Abstract
The first wave of personal genomes documents how no single individual genome contains the full complement of functional genes. Here, we describe the extent of variation in gene and pseudogene numbers between individuals arising from inactivation events such as premature termination or aberrant splicing due to single-nucleotide polymorphisms. This highlights the inadequacy of the current reference sequence and gene set. We present a proposal to define a reference gene set that will remain stable as more individuals are sequenced. In particular, we recommend that the ancestral allele be used to define the reference sequence from which a core human reference gene annotation set can be derived. In addition, we call for the development of an expanded gene set to include human-specific genes that have arisen recently and are absent from the ancestral set.
Keywords: ancestral allele, gene annotation, loss of function, nonsense SNP, personal genomics, pseudogene
Variations in the era of personal genomes
Over the last decade, technological advances have led to the generation of an unprecedented amount of human genomic data and dramatically increased our knowledge of variability in the human genome. The first draft sequence of the human genome was a composite haploid sequence assembled from several individuals (Lander et al. 2001). The first diploid genome of a single individual was published in 2007 (Levy et al. 2007). Since then, next-generation sequencing technologies have begun to provide whole-genome sequence information at an accelerated pace, and several personal genomes have been published, with many more to follow (Levy et al. 2007; Wang et al. 2008; Wheeler et al. 2008; Ahn et al. 2009; Kim et al. 2009; McKernan et al. 2009; Pushkarev et al. 2009; Drmanac et al. 2010; Lunshof et al. 2010; Lupski et al. 2010; Rasmussen et al. 2010; Schuster et al. 2010).
While the initial human genome was assembled and annotated as “the reference,” the recent personal genome sequences provide us with a glimpse of the extent of human genetic variation. This genetic variability manifests itself not only in single-nucleotide polymorphisms (SNPs), but also in insertion/deletions (indels) and copy number variations (CNVs) of blocks of varying lengths in the genome (Conrad et al. 2006; Mills et al. 2006; Redon et al. 2006; Korbel et al. 2007).
The availability of sequences from tens of personal genomes has revealed considerable variation between individuals. This is an opportune time to think about genome annotation in a new context in that the new data enable us to redefine the reference gene sequences and the reference gene set. With personal genome sequencing endeavors rapidly moving ahead, it is vital to establish a stable foundation to facilitate the interpretation of further personal genome sequences.
Current status of gene annotation: impact of genetic variability
The human reference genome is a haploid sequence derived as a composite from multiple individuals. Current gene annotations are based on this reference. One problem with genome annotations is that they are historically heavily biased toward protein-coding genes. While the rationale behind this is based on the well-understood importance of protein-coding genes, identification of pseudogenes and events that may lead to gene inactivation have been largely ignored in the automatic annotation process. Moreover, genetic variations that can affect gene annotations have not been systematically integrated into annotation pipelines. This problem is most severe for interindividual variations that affect genes in such a way that a locus is a functional gene in some people but a pseudogene in others.
Besides the bias toward annotating coding genes, there are other factors such as assembly errors, base-calling errors, and rare alleles in the reference genome that can contribute to erroneous gene annotation. Another difficulty in using the current human genome as the reference is that it does not represent the enormous genomic diversity in the human population.
The definitions of the terms “gene” and “pseudogene” have broadened in the post-genomic era (Gerstein and Zheng 2006; Gerstein et al. 2007; Zhang et al. 2010). In this Perspective, we define genes as protein-coding loci and pseudogenes as protein-coding loci that may become nonfunctional due to loss-of-function (LOF) variations such as nonsense SNPs or SNPs that affect canonical splice sites. At one extreme, LOF events can affect one single individual in the population, perhaps leading to the deactivation of an important gene and giving rise to a disease in that individual. However, sometimes a LOF mutation becomes more common in the population and may even become fixed (MacArthur and Tyler-Smith 2010). Thus, LOF variations can affect genes in various ways: Some genes will get inactivated, while others may retain some or all of their function; still others may be either on the way to pseudogenization or evolving into genes with new or related functions.
LOF events include SNPs that introduce premature STOP codons (nonsense SNPs) and lead to truncation of the protein, SNPs in splice sites, and indels and CNVs that can lead to changes in gene expression and function. A recent estimate of the number of genes in the human genome draws attention to the fact that the number of genes will vary between individuals due to CNVs (Pertea and Salzberg 2010). An analysis of CNVs of three personal genomes shows that, on average, 73–87 genes vary in copy number between two individuals (Alkan et al. 2009).
A careful analysis of human gene annotation suggested that the human genome apparently contained 1177 orphan ORFs that are not conserved across species (Clamp et al. 2007). While the majority of these were shown to be spurious genes, 168 candidates were identified as potential human-specific genes present mostly in duplicated regions. It has been postulated that new genes derived from segmental duplications evolve rapidly and may contribute to human-specific cell signaling pathways, as they are often related to cell proliferation, immunity, and inflammation responses (Stahl and Wainszelbaum 2009).
