Abstract
Dynamic regulation of histone modifications is critical during development, and aberrant activity of chromatin-modifying enzymes has been associated with diseases such as cancer. Histone demethylases have been shown to play a key role in eukaryotic gene transcription; however, little is known about how their activities are coordinated in vivo to regulate specific biological processes. In Drosophila, two enzymes, dLsd1 (Drosophila ortholog of lysine-specific demethylase 1) and Lid (little imaginal discs), demethylate histone H3 at Lys 4 (H3K4), a residue whose methylation is associated with actively transcribed genes. Our studies show that compound mutation of Lid and dLsd1 results in increased H3K4 methylation levels. However, unexpectedly, Lid mutations strongly suppress dLsd1 mutant phenotypes. Investigation of the basis for this antagonism revealed that Lid opposes the functions of dLsd1 and the histone methyltransferase Su(var)3–9 in promoting heterochromatin spreading at heterochromatin–euchromatin boundaries. Moreover, our data reveal a novel role for dLsd1 in Notch signaling in Drosophila, and a complex network of interactions between dLsd1, Lid, and Notch signaling at euchromatic genes. These findings illustrate the complexity of functional interplay between histone demethylases in vivo, providing insights into the epigenetic regulation of heterochromatin/euchromatin boundaries by Lid and dLsd1 and showing their involvement in Notch pathway-specific control of gene expression in euchromatin.
Keywords: Drosophila, histone demethylases, H3K4me, H3K9me, heterochromatin, Notch signaling
In eukaryotic cells, genomic DNA is packaged with histone and nonhistone proteins to form chromatin. The combined action of chromatin remodeling activities and histone-modifying enzymes dynamically regulates the chromatin status and has been implicated in the control of numerous biological processes, including transcription, cell division, differentiation, and DNA repair. Furthermore, aberrant activity of chromatin-modifying enzymes has been strongly associated with diseases such as cancer (Chi et al.2010).
The methylation pattern of histones is believed to define transcriptionally active and inactive chromatin domains. For example, methylation at Lys 9 of histone H3 (H3K9) is associated with silenced chromatin (heterochromatin), while methylation of Lys 4 of histone H3 (H3K4) is an important mark of actively transcribed genes (Bannister and Kouzarides 2005; Martin and Zhang 2005; Trewick et al. 2005; Wysocka et al. 2005). H3K4 can be mono-, di-, or trimethylated, and several histone methyltransferases act at this residue, including MLL1, which is frequently translocated in leukemias (Krivtsov and Armstrong 2007). Levels of H3K4 trimethylation peak around promoters of actively transcribed genes (Bernstein et al. 2005), while H3K4me2 is highest just downstream from transcriptional start sites, and monomethylation is more dispersed throughout the body of genes (Liu et al. 2005). In addition to the promoter regions, these modifications are also detected in intergenic regions, and H3K4me1 signals have been correlated with functional enhancers (Roh et al. 2004; Heintzman et al. 2007). These observations suggest that different levels of methylation may have different functional consequences, and that the transition from mono- to di- to trimethyl moieties is likely to be a critical and highly regulated event.
Until recently, histone methylation was regarded as an irreversible modification. The discovery of the first histone lysine demethylase LSD1/KDM1 (lysine-specific demethylase 1), and subsequently of a second family of histone demethylases, including members of the Jmj domain-containing proteins, suggested a more dynamic regulation of the methylation state of histones. These discoveries raise several fundamental questions: How are the activities of histone methyltransferases and demethylases coordinated to establish the appropriate patterns of methylation, and how are these patterns regulated to allow accurate chromatin dynamics and gene expression cascades contributing to animal development and homeostasis? Very little is known about the dynamic regulation of histone methylation by methyltransferases and demethylases in vivo, and even less is known about the signaling pathways that control their activity.
We and others generated mutants for the Drosophila ortholog of the histone demethylase LSD1 (dLsd1). Like its mammalian counterpart, dLsd1 specifically demethylates H3K4me2 and H3K4me1 residues, indicating functional conservation (Rudolph et al. 2007). We showed that dLsd1 mutation affects male viability as well as specific developmental processes such as wing development and oogenesis (Di Stefano et al. 2007). Furthermore, mutant alleles of dLsd1 strongly suppress positional effect variegation (PEV), indicating that dLsd1 contributes to maintaining the balance between euchromatin and heterochromatin (Di Stefano et al. 2007; Rudolph et al. 2007). Taken together, these studies showed that dLsd1 plays a crucial role in chromatin regulation during Drosophila development, and that its depletion impacts specific developmental processes.
An important question arising from these initial studies is how dLsd1 cooperates with other chromatin-associated proteins to dynamically control chromatin during animal development. We reasoned that, while many different chromatin regulators might have activities that could directly or indirectly interconnect with dLsd1, the clearest functional interactions would likely be seen with other enzymes that act on H3K4 methylation. In Drosophila, the ortholog of the Jarid 1 family of JmjC domain-containing proteins, Lid (little imaginal discs), has been shown to specifically demethylate H3K4me2 and H3K4me3 residues in vitro and H3K4me3 in vivo (Eissenberg et al. 2007; Lee et al. 2007; Secombe et al. 2007; Lloret-Llinares et al. 2008). Therefore, as a starting point, we generated flies that are mutant for both dLsd1 and Lid. Consistent with their enzymatic activities, we find that Lid; dLsd1 mutant flies display an increased level of H3K4 methylation. However, intriguingly, Lid mutations strongly suppress dLsd1 mutant phenotypes. An analysis of the basis of this antagonism reveals that the interplay of Lid with dLsd1 occurs through distinct mechanisms in different contexts. For example, Lid opposes the function of dLsd1 and the H3K9 methyltransferase Su(var)3–9 at heterochromatin–euchromatin boundaries while cooperating with dLsd1 in regulating certain Notch target genes in euchromatic contexts. These findings illustrate the complexity of functional interactions between demethylases in vivo. To understand the functional interplay between these enzymes in animal development, we must therefore not only consider the biochemical properties of the enzymes, and the chromatin context of their targets, but also understand how their activities impact the status of key signaling pathways that define the cellular response, such as the Notch pathway.
Results
Lid suppresses phenotypes associated with dLsd1 mutation
To understand the function of the dLsd1 demethylase, it is essential to determine how its activities are integrated with other chromatin-associated proteins, since it is the concerted action of multiple enzymes that enables chromatin states to be controlled dynamically in vivo. One protein likely to impact dLsd1 function in Drosophila melanogaster is Lid. Lid and dLsd1 have both been shown to demethylate histone H3K4 methyl residues. dLsd1 acts specifically on mono- and dimethyl residues, while Lid can act on di- and trimethyl residues. To study the combinatorial contributions of H3K4 demethylases in vivo, we generated compound mutants for Lid and dLsd1. Given their enzymatic activities, we expected that Lid; dLsd1 double-mutant flies would likely show a synergistic increase in the levels of H3K4 methylation. If these proteins act cooperatively, then the combined mutation of the two H3K4 demethylases would be expected to cause defects that were more severe than the single mutants.
