 h at 37°C and 5 min at 85°C (18S RNA). For miR-122, 50 nM stem–loop RT primer (CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAAACAC) was used instead of random hexamers, and incubation was 30 min at 16°C, 30 min at 42°C, and 5 min at 85°C. QPCR was performed with SYBR Green Master Mix Reagent (Invitrogen) on an IQ cycler (Bio-Rad). Oxct1 and Hmgcs2 were normalized to beta-actin and miR-122 to 18S RNA.
Acknowledgments
This study was supported by research grants from K.U. Leuven (GOA/2004/11, GOA/2005/04, GOA/2009/10, and CoE EF/05/007), Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO grant G.0733.09N), the Belgian Science Policy (Interuniversity Attraction Poles Program (PAI 6/20 and 6/40), the D.G. Higher Education and Scientific Research of the French Community of Belgium, the Juvenile Diabetes Research Foundation (JDRF grants 2006-182, 2007-685), and the National Institutes of Health (NIH R01 DK 66056). P.G. is Research Director at the FNRS, and I.L. holds a fellowship from Télévie (Belgium). We thank Joris Van Arensbergen for providing the data used in Supplemental Figure S6 and Sabine Cordi for technical assistance.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE24207 and GSE24940.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109173.110.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van OE 2007. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem 282: 19575–19588 [DOI] [PubMed] [Google Scholar]
- Berger RL 1982. Multi-parameter hypothesis-testing and acceptance sampling. Technometrics 24: 295–300 [Google Scholar]
- Berger RL, Hsu JC 1996. Bioequivalence trials, intersection-union tests and equivalence confidence sets. Stat Sci 11: 283–302 [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C 2008. The microRNA.org resource: Targets and expression. Nucleic Acids Res 36: D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner-Weir S 2000. Perspective: Postnatal pancreatic beta cell growth. Endocrinology 141: 1926–1929 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cahill GF Jr, Veech RL 2003. Ketoacids? Good medicine? Trans Am Clin Climatol Assoc 114: 149–161 [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of Polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY 2003. Human housekeeping genes are compact. Trends Genet 19: 362–365 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P, et al. 2001. A compendium of gene expression in normal human tissues. Physiol Genomics 7: 97–104 [DOI] [PubMed] [Google Scholar]
- Kostianovsky AM, Maier LM, Baecher-Allan C, Anderson AC, Anderson DE 2007. Up-regulation of gene related to anergy in lymphocytes is associated with Notch-mediated human T cell suppression. J Immunol 178: 6158–6163 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di CL, Shiekhattar R 2007. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Li E 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3: 662–673 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- MacKenzie DA, Schartner J, Lin J, Timmel A, Jennens-Clough M, Fathman CG, Seroogy CM 2007. GRAIL is up-regulated in CD4+ CD25+ T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype. J Biol Chem 282: 9696–9702 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missaghian E, Kempná P, Dick B, Hirsch A, Alikhani-Koupaei R, Jégou B, Mullis PE, Frey BM, Flück CE 2009. Role of DNA methylation in the tissue-specific expression of the CYP17A1 gene for steroidogenesis in rodents. J Endocrinol 202: 99–109 [DOI] [PubMed] [Google Scholar]
- Nielsen JE, Lindegaard ML, Friis-Hansen L, Almstrup K, Leffers H, Nielsen LB, Rajpert-De ME 2010. Lipoprotein lipase and endothelial lipase in human testis and in germ cell neoplasms. Int J Androl 33: e207–e215 [DOI] [PubMed] [Google Scholar]
- Otonkoski T, Kaminen N, Ustinov J, Lapatto R, Meissner T, Mayatepek E, Kere J, Sipila I 2003. Physical exercise-induced hyperinsulinemic hypoglycemia is an autosomal-dominant trait characterized by abnormal pyruvate-induced insulin release. Diabetes 52: 199–204 [DOI] [PubMed] [Google Scholar]