Consequences of gene inactivation events
Gene inactivation events can have widely varying effects on human phenotypes. LOF due to nonsense mutations has been implicated as disease-causing in ∼15%–30% of monogenic inherited diseases such as cystic fibrosis, hemophilia, retinitis pigmentosa, and Duchenne muscular dystrophy (Mort et al. 2008). However, there are also examples where such events appear to have been evolutionarily advantageous, resulting in the LOF allele increasing in frequency in the human population through positive natural selection. For instance, both the ACTN3 and CASP12 genes contain nonsense SNPs leading to premature STOP codons that result in the presence of both the active and inactive forms of the genes in the human population. The stop variant in CASP12 is the most common allele in all human populations, and is close to a frequency of 100% in many Eurasian populations; this inactive form is associated with increased resistance to sepsis (Xue et al. 2006). Likewise, the stop variant in ACTN3 has reached a frequency of ∼50% throughout Eurasia but is rare in African populations, and is associated with reduced muscle strength and enhanced endurance athletic performance in humans and a knockout mouse (MacArthur et al. 2007).
Advantageous LOF variants are likely to be the exception rather than the rule, but the sheer scale of LOF variation in the human genome suggests that many of these variants do not cause severe disease. One genotyping study analyzed 805 putative nonsense SNPs from the database of SNPs (dbSNPs) (Sherry et al. 2001) in 56 worldwide populations, and identified 169 genes containing nonsense variations in apparently healthy individuals, some of which were present at a high frequency in one or more populations (Yngvadottir et al. 2009). This study also showed that two individuals would differ by ∼24 genes because of nonsense SNPs. Ninety-nine genes were homozygously inactivated in one or more individuals. The analysis was necessarily limited in scope by the availability of candidate polymorphisms, but demonstrated that many genes are nonfunctional in some fraction of healthy individuals. Variation in the number of functional genes between people means that no individual genome possesses the full complement of functional human genes.
SNPs and gene inactivation
In this Perspective, we focus on nonsense SNPs and SNPs in splice sites because these are more clearly defined at the sequence level than many CNVs and indels, and their consequences are easier to predict from sequence data alone. Nevertheless, several complexities associated with such predictions need to be considered.
Nonsense SNPs
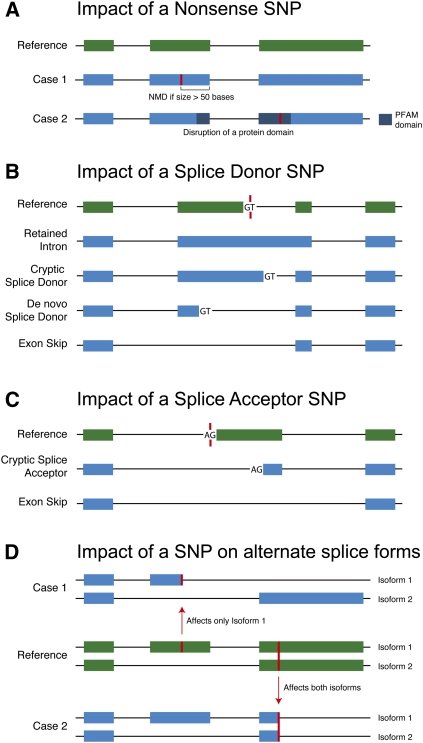
A premature STOP codon might affect all transcripts of a protein, or only some isoforms. When a nonsense SNP affects all transcripts of a gene, we can predict loss of gene function if the truncation makes it a candidate for nonsense-mediated decay (NMD), as shown in Figure 1A (Matsuda et al. 2008). Although transcripts that contain a premature STOP codon at distances >50 base pairs (bp) upstream of the last exon–exon junction are likely to be degraded by the NMD pathway, it is known that ∼5%–25% of residual mRNA remains (Isken and Maquat 2007). If a transcript containing a premature STOP is not degraded by NMD, the function of the protein may still be affected if the truncation removes a functional or structural domain, as shown in Figure 1A. Thus, it is nontrivial to predict whether a nonsense SNP will inactivate a gene.
Figure 1.
Consequences of nonsense SNPs and SNPs in canonical splice sites. The SNP is indicated by a red line in A and D, and a SNP in either of the canonical splice site positions is indicated by a red line in B and C.