We showed previously that inactivation of dLsd1 in Drosophila results in specific developmental defects. Homozygous mutation of dLsd1 leads to a held-out wing phenotype, male lethality, and defects in oogenesis (Di Stefano et al. 2007). Further analysis of dLsd1ΔN mutant flies revealed an additional defect in the wing. Approximately 36% of dLsd1ΔN mutants exhibit ectopic vein tissue emanating from the posterior cross-vein (pcv), posterior to the L5 longitudinal vein (Fig. 1A,A′). This phenotype, which has already been observed in mutants of chromatin remodeling complex components, such as Snr1 (Marenda et al. 2003), indicates that dLsd1 has a role in the repression of vein development in intervein cells.
Figure 1.
Lid10424 mutation suppresses phenotypes associated with dLsd1ΔN loss-of-function mutation. (A) Loss of dLsd1 results in extra wing vein tissue emanating from the pcv (indicated by arrows; closer view in A′). (B) This phenotype is rescued by heterozygous mutation of Lid (indicated by arrows; closer view in B′). (C,D) Lid mutation suppresses the held-out wing phenotype observed in homozygous dLsd1 mutant flies. (E) Schematic representation of a wild-type ovariole, including the germarium and egg chambers (stages 1–3). Germline stem cells (GSC) reside at the tip of the germarium in a microenvironment created by the cap cells (CC) and the terminal filaments (TF). The differentiating daughter cell of a germline stem cell is the cystoblast (CB), which moves posterior and becomes encompassed by inner germarian sheath cells (IGS). Cystoblasts divide four times and generate germline cysts of 16 cells, which, after passing region 2 of the germarium, become surrounded by follicle cell precursors (FCP). Follicle cell precursors are generated by somatic stem cells (SSC) and differentiate into follicle cells (FC), polar cells (PC), and stalk cells (SC). Among the 16 germ cells, one differentiates into the oocyte (O), and the remaining 15 become nurse cells (NC). (F,G,H) Lid10424 mutations partially rescue dLsd1ΔN mutant follicle cell defects. Wild-type (w1118), dLsd1ΔN homozygous, and Lid10424/+; dLsd1ΔN/dLsd1ΔN ovaries were stained with YOYO-1 (green) and anti-Fasciclin-III to outline somatic cells. (I,J,K) Lid10424 mutation partially rescues dLsd1ΔN mutant defects in the germline. Wild-type (w1118), dLsd1ΔN homozygous, and Lid10424/+; dLsd1ΔN/dLsd1ΔN ovaries were stained with TOTO (blue) and anti-orb to visualize the oocyte (indicated by arrows). Cell outlines and ring canals were visualized by phalloidin staining of the actin cytoskeleton (red).
Remarkably, combining homozygous dLsd1 mutants with a loss-of-function mutation of Lid (Lid10424) results in a strong dominant suppression of the dLsd1ΔN wing vein phenotype (Fig. 1A,B; Table 1). The Lid mutant was further tested against the dLsd1ΔN allele in a second independent assay using the held-out wing phenotype. When we quantified the number of flies with held-out wings, we found that heterozygous mutation in Lid reduces the penetrance of this phenotype from 87% of dLsd1 homozygous mutant flies to 36% (Fig. 1C,D; Table 1).
Table 1.
Lid 10424 supresses dLsd1ΔN wing phenotypes
(N=) The number of wings counted, in the case of the modified wing phenotype, and the number of flies counted, in the case of the held-out wing phenotype.
Each of these wing phenotypes resulting from dLsd1 mutation was markedly suppressed by mutant alleles of Lid (Fig. 1A–D; Table 1). To determine whether this suppression was limited to the wing, we tested the Lid mutant allele for its ability to modify the highly penetrant dLsd1ΔN oogenesis defect (Di Stefano et al. 2007). To gain insights into the specific effects of Lid mutation on the dLsd1ΔN-dependent oogenesis defect, wild-type and mutant ovaries were stained with antibodies specific for ovarian cell markers. The Drosophila ovary consists of ∼14–16 ovarioles, which contain strings of developing egg chambers of progressive age with a germarium at the anterior tip (Fig. 1E; Kirilly and Xie 2007). As a consequence of dLsd1 mutation, oogenesis arrests very early, with dLsd1 homozygous mutant ovaries consisting of germaria that do not bud off egg chambers. Labeling the ovaries with an antibody to Fasciclin III (Fas III), a membrane-associated protein present in prefollicle and follicle cells, shows that the number of these cell types is strongly reduced in dLsd1ΔN mutant ovaries (Fig. 1G) compared with wild-type ovaries (Fig. 1F), and that these cells fail to completely encapsulate the germline cyst. In the partially encapsulated cysts that do exist, oocyte determination/localization appears abnormal, as judged by nuclei staining and Orb accumulation (Fig. 1J). In many cases, one mutant copy of Lid partially rescued these defects, as shown by the presence of egg chambers at later stages of development, an increased number of cells positively staining for Fas III (Fig. 1H), and a more localized accumulation of Orb protein in the oocyte (Fig. 1K). To ensure that this suppression is due specifically to Lid inactivation, we tested two independently generated P-element insertions, both within the Lid gene, and found similar results (Supplemental Fig. S1).
To further confirm this interaction, we tested the effect of Lid mutation on another phenotype associated with dLsd1 loss of function. Previously, we showed that homozygous mutation of dLsd1 dramatically reduces male viability; in this study, we found that Lid10424 dominantly rescues dLsd1ΔN-dependent male lethality (from 1%–12% viability) (Table 2; Supplemental Table S1). Taken together, this set of observations shows that Lid antagonizes dLsd1 function in vivo. This antagonism is a general phenomenon that is evident in multiple tissues.
Table 2.
Effect of Lid 10424 and dLsd1ΔN mutation on gender distribution
Percentage of males and females of the indicated genotypes obtained from Lid10424/CyO; dLsd1ΔN/TM3 X Lid10424/CyO; dLsd1ΔN/TM3. A total of at least 106 flies were counted for each genotype.
To exclude the possibility that any general manifestation of abnormal chromatin regulation could suppress dLsd1 mutant phenotypes, we carried out a candidate genetic screen among components of chromatin remodeling complexes, Trx-G complexes, and PcG complexes for dominant modification of the homozygous dLsd1ΔN wing and ovary phenotypes. The extra wing vein phenotype observed in dLsd1 mutant flies was unaffected by mutant alleles of ash2, Trl, and Psc, or by mutations in Bap180, a specific component of the P-BAP complex (Supplemental Table S2). Hypomorphic, amorphic, null, and loss-of-function mutations in most BAP complex subunits (mor, osa, and brm) enhanced, rather than suppressed, the severity of the extra wing vein phenotype of dLsd1ΔN mutants (Supplemental Fig. S2; Supplemental Table S2), suggesting that dLsd1 and the BAP complex may cooperate in the regulation of intervein-specific gene expression. In addition to Lid alleles, the extra vein phenotype of dLsd1ΔN mutants was also strongly suppressed by Snr1 mutations. However, unlike the Lid alleles, which gave a consistent pattern of interaction across all of the dLsd1 mutant phenotypes, these other alleles that we tested had variable effects on different aspects of the dLsd1 mutant phenotype (Supplemental Table S2). We conclude that the ability of Lid alleles to consistently suppress the effects of dLsd1 mutation is an unusual property that is not seen with other general regulators of chromatin structure.