- Otonkoski T, Jiao H, Kaminen-Ahola N, Tapia-Paez I, Ullah MS, Parton LE, Schuit F, Quintens R, Sipila I, Mayatepek E, et al. 2007. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am J Hum Genet 81: 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzi N, Prieto I, Kremer L, Perez Jurado LA, Valero C, Del MJ, Martinez A, Barbero JL 2000. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J 14: 581–592 [DOI] [PubMed] [Google Scholar]
- Plumb-Rudewiez N, Clotman F, Strick-Marchand H, Pierreux CE, Weiss MC, Rousseau GG, Lemaigre FP 2004. Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3α/Foxa1 gene by TGF-β in mouse liver. Hepatology 40: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Pullen TJ, Khan AM, Barton G, Butcher S, Sun G, Rutter G 2010. Identification of genes selectively disallowed in the pancreatic islet. Islets 2: 89–95 [DOI] [PubMed] [Google Scholar]
- Quintens R, Hendrickx N, Lemaire K, Schuit F 2008. Why expression of some genes is disallowed in beta-cells. Biochem Soc Trans 36: 300–305 [DOI] [PubMed] [Google Scholar]
- Quoix N, Cheng-Xue R, Guiot Y, Herrera PL, Henquin JC, Gilon P 2007. The GluCre-ROSA26EYFP mouse: A new model for easy identification of living pancreatic alpha-cells. FEBS Lett 581: 4235–4240 [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W 2007. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol 311: 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuit F, De Vos A, Farfari S, Moens K, Pipeleers D, Brun T, Prentki M 1997. Metabolic fate of glucose in purified islet cells. Glucose-regulated anaplerosis in beta cells. J Biol Chem 272: 18572–18579 [DOI] [PubMed] [Google Scholar]
- Schuit F, Moens K, Heimberg H, Pipeleers D 1999. Cellular origin of hexokinase in pancreatic islets. J Biol Chem 274: 32803–32809 [DOI] [PubMed] [Google Scholar]
- Sekine N, Cirulli V, Regazzi R, Brown LJ, Gine E, Tamarit-Rodriguez J, Girotti M, Marie S, MacDonald MJ, Wollheim CB, et al. 1994. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J Biol Chem 269: 4895–4902 [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Serra D, Bellido D, Asins G, Arias G, Vilaro S, Hegardt FG 1996. The expression of mitochondrial 3-hydroxy-3-methylglutaryl-coenzyme-A synthase in neonatal rat intestine and liver is under transcriptional control. Eur J Biochem 237: 16–24 [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N 2006. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci 103: 2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick-Marchand H, Morosan S, Charneau P, Kremsdorf D, Weiss MC 2004. Bipotential mouse embryonic liver stem cell lines contribute to liver regeneration and differentiate as bile ducts and hepatocytes. Proc Natl Acad Sci 101: 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F 2008. Using ribosomal protein genes as reference: A tale of caution. PLoS ONE 3:e1854 doi: 10.1371/journal.pone.0001854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F 2006. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics 7: 31 doi: 10.1186/1471-2164-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen J, García-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, Skoudy AL, Palassini M, Heimberg H, Ferrer J 2010. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res 20: 722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun K, Hoijtink H, Thorrez L, Van Lommel L, Schuit F, Van Mechelen I 2009. Testing the hypothesis of tissue selectivity: The intersection-union test and a Bayesian approach. Bioinformatics 25: 2588–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van EA, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X 2006. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res 34: 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M 2000. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics 2: 143–147 [DOI] [PubMed] [Google Scholar]
- Zhao C, Wilson MC, Schuit F, Halestrap AP, Rutter GA 2001. Expression and distribution of lactate/monocarboxylate transporter isoforms in pancreatic islets and the exocrine pancreas. Diabetes 50: 361–366 [DOI] [PubMed] [Google Scholar]