SNPs in splice sites
Variations in canonical splice sites may affect splicing and lead to gene inactivation. We consider the two consensus bases in the internal intronic sequences flanking coding exons as the canonical splice sites (5′-GT … AG-3′). The effects of splice SNPs on gene structure and function are not obvious, and the consequences of such changes are complex (Krawczak et al. 1992; Chillon et al. 1995; Baralle and Baralle 2005). For example, a SNP at the splice donor site can lead to one of the following outcomes, as shown in Figure 1B: (1) The spliceosome does not recognize the donor site, and hence the intron is not excised. This leads to an mRNA with a retained intron. (2) A downstream cryptic donor splice site could be revealed and result in the production of a novel mRNA. (3) An upstream de novo donor splice site could be activated and also lead to the production of a novel mRNA. (4) Exon-skipping could occur as a result of a weak accepter site in the splice site preceding the donor site and/or due to the use of new intronic cryptic splice sites. Several instances of exon-skipping have been reported to result from donor splice site mutations (Aoshima et al. 1996; Chambliss et al. 1998).
Similarly, a SNP at the splice accepter site can affect splicing in several ways. A commonly seen effect is the skipping of the exon 3′ of the accepter site because the accepter site is not recognized (Fig. 1C). In all of these different splicing scenarios, several outcomes are possible. The change in splicing pattern may result in the inclusion of a premature STOP codon either directly or via a frameshift, and in turn lead to NMD or the truncation of a functional or structural domain. Alternatively, the change in gene structure may lead to a plausible full-length coding sequence that nevertheless misfolds due to insertion or deletion of additional amino acids, or the splice variant may produce a stable full-length protein that is structurally or functionally distinct from the “canonical” coding sequence encoded by the locus.
SNPs in alternate isoforms
The impact of SNPs that affect only some isoforms of a gene is even more difficult to interpret. Figure 1D illustrates the effect of a SNP on two isoforms of a gene. Only isoform 1, consisting of three exons, is affected when the SNP is in exon 2. On the other hand, when the SNP is in the third exon, both isoforms are affected. Transcript complexity is an often-overlooked aspect in genome analysis despite the fact that alternatively spliced isoforms are common. It is known that different isoforms of protein-coding genes may be expressed in different cell types. Therefore, SNPs affecting only some transcripts could have profound tissue-specific effects. It has been shown that SNP variation among transcripts in B cells is associated with loci related to four autoimmune diseases (Fraser and Xie 2009).
In the above discussion, we elaborated on the effects of SNPs on gene function. While it is clear that SNPs can lead to inactivation of genes, prediction of gene inactivation is not easy and requires experimental validation. Despite these uncertainties, some of the SNPs in the categories above lead to LOF and thus affect gene annotation.
Survey of personal genomes for impact of gene inactivation on gene annotation
Most of the examples discussed above were known before the current era of personal genome data. However, the availability of large numbers of individual whole-genome sequences now allows us to get an initial sense of the extent of genetic variation discovered by personal genome sequencing and its inferred impact on protein-coding gene annotation.
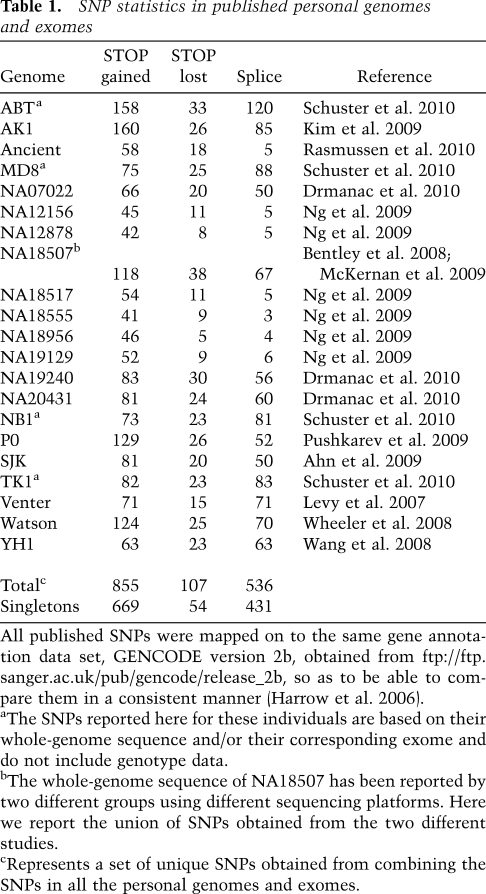
We surveyed the sequence variations in 21 recently published personal genomes and exomes for nonsense SNPs and SNPs in canonical splice sites using a uniform gene annotation (Table 1). All of the genomes contain many nonsense SNPs, ranging from 41 to 160 per individual (Table 1). A large proportion of these SNPs are seen in only one individual (singletons) (Table 1).
Table 1.
SNP statistics in published personal genomes and exomes
All published SNPs were mapped on to the same gene annotation data set, GENCODE version 2b, obtained from ftp://ftp.sanger.ac.uk/pub/gencode/release_2b, so as to be able to compare them in a consistent manner (Harrow et al. 2006).
aThe SNPs reported here for these individuals are based on their whole-genome sequence and/or their corresponding exome and do not include genotype data.
bThe whole-genome sequence of NA18507 has been reported by two different groups using different sequencing platforms. Here we report the union of SNPs obtained from the two different studies.
cRepresents a set of unique SNPs obtained from combining the SNPs in all the personal genomes and exomes.