dLsd1 and Lid mutations cooperatively increase the global level of H3K4 methylation
Given that both Lid and dLsd1 have been shown to have demethylase activity toward methylated H3K4 residues, we performed an immunoblotting analysis to assess whether the combined mutation of Lid and dLsd1 does indeed result in increased global levels of H3K4 methylation. As predicted from the biochemical studies, we found substantially increased levels of H3K4 monomethylation and dimethylation in dLsd1ΔN homozygous mutant adult flies compared with wild type, whereas the global levels of H3K4 trimethylation were unchanged in these mutants. Mutation of Lid, as expected, impacted H3K4 di- and trimethylation and, most likely through an indirect mechanism, also resulted in higher H3K4 monomethylation levels. Consistent with their function as H3K4 demethylases, the combined mutation of dLsd1 and Lid resulted in a significant increase in mono-, di-, and tri-H3K4 methylation in adult flies, while the global level of H3 acetylation was not affected (Fig. 2A).
Figure 2.
dLsd1 and Lid mutations cooperatively increase the global level of histone H3K4 methylation. (A) Increase in histone H3K4 methylation level in double-mutant adult flies. Immunoblots of wild-type (wt) versus HDM mutant lysates from adult flies were probed with antibodies specific for mono-, di-, and trimethyl H3K4 and pan-acetyl H3; anti-H3 was used as a loading control. (B–G) H3K4 methylation levels are increased in polytene chromosomes of double mutants. Staining of polytene chromosome with an antibody specific for H3K4me2 (red) and with YOYO-1 (green). Reduced levels of Lid in combination with loss of dLsd1 results in a chromosome-wide increase in H3K4me2 levels.
To attest that methylation changes occur on H3 loaded on chromosomes, and to visualize these changes on the genome, we examined the effect of combined mutation of Lid and dLsd1 on the levels of H3K4 dimethylation by immunostaining polytene chromosomes dissected from larvae. This staining showed a general increase of H3K4 dimethylation in double-mutant flies compared with the wild-type counterparts (Fig. 2B–G), while the levels of H3 are similar in all of the genotypes analyzed (Supplemental Fig. S3). Thus, the less severe phenotype of double-mutant flies is associated with a higher overall level of H3K4 methylation throughout the genome.
dLsd1 and Lid antagonistically control heterochromatin spreading in PEV
The apparent contradiction between biochemical and genetic data is very intriguing: The biochemical data, showing a global increase of H3K4 methylation in the double mutants, suggest that Lid and dLsd1 cooperate, whereas the less severe phenotype of dLsd1 mutants in a heterozygous Lid background indicates that the two proteins can also function antagonistically.
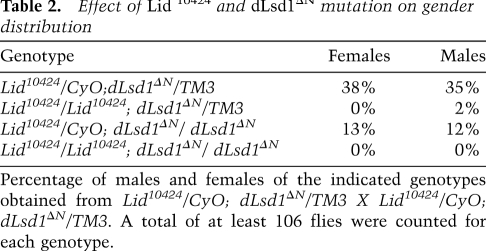
To understand the basis for this antagonism, we initially investigated how the combined mutation of Lid and dLsd1 affects chromatin homeostasis. First, we examined the effects of these mutations on PEV. PEV is the mosaic pattern of gene silencing observed as the consequence of an abnormal juxtaposition of a gene next to a segment of heterochromatin. A prototypical example of PEV involves the Drosophila white gene (w). white is normally located at the distal tip of the X chromosome and is expressed in every ommatidium of the eye, resulting in a red eye phenotype. In wm4 mutants, a chromosomal inversion places white next to pericentric heterochromatin, resulting in mosaic expression of the white gene and patches of red and white tissue (Schotta et al. 2003).
dLsd1 has been implicated in the regulation of heterochromatin gene silencing on the basis of its function as a strong suppressor of PEV (Di Stefano et al. 2007; Rudolph et al. 2007). While dLsd1 mutation dominantly suppresses variegation in a wm4h background (Fig. 3; Di Stefano et al. 2007; Rudolph et al. 2007), Lid mutation, surprisingly, was shown to enhance variegation of the white locus (Fig. 3; Lloret-Llinares et al. 2008). This indicates opposing functions of dLsd1 and Lid in PEV regulation, and suggests a role for Lid in maintaining transcriptionally competent chromatin states at heterochromatin–euchromatin boundaries. Based on these findings, we hypothesized that reducing the level of Lid in a dLsd1 heterozygous mutant background might restore heterochromatin formation and contribute to the suppression of dLsd1 mutant phenotypes. To test this hypothesis, we determined the effects of Lid; dLsd1 double mutants on wm4h variegation. In accordance with our hypothesis, mutation of Lid suppressed the effect of dLsd1 mutation on PEV, suggesting that Lid and dLsd1 act in an antagonistic manner in heterochromatin gene silencing. To investigate the transcriptional status of the white gene in these mutant flies, we measured its expression in the heads by RT-qPCR analysis. We found that white expression is down-regulated in wm4h; Lid mutants and up-regulated in wm4h; dLsd1 mutants, while in double mutants its expression is more similar to wild-type levels (Fig. 3G; Supplemental Fig. S4), indicating that the positive role of dLsd1 in heterochromatin gene silencing is indeed opposed by Lid. Interestingly, the expression of the genes neighboring the w locus was only marginally affected by dLsd1 and Lid mutations, suggesting differences in the response to expansion or contraction of heterochromatin by different genes, with white being the most responsive in the head tissues.
Figure 3.
Lid and dLsd1 antagonistically control heterochromatin spreading in PEV. (A–D) The effect of a reduced dosage of Lid and dLsd1 on wm4h variegation compared with control flies is shown. Reducing the dosage of dLsd1 results in suppression of variegation (B), while reducing the levels of Lid results in enhancement (C); combined reduction of Lid and dLsd1 dosage has a minimal effect on variegation (D). (E) The effect of a reduced dosage of Lid and dLsd1 on T(2;3)Sbv variegation compared with control flies is shown. The effect was quantified by counting the number of stubble and wild-type bristles of the thorax. A minimum of 380 bristles per genotype was counted. (F) A schematic representation of the white locus at the wm4 inversion is shown. (G) dLsd1 and Lid mutations have opposite effects on white gene expression. Reducing the levels of dLsd1 results in increased expression of the white gene, while reducing the levels of Lid results in decreased expression as compared with wild-type flies. Combined reduction of Lid and dLsd1 dosage results in an intermediate effect compared with single mutants. (H) dLsd1 and Lid control H3K9 methylation levels at the white locus. ChIP analysis of H3K9me2 levels along the wm4h inverted region. Reducing the levels of dLsd1 results in decreased levels of H3K9me2 at the white locus, while reducing the levels of Lid results in increased H3K9me2, and a combined reduction of Lid and dLsd1 dosage results in levels of H3K9me2 comparable with wild-type flies.