The personal genome sequences have been obtained using different technologies and differing depths of sequence coverage. Therefore, the range of SNP numbers represents a combination of genuine diversity and experimental artifacts, with the latter being due to differences in sequencing platforms, depth of coverage, and different algorithms used for mapping reads to the genome and for calling genetic variants. In the following sections, we thus restricted the analysis to the subset of potential LOF SNPs found in both dbSNP129 and at least one of the personal genomes to obtain a more confident data set. We further narrowed the set to SNPs that affect all transcripts of a gene to obtain a SNP set with likely functional significance. After applying these criteria, the set of LOF variants contains 217 SNPs that introduce a premature STOP, 33 SNPs predicted to disrupt a STOP codon present in the reference, and 64 SNPs within canonical splice sites.
SNPs that introduce new STOP codons
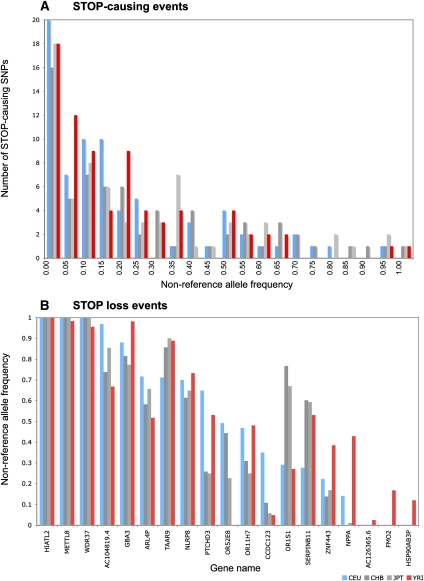
Of the 217 STOP-causing SNPs, 113 lead to truncated genes that are predicted to be targets for NMD. Figure 2A shows the frequency distribution of SNPs that lead to premature STOP codons in four different HapMap populations, and it is clear that the nonsense SNP is the major allele in a number of genes (Thorisson et al. 2005; Altshuler et al. 2010). Thus, in some cases, the gene variant containing the premature STOP codon is the predominant form present in most humans.
Figure 2.
(A) Frequency distribution of STOP-causing SNPs leading to premature truncations of proteins. Allele frequencies were obtained from HapMap data (Altshuler et al. 2010). The histograms are divided into bins of size 0.05 along the X-axis; each bin, except the first one, is inclusive of the lower bound and exclusive of the upper bound value. In the case of the interval 0–0.05, alleles with 0 frequency are not included. All other bins correspond to similar ranges. (B) Nonreference allele frequency at positions that lead to loss of a STOP codon in the human reference genome. Colors represent the HapMap populations: CEU, European (blue); CHB, Chinese (dark gray); JPT, Japanese (light gray); and YRI, Yoruban (red).
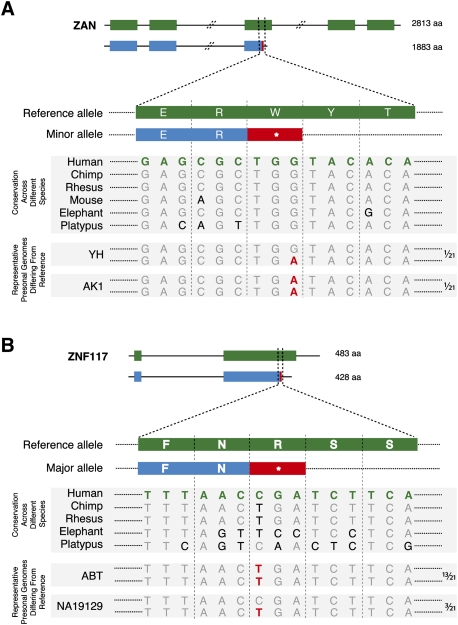
Figure 2A also shows that, while in most cases the frequency of the nonsense SNP is broadly similar across human populations, in some cases there is considerable variation. For example, Figure 3A shows the truncation of Zonadhesion (ZAN) at Trp 1883 due to the introduction of a premature STOP codon. However, while the frequency of the nonsense SNP for CEU and YRI is <3%, it is ∼50% for the Asian populations (CHB and JPT). We see a similar trend in the personal genomes where only the Asian individuals, AK1 and YH1, contain the STOP allele. It is easy to see that if the reference genome were that of an Asian individual, the truncated form might be annotated as the representative ZAN gene.
Figure 3.
SNPs resulting in premature STOP codons. (A) A G/A SNP, rs2293766, introduces a premature STOP codon at Trp 1883. The truncated form of ZAN is found predominantly in Asian populations at ∼50% frequency. The homozygous A/A genotype is seen in AK1 (Korean), and the heterozygous G/A genotype is seen in the YH (Chinese) personal genome. (B) A C/T SNP, rs1404453, results in truncation of ZNF117. The truncated form is conserved in other species and is the major allele in humans. The human reference genome sequence contains the minor allele C. The homozygous T/T genotype is seen in 13 (ABT, AK1, YH, Korean, NA07022, NA12156, NA12878, NA18517, NA18555, NA18956, NA20431, P0, and Venter) and the heterozygous C/T genotype is seen in three (NA19129, NA19240, and Yoruban) of the 21 personal genomes, as indicated on the right side of the figure. The SNP is labeled in red in the personal genome sequences.
Unlike ZAN, in the ZNF117 locus, the nonsense SNP occurs at a high frequency in all populations and also corresponds to the ancestral allele at this position (Fig. 3B). Thus the nonsense SNP in ZNF117 represents an example where the reference genome is different from the majority of analyzed humans.
SNPs that disrupt STOP codons
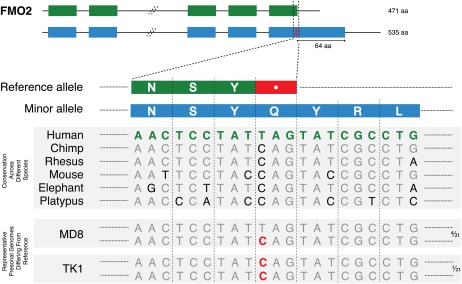
A nonsynonymous polymorphism at a STOP codon leads to elongation of the protein, as translation of mRNA continues until the next in-frame STOP codon is reached. In Figure 4, we illustrate an example of an annotated gene that likely represents a nonfunctional variant. In FMO2, a T/C SNP changes the annotated STOP codon to Glu 472 and extends the protein by 64 amino acids (Fig. 4). The truncated form, although not a candidate for NMD, does not appear to be catalytically active (Dolphin et al. 1998), and, although the functional allele is the longer version of FMO2, the shorter inactive form is annotated as if it were a functional gene.
Figure 4.
A T/C SNP, rs6661174, at the annotated STOP codon of FMO2 leads to loss of the STOP codon (TAG to CAG). This results in a 535-residue protein, 64 amino acids longer than the annotated FMO2 protein. The C allele is present in many mammalian species and is seen predominantly in the African genomes. The homozygous C/C genotype is seen in TK1 (Khoisan African genome), and the heterozygous T/C genotype is seen in four (MD8, NB1, NA19240, and Yoruban) of the 21 personal genomes, as indicated on the right side of the figure. The SNP is labeled in red in the personal genome sequences.
The functional allele exists in a minority of humans and is predominantly seen in the African population (Whetstine et al. 2000). Multiple sequence alignment of several mammalian species shows that the longer form is also present in other species (Fig. 4). There is thus a compelling argument that the minor allele encoding the longer, functional, and evolutionarily conserved protein should be annotated as the gene. A number of other examples where genes exist in both the active and potentially inactive forms in humans are shown in Figure 2B. In particular, the reference genome has a STOP codon early in the coding sequences of TAAR9 and SERPINB11.
Splice site SNPs
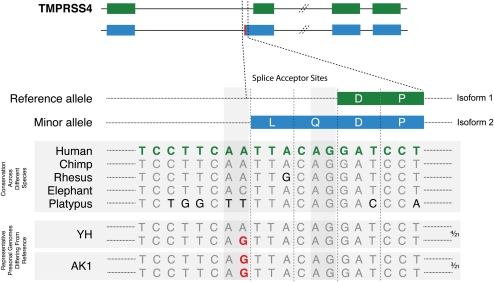
Of the 64 SNPs in splice positions from our SNP set, the reference genome has a noncanonical base at the annotated splice site in 13 cases. This may represent a variant or error in the reference, an incorrect splice site annotation, or a noncanonical but functional splice site. Figure 5 depicts a SNP A/G, rs2276122, where a new accepter splice site is created in intron 1 of TMPRSS4. This leads to alternative splicing at this newly formed accepter site, resulting in a protein that has two additional amino acids inserted in its sequence relative to the reference protein. Thus, the gene structure is changed relative to the reference due to a polymorphism, resulting in creation of an alternate accepter site.
Figure 5.
The SNP A/G, rs2276122, activates a cryptic splice site. A G at this position leads to creation of a new accepter site and insertion of two amino acids, leucine and glutamine, in isoform 2. The homozygous G/G is seen in three (AK1, NA12156, and NA20431) and the heterozygous A/G genotype is seen in four (YH, NA18956, P0, and Watson) of the 21 personal genomes, as indicated on the right side of the figure. The SNP is labeled in red in the personal genome sequences.