To test whether this effect on heterochromatin gene silencing is limited to the pericentric chromatin of the X chromosome or whether it is a more general effect, we measured the effect on PEV using the T(2;3)Sbv translocation. In these flies, the Stubble (Sb) mutation is juxtaposed to the centric heterochromatin of the second chromosome, resulting in mosaic flies with both stubble and normal bristles (Sinclair et al. 1983; Schotta et al. 2004). When T(2;3)Sbv was crossed to dLsd1ΔN, we observed a significant increase in the frequency of stubble bristles, indicating a suppression of variegation, while Lid mutation resulted in a decreased number of stubble bristles, consistent with an enhancement of PEV. We also found that combinatorial mutation of Lid and dLsd1 restored the number of stubble bristles to the wild-type level (Fig. 3E). The general effect on PEV illustrates the crucial requirement of Lid and dLsd1 for heterochromatin homeostasis, and suggests that Lid and dLsd1 affect heterochromatin antagonistically, possibly by controlling the status of histone methylation at heterochromatin–euchromatin boundaries.
Heterochromatin is characterized by elevated H3K9 methylation levels, and the main histone methyltransferase responsible for H3K9 methylation at pericentric chromatin is Su(var)3–9 (Peters et al. 2003; Ebert et al. 2004). dLsd1 has been shown to physically associate with Su(var)3–9 and to control Su(var)3–9-dependent spreading of H3K9 methylation along euchromatin (Rudolph et al. 2007). To study the effects of Lid mutation and of the combinatorial mutation of Lid and dLsd1 on H3K9 methylation at the boundary between euchromatin and heterochromatin, we performed chromatin immunoprecipitation (ChIP) for H3K9me2 across the locus of the wm4h rearrangement. In wm4h, the white-rough-est region is located next to pericentric heterochromatin, and variable spreading of H3K9 methylation results in heterochromatization and silencing of the genes located in this region. As reported previously, reducing the dosage of wild-type dLsd1 results in a significant decrease of H3K9me2 levels across the white-rough-est region (Fig. 3H; Rudolph et al. 2007). Consistent with the classification of Lid as an enhancer of variegation (Lloret-Llinares et al. 2008), we observed that reducing the dosage of Lid substantially increased the levels of H3K9me2 along the white-rough-est locus. Interestingly, reducing the dosage of both Lid and dLsd1 had no significant effect on H3K9me2 (Fig. 3H). An opposite effect is seen on the levels of H3K4me1 along the white-rough-est locus; dLsd1 mutants show high levels of H3K4me1, while Lid mutants exhibit a decrease in H3K4me1, and, in the double mutants, the levels are comparable with wild-type flies (Supplemental Fig. S5). The levels of H3K4me2 and H3K4me3 are negligible along this locus, and therefore do not seem to play a role in this process (Supplemental Fig. S5).
Taken together, these findings suggest that Lid is required to block dLsd1-mediated H3K4me1 demethylation and oppose Su(var)3–9-mediated spreading of H3K9me2 into euchromatin. Interestingly, at the boundaries between euchromatin and heterochromatin, the effect of Lid on the spreading of H3K9me2 is far more evident than any effect on the levels of H3K4me2 and H3K4me3.
Lid mutation results in increased levels of Su(var)3–9-dependent H3K9 methylation
To determine whether the spreading of heterochromatin observed in Lid mutants is dependent on Su(var)3–9, we combined wm4 with Lid and Su(var)3–9 mutant alleles. Reducing the dosage of Su(var)3–9 abolished the enhancement of variegation observed in Lid mutants alone (Fig. 4A–H). This result suggests that Lid is required to oppose Su(var)3–9 histone methyltransferase activity and prevent Su(var)3–9-dependent spreading of heterochromatin into neighboring euchromatic regions. If this interpretation is correct, then we would expect to see an increased bulk level of H3K9 methylation in the Lid mutants compared with wild-type flies. Evaluation of bulk H3K9 methylation levels by immunoblot analysis in Lid10424 homozygous and heterozygous flies revealed a significant increase in H3K9me2 levels compared with wild-type flies, while intermediate levels of H3K9me2 were observed in double mutants for Lid and dLsd1 (Fig. 4I; Supplemental Fig. S6).
Figure 4.
Enhancement of variegation due to Lid mutation is dependent on Su(var)3–9 dosage, and Lid mutation results in increased levels of H3K9 methylation. (A–H) The enhancer effect of Lid mutation on white variegation in wm4 is suppressed by reduction of Su(var)3–9 dosage. (I) H3K9 methylation levels are globally increased in Lid mutants. Immunoblot analysis of global levels of H3K9me2 in adult males of the indicated genotypes. Total histone H3 and acetyl-H3 levels are shown. (J–Q) H3K9 methylation levels are increased at the chromocenter of polytene chromosomes in mutants for the Lid allele. The top panel shows the staining of polytene chromosomes with an antibody specific for H3K9me2 (red) and with YOYO-1 (green), whereas the bottom panel shows anti-H3K9me2 staining alone. dLsd1 mutation results in reduced levels of H3K9me2 at the chromocenter, while Lid mutation results in an increase of H3K9me2 at the chromocenter compared with w1118. Reduced levels of Lid in combination with loss of dLsd1 results in H3K9me2 levels comparable with wild-type flies.
We then evaluated the distribution of H3K9me2 in salivary gland polytene chromosomes from third instar larvae. Immunostaining for H3K9me2 revealed that the increase in H3K9 methylation levels in Lid mutants is observed predominantly at the chromocenter, where pericentric heterochromatin is located. As reported previously (Rudolph et al. 2007), dLsd1 mutation resulted in decreased levels of H3K9me2 at the chromocenter, and we found that, in double mutants for Lid and dLsd1, the levels of H3K9me2 were comparable with those of wild-type flies (Fig. 4J–Q). Taken together, these results demonstrate that Lid and dLsd1 have opposite effects on the extent of H3K9 methylation, and have a significant impact on the spreading of heterochromatin at boundaries between euchromatin and heterochromatin. While Lid opposes the spreading, dLsd1 facilitates it, and, consequently, the combined mutation of Lid and dLsd1 restores the status quo by artificially resetting the balance between these two opposing activities.
Of the 450 genes predicted to be located in heterochromatic regions, ∼100 have been mapped to the heterochromatin of chromosome 2 (Hoskins et al. 2002; Corradini et al. 2003; Yasuhara et al. 2003). To rule out the possibility that the effects of Lid and dLsd1 on gene expression at chromatin boundaries is limited to the artificial PEV chromosomal context, we selected several genes located in either the heterochromatic region of chromosome 2R or the proximal euchromatic region, and determined their expression levels in wild-type, dLsd1 mutant, and Lid; dLsd1 mutant flies. Consistent with the derepression of the white gene in PEV upon dLsd1 mutation, we observed derepression of many genes located at the boundary between euchromatin and heterochromatin in dLsd1 mutants (Fig. 5; Supplemental Table S4). Reducing the dosage of Lid abolished this effect (Fig. 5), supporting the idea that Lid and dLsd1 indeed have antagonistic roles in the control of gene expression at chromatin boundaries. Interestingly, dLsd1 mutation also results in a reduction of expression levels of some heterochromatic genes compared with wild type, a phenotype that was not rescued by Lid mutation. Down-regulation of heterochromatic genes was observed previously in larvae carrying a mutation for an important determinant of Drosophila heterochromatin, HP1 (Lu et al. 2000), suggesting that HP1 might be required for proper expression of genes residing in heterochromatin. Our results suggest that dLsd1 might have a similar role. Taken together, these data show that dLsd1 is required for the silencing of some genes located at the euchromatin/heterochromatin boundaries, and that Lid opposes dLsd1 function in this context.
Figure 5.
Increased expression of genes at the 2R euchromatin–heterochromatin boundary in dLsd1 mutant flies. RT-qPCR analysis of the expression of the indicated genes in w1118 (wild-type), dLsd1 mutant, and Lid and dLsd1 double mutant ovaries is shown. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate standard error of the mean. Supplemental Table S4 displays the sequence coordinates and the position on the cytogenetic map of each of the genes used in this expression analysis.
dLsd1 directly regulates the expression of a subset of Notch target genes
Histone H3K4 methylation is highly enriched in euchromatin and is generally associated with active transcription. Since the spreading of H3K9 methylation was the key parameter in the interaction between Lid and dLsd1 at euchromatin/heterochromatin boundaries, we reasoned that the interplay between Lid and dLsd1 in the control of gene expression might be different at euchromatic loci where H3K9 methylation is not so paramount.
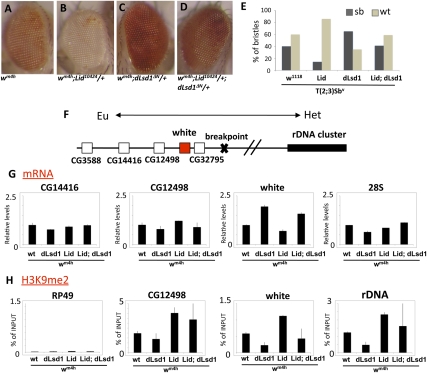
Recently, the Verrijzer laboratory (Moshkin et al. 2009) reported that Lid is involved in silencing of genes regulated by the Notch signaling pathway. To determine whether dLsd1 plays a role in the control of Notch target gene expression, we used RT-qPCR to compare the expression level of E(spl) Notch target genes in dLsd1 homozygous mutant flies with wild-type controls. The results showed a clear derepression of E(spl)m4, E(spl)m6, E(spl)m7, and E(spl)m8 genes in dLsd1 mutant flies (Fig. 6A). To test whether dLsd1 directly regulates E(spl) genes, we performed ChIP analysis with an antibody specific for dLsd1 and examined binding to the E(spl)m4, E(spl)m6, E(spl)m7, and E(spl)m8 enhancers and promoters in Drosophila S2 cells where these genes are repressed. Consistent with the in vivo gene expression data, dLsd1 binds strongly to the E(spl) locus. The dLsd1 ChIP signal was measured by comparison with preimmune serum and was absent at an unrelated control gene (RP49) (Fig. 6B). Additional confirmation of the specificity of the signal was provided by experiments showing that the ChIP signal was lost when dLsd1 was depleted using dsRNA. These data indicate that dLsd1 binds directly to the E(spl) locus and, like Lid, represses the expression of Notch target genes.
Figure 6.
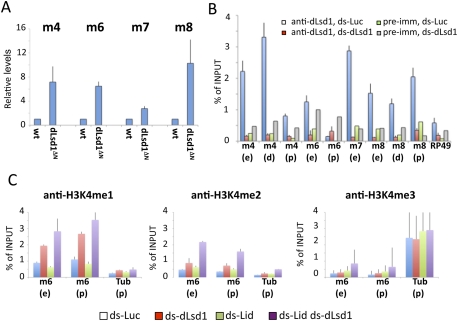
dLsd1 directly regulates Notch target gene expression by controlling histone methylation levels. (A) dLsd1 is required for repression of Notch target genes. RT-qPCR analysis of the expression of E(spl) genes in w1118 (wild-type, wt) and dLsd1 mutant flies. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate the standard error of the mean. (B) dLsd1 binds directly to the E(spl) locus. ChIP analysis of dLsd1 binding across the E(spl) locus using an anti-dLsd1-specific antibody in S2 cells incubated with dsRNA against Luciferase (mock) and dLsd1 to control for specificity. Preimmune serum was used as an additional control for specificity. ChIP data are the result of three independent immunoprecipitations. Error bars indicate standard deviation. Each gene is represented by two or three fragments: enhancer (e), distal (d), and proximal (p) to the transcriptional start site. (C) dLsd1 and Lid cooperatively control histone methylation levels at Notch target genes. Cross-linked chromatin was isolated from S2 cells incubated with dsRNA against the indicated mRNAs, and ChIP analysis was performed using antibodies specific for H3K4me1, H3K4me2, and H3K4me3. Error bars indicate standard deviation.
Because dLsd1 is an H3K4 demethylase, we asked whether dLsd1 might silence Notch target gene expression by directly demethylating H3K4me1 and H3K4me2. Depletion of dLsd1 in S2 cells by dsRNA resulted in increased levels of H3K4me1 and H3K4me2 at the m6 promoter and enhancer region but not at the tubulin promoter, used as a control (Fig. 6C). Consistent with our immunoblot analysis and polytene chromosome staining, upon double depletion of dLsd1 and Lid, we observed a greater increase in H3K4me1 and H3K4me2 compared with single depletions and an increase in H3K4me3 (Fig. 6C). The elevated methylation levels are accompanied by derepression of m6 expression in S2 cells upon single and double depletion of dLsd1 and Lid (Supplemental Fig. S7).
These results suggest that both dLsd1 and Lid suppress the expression of Notch target genes by removing H3K4 methyl marks associated with active transcription at E(spl) regulatory elements. In cells where Notch targets are repressed, dLsd1 and Lid seem to act synergistically, and their interaction is different from the antagonism seen at euchromatin–heterochromatin boundaries.