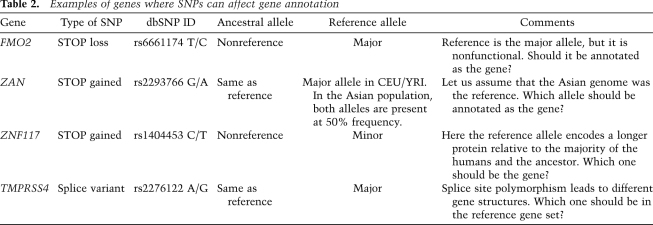
In Table 2, we summarize the examples discussed above that could potentially inactivate or alter gene structure and consequently affect gene annotation. The table highlights several issues in gene annotation in the context of SNPs. It is clear that sequence variations need to be taken into consideration for gene annotation purposes. Here we pose a key question: What sequence should be used as the reference for gene annotation, given that some genes are nonfunctional in some humans?
Table 2.
Examples of genes where SNPs can affect gene annotation
Defining the reference sequence
Here we present five models for selecting the reference allele: (1) the allele present in the current reference, (2) the allele present in a chosen high-quality individual genome, (3) the most common human allele, (4) the ancestral allele, and (5) the maximum coding potential allele.
Current reference allele
The current human reference is a composite of sequences from multiple individuals. While it does not represent any single individual, it is also not representative of the diversity that we see from the personal genome sequences. There is also an inherent bias toward annotating genes that are functional in the reference genome, resulting in events that may lead to gene inactivation being largely overlooked in automatic annotation processes. We earlier described cases in which the reference allele is the minor allele and therefore is not representative of the majority of all humans. Thus, annotation based on the current human reference genome does not provide an accurate and complete set of the genes found across all human populations. We continue to use the initial human genome as the reference sequence, despite it being an arbitrary choice, because it provides us with a convenient high-quality assembly and annotation.
Allele from a single individual's genome
Alternatively, a single individual genome sequence could be used as the reference. One advantage of using an individual genome would be that it includes haplotypes that are actually present in the population; the critical disadvantage is that any one individual represents an arbitrary choice and would not be representative of the diversity of gene content seen in the human population. In addition, choices would have to be made between the two alleles at heterozygous positions, including between active and inactive forms of a gene. Another issue with using an individual genome is that, while we assume that the genome is representative of a healthy individual, one cannot rule out the possibility that some genes may represent a disease state even though the disease was not apparent in the individual sequenced. Thus, there is a potential to contaminate the reference gene set with genes representative of a disease state (e.g., fusion genes from an undetected cancer).
Most common human allele as the basis of annotation
Reference alleles at polymorphic sites could be defined using the most common allele obtained from alignment of multiple individual genome sequences. The problem with this approach is that, in many cases, both allelic variants at a given position may occur at similar frequencies, making it difficult to pick the most frequent reference allele. This issue is compounded by potential biases in the choice of individuals sequenced; it would result in a relatively unstable reference gene set, as new sequenced genomes would alter the observed frequency of alleles and thus result in inclusion or exclusion of different alleles in the reference gene sequence over time.
Ancestral allele as the basis of annotation
The ancestral allele at any variable site in the genome is defined as the allele present in the common ancestor of all humans, a state that can typically be inferred confidently through comparison with outgroup species, usually nonhuman primates such as chimpanzees or macaques, at the majority of locations in the genome where such comparative data are available.
The primary benefit of using ancestral state to define the reference allele is that it represents a fundamental attribute of a polymorphism rather than an arbitrarily defined one. However, this approach also raises challenges: Incomplete genome sequences of nonhuman species mean that ancestral state cannot be accurately assigned for a small but nontrivial fraction of human polymorphisms, and some positions (hot spots) mutate so frequently that ancestral state information is lost.
We anticipate that the issue of incomplete comparative data will be largely resolved as additional nonhuman primates (including from fossils) are sequenced and the genome assemblies of currently sequenced species are improved. In the case of LOF variants, it would be possible to use the functional allele as a proxy for ancestral state when it cannot be unambiguously assigned.
However, inferring ancestral state will remain complicated for some regions of the genome such as highly repetitive, recently duplicated, and copy number variant regions. For example, the chromosome 17q21.31 MAPT inversion polymorphism, enriched in Europeans, contains a 970-kb region that has been predicted to be inverted at least three times independently in the human, chimpanzee, and orangutan lineages (Zody et al. 2008). For such complex cases, ancestral state inference will require high-quality, long-read sequence data from multiple primate species, along with careful manual annotation of the resulting assemblies.
It is also difficult to infer ancestral state for some genes and gene families that have undergone human-specific expansion, such as olfactory receptors, defensins, and zinc finger transcription factor genes (Newman and Trask 2003; Schneider et al. 2005; Hamilton et al. 2006; Groth et al. 2010). In some cases, the assignment of ancestral alleles may not be possible, as orthologs may be difficult to identify. However, in many cases, it will still be possible to reconstruct the ancestral state using a combination of human variation data and the sequence of closely related genes.