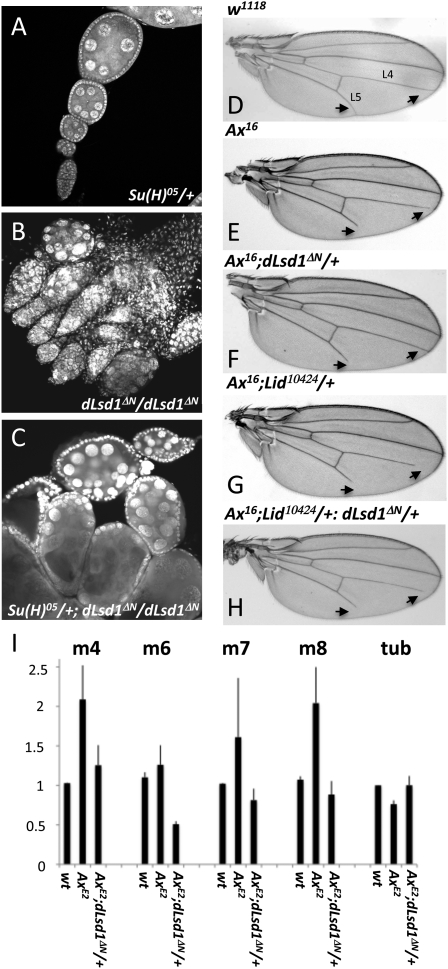
dLsd1 genetically interacts with the Notch signaling pathway
To determine whether the levels of Notch signaling make a functionally significant contribution to the dLsd1 mutant phenotype, we examined the effect of introducing heterozygous mutation for Su(H)05, the transcriptional mediator of Notch signaling, into flies that were homozygous for dLsd1ΔN. Remarkably, the results show that Su(H)05 dominantly suppressed the oogenesis defects of dLsd1ΔN/dLsd1ΔN mutant flies (Fig. 7A–C). We infer not only that dLsd1 is present at Notch target genes, but also that altered regulation of Notch signaling is a very important component of the dLsd1 mutant phenotype. Indeed, we noticed the presence of other phenotypes in dLsd1 homozygous mutants, which have been associated with mutations in the Notch pathway, such as veins broadened into deltas at the wing junction margins (Fig. 1A; Supplemental Fig. S8). As expected from the evidence that dLsd1 contributes to the repression of Notch targets, heterozygous mutations in dLsd1 suppress the dominant wing notching associated with loss-of-function alleles of Notch (P Mulligan, F Yang, L Di Stefano, J Ji, J Nishikawa, Q Wang, M Kulkarni, H Najafi-Shoushtari, R Mostosvalsky, S Gygi, et al., in prep.).
Figure 7.
dLsd1 genetically interacts with various components of the Notch signaling pathway. (A–C) Su(H)05 partially suppresses the oogenesis defect in dLsd1 homozygous mutant flies. (D–H) dLsd1 mutation suppresses truncations of the L4 and L5 wing veins in flies heterozygous for the Ax16 gain-of-function Notch mutation, while Lid mutation enhances it. Shown are representative examples of wings of the indicated genotypes; arrows mark the distal part of L4 and L5 wing veins where the phenotype is evident. (I) dLsd1 is required for activation of Notch target genes. RT-qPCR analysis of the expression of E(spl) genes in w1118 (wild-type, wt), AxE2, and AxE2, dLsd1 heterozygous mutant males. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate standard error of the mean.
More extensive genetic tests revealed that Lid and dLsd1 differ in their interaction with the Notch pathway. When we crossed dLsd1 mutants to gain-of-function alleles of Notch (Abruptex; AxE2 and Ax16), we noted that dLsd1 alleles failed to enhance the L4 and/or L5 wing defects of the Ax alleles, as observed with Lid mutant alleles (Fig. 7D–G; quantification in Supplemental Table S5–S7), and as would be expected if dLsd1 solely repressed Notch target genes. Instead, mutant alleles of dLsd1 suppressed the Abruptex phenotype. This indicates that, in the context of a hyperactive Notch signaling pathway, dLsd1 contributes positively to the activation of Notch targets. To test this idea, we measured the expression of E(spl) genes in AxE2 hemizygous mutants. Consistent with our hypothesis, we found that these targets are up-regulated in hemizygous AxE2 mutants, and that the effect of heterozygous mutation of dLsd1 is to restore wild-type levels of gene expression (Fig. 7I; Supplemental Fig. S9).
We infer that, in cells where Notch is inactive, such as S2 cells, Lid and dLsd1 act cooperatively to keep Notch target genes repressed. However, in the context of activated Notch signaling, such as occurs in Abruptex mutants, dLsd1 switches from being a repressor to an activator, promoting the expression of these targets. Thus, in the context of activated Notch, dLsd1 acts in opposition to Lid. In support of this hypothesis, we note that the effects of mutant Lid and dLsd1 alleles on Ax phenotypes essentially cancel one another out, and the double mutation has, in general, an intermediate effect on phenotypes associated with gain-of-function Ax alleles (Fig. 7H; Supplemental Table S5–S7). Taken together, these genetic studies suggest that dLsd1 has a significant, context-dependent, role in modulating Notch signaling, and that mutant alleles that act in the Notch pathway can partly suppress dLsd1 mutant phenotypes.
Discussion
Molecular studies have identified an increasingly large number of histone-modifying enzymes, and biochemical assays readily allow these proteins to be classified, but the more difficult and more important challenge is to understand how these various enzymatic activities are integrated, in vivo, to control biological processes. We examined the effects of combining mutations in the two H3K4 demethylases Lid and dLsd1 in Drosophila. Our studies, performed in vivo, show that the interplay between Lid and dLsd1 is dependent on the chromatin context and active signaling pathways. Our results show a consistent pattern of genetic interactions between Lid and dLsd1 that is evident in multiple tissues and phenotypes. Unexpectedly, despite their activity as histone H3K4 demethylases, these proteins function antagonistically in a number of functional and developmental contexts. For example, dLsd1 and Lid have opposing functions in the establishment of euchromatin and heterochromatin boundaries. At these locations, the antagonism does not seem to stem from the effects of Lid on H3K4 methylation, but rather from its indirect effects on the spreading of H3K9me2. In addition, while our data show that both Lid and dLsd1 can repress Notch targets within euchromatin when Notch signaling is not active, and that Notch signaling is an important component of the dLsd1 mutant phenotype, genetic evidence supports the hypothesis that Lid and dLsd1 have antagonistic functions in the context of activated Notch signaling. This complex pattern of interactions illustrates that the functional interplay between demethylases, and most likely between other types of chromatin-associated proteins, cannot be rationalized into a single generic model. The evidence that dLsd1 can switch from being a negative regulator of Notch target genes to a positive regulator adds an extra layer of complexity to the interplay between Lid and dLsd1, and strongly supports the concept that the activity of histone demethylases is highly regulated and context-dependent.
Antagonistic roles of Lid and dLsd1 at chromatin boundaries
Our genetic and biochemical data support a model for the creation and maintenance of heterochromatin boundaries, proposed by Reuter and colleagues (Rudolph et al. 2007), in which dLsd1 promotes deacetylation of H3K9 by RPD3 and subsequent methylation of H3K9 by Su(var)3–9, thereby facilitating spreading of heterochromatin. In addition, we show an increase in H3K4me1 at the white-rough-est locus in dLsd1 mutant flies, suggesting that active demethylation of H3K4me1 by dLsd1 is an important step in the establishment of heterochromatin. Furthermore, we find that Lid antagonizes dLsd1 function by promoting euchromatin formation, and that the spreading of heterochromatin seen in Lid mutants is dependent on dLsd1 and Su(var)3–9 activities. Consistent with this notion, H3K9 methylation levels are increased in Lid mutant flies compared with control at the white-rough-est locus and in pericentric heterochromatin. Interestingly, the levels of H3K4me2 and H3K4me3 at the white-rough-est locus are very low and increase only marginally upon Lid mutation, suggesting that Lid function in this context is independent of its histone H3K4 demethylase activity. Previously, Lid had been reported to facilitate activation of Myc target genes in a demethylase-independent manner (Secombe et al. 2007), and to antagonize Rpd3 histone deacetylase function (Lee et al. 2009); moreover, mutation of Lid has been shown to cause a decrease of H3K9 acetylation levels (Lloret-Llinares et al. 2008). It is therefore tempting to speculate that Lid opposes the spreading of heterochromatin, independent of its function as a histone H3K4 demethylase, by antagonizing the activity of the dLsd1/Su(var)3–9/Rpd3 complex. This antagonism would explain why, in double mutants for dLsd1 and Lid, the balance between euchromatin and heterochromatin is artificially reset to wild-type levels. Consistently, reorganization of chromatin domains observed in dLsd1 mutant flies affects the expression of genes located at the 2R euchromatin–heterochromatin boundary, an effect that is reversed by mutation of Lid.
dLsd1 is a modulator of the Notch signaling pathway
Given the predominant presence of H3K4 methylation in euchromatin and its important role in determining the transcription status of a gene, we were interested in establishing the nature of the interplay between Lid and dLsd1 in a euchromatic context. Previous studies had implicated Lid as a crucial factor in the silencing of Notch target genes (Moshkin et al. 2009). Our study shows a cooperative role for Lid and dLsd1 in repressing Notch target gene expression, and suggests that they contribute to repression by maintaining low levels of H3K4 methylation. Repression of Notch target genes is essential for the establishment of Notch-inhibited cell fates (Fiuza and Arias 2007), suggesting that Lid and dLsd1 could play a role in proper cell fate specification during Drosophila development. Interestingly, the role of dLsd1 does not seem to be limited to repression of Notch target genes. Indeed, our genetic analysis suggests that, in a context in which the Notch signaling pathway is active, dLsd1 switches from a repressor to an activator role. Such a dual role had already been described for Su(H), whose switch from a repressor to an activator has been suggested to be mediated through an exchange of associated proteins (Bray 2006). Similarly, in mammalian cells, studies have shown that LSD1 activity can be modulated by changes in composition of the complexes present at the Gh promoter, and, depending on the cell type (somatotroph or lactotroph), LSD1 can act as either an activator or a repressor (Wang et al. 2007). Therefore, a possible explanation for our data is that, depending on the complexes available, dLsd1 can switch from being a repressor to acting as an activator of Notch target genes. Alternatively, dLsd1 mutation could promote derepression of negative regulators of Notch activity, or could directly modulate Notch activity by demethylating crucial components of the Notch-activating complex. Further studies are required to distinguish between these possibilities.
These results provide the basis for future studies aimed at investigating whether the dual role of dLsd1 in modulating Notch signaling is conserved in mammals. In mice, LSD1 has been shown to repress the Notch target Hey1 in late stages of pituitary development (Wang et al. 2007), suggesting that its ability to regulate Notch target genes is conserved. This pathway-specific function of LSD1 could potentially be exploited to create novel strategies to manipulate Notch-mediated carcinogenesis.
Collectively, these results reveal an intricate interplay between the histone demethylases Lid and dLsd1 in the control of higher-order chromatin structure at euchromatin and heterochromatin boundaries affecting developmental gene silencing. They also demonstrate an involvement of dLsd1 and Lid in Notch pathway-specific control of gene expression in euchromatin, and support the idea that, depending on the context, Lid and dLsd1 can favor either transcriptional activation or transcriptional repression.
Materials and methods
Genetic analysis
The following fly stocks were obtained from the Bloomington stock collection: lid10424, lidk06801, ash2EY03971, osa2, brm2, mor1, Snr101319, Trls2325, esc1, kis13416, Canton S, w1118, T(2;3)Sbv, In(1)wm4h, and In(1)wm4; Su(var)3–91. brm1, brm5, osa1, mor2, and mor6 alleles were kindly provided by Dr. James Kennison. Bap180Δ86 and Bap170Δ65 alleles were kindly provided by Dr. Jessica Treisman. Psch28 and Su(z)24 were kindly provided by Dr. Chao Ting Wu. AxE2, Ax16, and Su(H)05 were kindly provided by Dr. Spyros Artanavis-Tsakonas. Flies were grown on standard Drosophila medium and maintained at 25°C. Recombination of the dLsd1ΔN allele with other third chromosome mutants was performed by standard means. Candidate recombinant lines were verified by testing for noncomplementation crosses with other alleles, or by using allele-specific PCR assays. The held-out wings phenotype was scored if flies had both wings extended. We categorized the rescue of the dLsd1ΔN-dependent oogenesis defects into moderate and weak rescues. The moderate and weak descriptors were used for those lines that showed some degree of structural organization compared with dLsd1 mutant ovaries. For the ISWIK159R interaction test, yw; eyGAL4; UAS-ISWIK159R/T(2:3) virgins were crossed with the dLsd1ΔN/TM3. The F1 progeny that did not carry any balancer were scored based on the severity of eye phenotype, as in Burgio et al. (2008). The effect of dLsd1 and Lid mutations on white variegation was studied by crossing females homozygous for the In(1)wm4h allele with males heterozygous for dLsd1 and Lid mutations. The effects on eye color variegation of the male progeny were quantified by measuring the relative expression of the white gene and by observing the relative red eye pigment content. The effects on stubble variegation were studied by crossing females heterozygous for the T(2;3)Sbv allele with males heterozygous for dLsd1 and Lid mutations. The effect was quantified by counting the number of stubble and wild-type bristles. The bristles examined were the dorsocentral, anterior scutellar, and posterior scutellar.
Immunostaining
Immunostaining of Drosophila ovaries was performed as described in Di Stefano et al. (2007). Ovaries were stained with Rhodamine-Phalloidin (Molecular Probes) and the following antibodies: anti-Fasciclin III (7G10), anti-spectrin (3A9), and anti-ORB (4H8), obtained from the Developmental Studies Hybridoma Bank. DNA was stained using YOYO-1 iodide (Molecular Probes), and ovaries were mounted for confocal microscopic imaging.
For immunostaining of polytene chromosomes, salivary glands were dissected from third instar larvae. Glands were fixed in solution 2 (3.7% formaldehyde, 1% Triton X-100/PBS at pH 7.5) and squashed in solution 3 (3.7% formaldehyde, 50% acetic acid). Chromosomes were incubated overnight at 4°C with either anti-H3K9me2 (ab1220), anti-H3 (ab1791), or anti-H3K4me2 (ab32356), followed by incubation with Cy3-conjugated secondary antibody (Molecular Probes) for 2 h at 4°C. To stain the DNA, the chromosomes were incubated with YOYO-1 iodide (Molecular Probes) for 10 min at room temperature. Preparations were examined with confocal laser-scanning microscopy. The levels of histone methylation in wild-type and mutant polytene chromosomes were compared by processing and analyzing the samples in the same experiment under identical conditions, and pictures were taken using identical exposure times (Srinivasan et al. 2005). Multiple polytene chromosomes from at least three squashes were analyzed, and representative examples are shown.