Maximum coding potential allele as the basis for annotation
We define the maximum coding potential allele as a functional allele present in any one individual. In this all-inclusive reference sequence, the functional allele of a gene would be present in the reference genome even if it has been seen in only one individual, with every other sequenced sample carrying a pseudogene.
The benefit of this approach is that it is comprehensive: By including all potentially functional genes, the inclusive reference set would incorporate as much potentially functional coding sequence as possible—a feature of major interest to biological and medical researchers. However, this method ignores the evolutionary impact of gene inactivation by including a gene as functional even though it has become inactivated in a majority of populations and individuals. Moreover, there is a danger of including potential disease alleles in the reference.
Distinguishing benign LOF variants in apparently healthy humans from disease-causing variants will be a challenging task given that some rare variants, especially heterozygous ones, might represent hitherto undiagnosed or future disease states. Recessive disease-causing variants are benign in the heterozygous state but may result in severe Mendelian disease when present in the homozygous state. Nonetheless, it is likely that these marginal cases can be resolved through careful manual annotation (Harrow et al. 2006).
In choosing between these models, an additional factor to consider is that the model is applicable to other species. While several large-scale sequencing projects of other genomes are under way, some will not be performed at the same scale as for humans, in terms of both the sequencing accuracy and the number of individuals sampled. Careful manual analysis for other large-scale genome annotation projects may not always be feasible. For consistent genome annotation across species, the human reference annotation should be based on criteria that make it a suitable model for comparative genome annotation and analyses. We also note that, in many regions, there will be a need for more than one reference sequence, especially for genomic locations such as the HLA cluster, where multiple highly divergent haplotypes coexist in the population. In this case, the MHC Haplotype Project (Horton et al. 2008) annotated eight haplotypes and chose a single haplotype, PGF, as the reference MHC sequence simply because this is the longest MHC haplotype.
Concluding remarks
The tremendous growth in human genome sequence data in the last decade has clearly revealed the challenges of defining a single reference genome and reference gene set due to interindividual variability. However, resolving these challenges will be crucial if we are to have a reference gene set that provides maximum power for studies using large-scale sequencing to identify human functional variation.
Two desirable qualities of a reference gene set are that it (1) contains as much of the functional protein-coding sequence in the genome as possible, and (2) be relatively stable over time; i.e., unlikely to change frequently as more personal genome sequences become available. Weighing the benefits and limitations of each of our models, we propose that the reference sequence be derived on the basis of the ancestral allele, and the reference gene annotation be based on this sequence.
The ancestral allele model is nonarbitrary, should be relatively stable over time, and is applicable to genome annotation of other species. However, a drawback of this model is that information about some human-specific changes, such as duplications that have arisen since the most recent common ancestor of a region of the genome and remain polymorphic, will be excluded. Therefore, we propose that a second “expanded” human gene set be defined to include such cases in addition to the core reference gene set. Representing this rich knowledge base will require improved visualization platforms within existing browsers as well as comprehensive manual annotation.
While the focus of this Perspective has been on protein-coding genes, it should be noted that a reference sequence derived from the ancestral allele provides a consistent framework for annotating both coding and noncoding regions of a genome. Referencing the noncoding regions of the genome based on the simple criteria outlined in the earlier section will not be straightforward, as these regions are more variable than coding regions and there can be tremendous diversity in some duplicated regions that are both lineage- and individual-specific (Ewing and Kazazian 2010).
We can extend the concepts introduced here for the reference gene set to the entire human genome sequence. Specifically, we can represent all polymorphic positions in the reference genome with the most likely ancestral allele, wherever this state can be inferred. Under this scenario, the reference sequence would correspond to a reconstruction of the haplotype present in the most recent common ancestor of all humans. Such a choice provides a uniform and consistent basis for choosing one variant over another at every variable position, avoids biases arising from an arbitrary reference genome, and would provide a coherent framework for the next generation of medical and functional genomics.
Acknowledgments
S.B. thanks Declan Clarke and Jing Wang for thorough reading of the manuscript. We thank the referees for their insightful comments. This work was supported by grants from the NIH and the Albert L. Williams Professorship funds. D.G.M. and C.T.S. are supported by The Wellcome Trust.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1968411.
References
- Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim BC, Kim SY, Kim WY, Kim C, Park D, et al. 2009. The first Korean genome sequence and analysis: Full genome sequencing for a socio-ethnic group. Genome Res 19: 1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 41: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshima M, Nunoi H, Shimazu M, Shimizu S, Tatsuzawa O, Kenney RT, Kanegasaki S 1996. Two-exon skipping due to a point mutation in p67-phox–deficient chronic granulomatous disease. Blood 88: 1841–1845 [PubMed] [Google Scholar]
- Baralle D, Baralle M 2005. Splicing in action: Assessing disease causing sequence changes. J Med Genet 42: 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss KL, Hinson DD, Trettel F, Malaspina P, Novelletto A, Jakobs C, Gibson KM 1998. Two exon-skipping mutations as the molecular basis of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria). Am J Hum Genet 63: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillon M, Dork T, Casals T, Gimenez J, Fonknechten N, Will K, Ramos D, Nunes V, Estivill X 1995. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811+1.6kbA→G, produces a new exon: High frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am J Hum Genet 56: 623–629 [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK 2006. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38: 75–81 [DOI] [PubMed] [Google Scholar]
- Dolphin CT, Beckett DJ, Janmohamed A, Cullingford TE, Smith RL, Shephard EA, Phillips IR 1998. The flavin-containing monooxygenase 2 gene (FMO2) of humans, but not of other primates, encodes a truncated, nonfunctional protein. J Biol Chem 273: 30599–30607 [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81 [DOI] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH Jr 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 20: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Xie X 2009. Common polymorphic transcript variation in human disease. Genome Res 19: 567–575 [DOI] [PubMed] [Google Scholar]
- Gerstein M, Zheng D 2006. The real life of pseudogenes. Sci Am 295: 48–55 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M 2007. What is a gene, post-ENCODE? History and updated definition. Genome Res 17: 669–681 [DOI] [PubMed] [Google Scholar]
- Groth M, Wiegand C, Szafranski K, Huse K, Kramer M, Rosenstiel P, Schreiber S, Norgauer J, Platzer M 2010. Both copy number and sequence variations affect expression of human DEFB4. Genes Immun 11: 458–466 [DOI] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L 2006. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res 16: 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. 2006. GENCODE: Producing a reference annotation for ENCODE. Genome Biol 7: S4 doi: 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, et al. 2008. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics 60: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, et al. 2009. A highly annotated whole-genome sequence of a Korean individual. Nature 460: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet 90: 41–54 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. 2007. The diploid genome sequence of an individual human. PLoS Biol 5: e254 doi: 10.1371/journal.pbio.0050254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunshof JE, Bobe J, Aach J, Angrist M, Thakuria JV, Vorhaus DB, Hoehe MR, Church GM 2010. Personal genomes in progress: From the human genome project to the personal genome project. Dialogues Clin Neurosci 12: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, et al. 2010. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 362: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Tyler-Smith C 2010. Loss-of-function variants in the genomes of healthy humans. Hum Mol Genet 19: R125–R130 doi: 10.1093/hmg/ddq365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Seto JT, Raftery JM, Quinlan KG, Huttley GA, Hook JW, Lemckert FA, Kee AJ, Edwards MR, Berman Y, et al. 2007. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet 39: 1261–1265 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Sato H, Maquat LE 2008. Studying nonsense-mediated mRNA decay in mammalian cells. In Methods in enzymology vol. 449 (ed. Maquat LE, Kiledjian M), pp. 177–201 Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. 2009. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 19: 1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE 2006. An initial map of insertion and deletion (indel) variation in the human genome. Genome Res 16: 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA 2008. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29: 1037–1047 [DOI] [PubMed] [Google Scholar]
- Newman T, Trask BJ 2003. Complex evolution of 7E olfactory receptor genes in segmental duplications. Genome Res 13: 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Salzberg SL 2010. Between a chicken and a grape: Estimating the number of human genes. Genome Biol 11: 206 doi: 10.1186/gb-2010-11-5-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev D, Neff NF, Quake SR 2009. Single-molecule sequencing of an individual human genome. Nat Biotechnol 27: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, et al. 2010. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JJ, Unholzer A, Schaller M, Schafer-Korting M, Korting HC 2005. Human defensins. J Mol Med 83: 587–595 [DOI] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 463: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl PD, Wainszelbaum MJ 2009. Human-specific genes may offer a unique window into human cell signaling. Sci Signal 2: pe59 doi: 10.1126/scisignal.289pe59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD 2005. The International HapMap Project Web site. Genome Res 15: 1592–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. 2008. The diploid genome sequence of an Asian individual. Nature 456: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Yueh MF, McCarver DG, Williams DE, Park CS, Kang JH, Cha YN, Dolphin CT, Shephard EA, Phillips IR, et al. 2000. Ethnic differences in human flavin-containing monooxygenase 2 (FMO2) polymorphisms: Detection of expressed protein in African-Americans. Toxicol Appl Pharmacol 168: 216–224 [DOI] [PubMed] [Google Scholar]
- Xue Y, Daly A, Yngvadottir B, Liu M, Coop G, Kim Y, Sabeti P, Chen Y, Stalker J, Huckle E, et al. 2006. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet 78: 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yngvadottir B, Xue Y, Searle S, Hunt S, Delgado M, Morrison J, Whittaker P, Deloukas P, Tyler-Smith C 2009. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am J Hum Genet 84: 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M 2010. Identification and analysis of unitary pseudogenes: Historic and contemporary gene losses in humans and other primates. Genome Biol 11: R26 doi: 10.1186/gb-2010-11-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. 2008. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 40: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]