Real-time qPCR
Total RNA was prepared using Trizol (Invitrogen). RT–PCR was performed using TaqMan Reverse Transcription reagents (PE Applied Biosystems) according to the instructions provided by the manufacturer. Real-time qPCR was performed using the Roche LightCycler 480 system. Relative levels of specific mRNAs were determined using the SYBR Green I detection chemistry system (Roche). Quantification was performed using the comparative CT method as described in the manufacturer procedures manual. RP49 was used as normalization control. Primer sequences are available on request.
ChIP
Flies were fixed in 1.8% formaldehyde for 15 min at room temperature, resuspended in lysis buffer (140 mM NaCl, 15 mM HEPES at pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.5 mM DTT, 0.1% sodium deoxycholate, 0.05% SDS, 0.5% N-lauroylsarcosine, protease inhibitors), and lysed by sonication. The lysates were cleared by centrifugation, preabsorbed by incubation with protein G and A sepharose beads (GE Healthcare), and incubated overnight at 4°C with 2 μg of one of the following antibodies: anti-H3 (ab1791), anti-H3K4me1 (ab8895), anti-H3K4me2 (ab32356), anti-H3K4me3 (ab8580, ab1012), or anti-H3K9me2 (ab1220), and 8 μL of anti-dLsd1 (described in Di Stefano et al. 2007). Antibody complexes were recovered with a mixture of protein A and G sepharose. After extensive washes, immunocomplexes were eluted from the beads and cross-link-reversed, and the DNA was recovered by phenol/chloroform extraction and ethanol precipitation. DNA was resuspended in 150 μL of water, and 7.5 μL was used for real-time qPCR reactions.
S2 cells were fixed in 1.8% formaldehyde for 15 min at room temperature, resuspended in hypertonic buffer A (300 mM sucrose, 2 mM Mg acetate, 3 mM CaCl2, 10 mM Tris at pH 8.0, 0.1% Triton X-100, 0.5 mM DTT), incubated for 5 min on ice, and dounced 20 times with a dounce homogenizer (tight pestle, Wheaton). Nuclei were collected by centrifuging at 720g for 5 min at 4°C. The pellets were washed twice in buffer A and then resuspended in buffer D (25% glycerol, 5 mM Mg acetate, 50 mM Tris at pH 8.0, 0.1 mM EDTA, 5 mM DTT). Chromatin was collected by centrifuging at 720g for 5 min at 4°C. The pellets were washed twice in buffer D and then resuspended in buffer MN (60 mM KCl, 15 mM NaCl, 15 mM Tris at pH 7.4, 0.5 mM DTT, 0.25 M sucrose, 1.0 mM CaCl2). Nuclei were digested with 100 U of MNase (USB) and diluted in buffer MN for 30 min at room temperature. Reactions were stopped with the addition of EDTA and SDS to final concentrations of 12.5 mM and 0.5%, respectively. The samples were then processed for ChIP as described above.
Cell culture and dsRNA
S2 cells were cultured at 25°C in Schneider's insect medium (Sigma; 10% fetal bovine serum) and treated with dsRNA as described (Stevaux et al. 2002).
Immunoblot analysis and antibodies
Protein extracts were obtained by acid extraction. Briefly, flies were resuspended in lysis buffer (50 mM Tris at pH 8.0, 0.4% NP40, 300 mM NaCl, 10 mM MgCl2, 0.5 mM DTT, 1.5 mM PMSF) and homogenized using a douncer with type A pestle. Lysates were incubated with 0.2 M HCl for 2 h on ice. Lysates were then centrifuged and the supernatant was dialyzed overnight using Slide-a-Lyser cassettes (Pierce) according to the manufacturer's procedure. Immunoblots were performed using standard procedures. The blots were probed using antibodies specific for anti-H3 (ab1791), anti-H3K4me1 (ab8895), anti-H3K4me2 (ab32356), anti-H3K4me3 (ab8580), anti-H3K9me2 (ab1220), and anti-acetyl-H3 (Millipore, #06-599).
Acknowledgments
We thank Dr. Spyros Artanavis-Tsakonas, Dr. James Kennison, Dr. Jessica Treisman, and Dr. Chao Ting Wu for providing fly stocks, and members of the Dyson laboratory for technical assistance and helpful discussion. We thank Dr. Kristian Helin for critical reading of the manuscript. This study was supported by NIH grants GM81607, GM53203, and CA64402 to N.J.D., and GM071449 to A.M.N. L.D.S. was supported by a fellowship from the Leukemia and Lymphoma Society. J.A.W. was supported by a grant from the DoD (W81XWH-09-1-0487). G.B. was supported by a FIRC Fellowship, and D.C. was supported by Fondazione Telethon, Giovanni Armenise Harvard Foundation, MIUR-PRIN, HFSP, AIRC, and EMBO-YIP. N.J.D. is the Saltonstall Scholar of the Massachusetts General Hospital Cancer Center.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1983711.
Supplemental material is available for this article.
References
- Bannister AJ, Kouzarides T 2005. Reversing histone methylation. Nature 436: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bray SJ 2006. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Burgio G, La Rocca G, Sala A, Arancio W, Di Gesù D, Collesano M, Sperling AS, Armstrong JA, van Heeringen SJ, Logie C, et al. 2008. Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet 4: e1000089 doi: 10.1371/journal.pgen.1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini N, Rossi F, Verni F, Dimitri P 2003. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112: 26–37 [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N 2007. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol 17: 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18: 2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A 2007. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol 14: 344–346 [DOI] [PubMed] [Google Scholar]
- Fiuza UM, Arias AM 2007. Cell and molecular biology of Notch. J Endocrinol 194: 459–474 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, et al. 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: RESEARCH0085 doi: 10.1186/gb-2002-3-12-research0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Xie T 2007. The Drosophila ovary: An active stem cell community. Cell Res 17: 15–25 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833 [DOI] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2007. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol 14: 341–343 [DOI] [PubMed] [Google Scholar]
- Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2009. The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol Cell Biol 29: 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328 doi: 10.1371/journal.pbio.0030328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F 2008. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res 36: 2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenda DR, Zraly CB, Feng Y, Egan S, Dingwall AK 2003. The Drosophila SNR1 (SNF5/INI1) subunit directs essential developmental functions of the Brahma chromatin remodeling complex. Mol Cell Biol 23: 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP 2009. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID–RPD3 complexes during NOTCH silencing. Mol Cell 35: 782–793 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K 2004. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol 22: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol Cell 26: 103–115 [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14: 67–75 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DAR, Mottus RC, Grigliatti TA 1983. Genes which suppress position-effect variegation in Drosophila melanogaster are clustered. Mol Gen Genet 191: 326–333 [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132: 1623–1635 [DOI] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J 21: 4927–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, McLaughlin PJ, Allshire RC 2005. Methylation: Lost in hydroxylation? EMBO Rep 6: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446: 882–887 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Milne TA, Allis CD 2005. Taking LSD 1 to a new high. Cell 122: 654–658 [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Marchetti M, Fanti L, Pimpinelli S, Wakimoto BT 2003. A strategy for mapping the heterochromatin of chromosome 2 of Drosophila melanogaster. Genetica 117: 217–226 [DOI] [PubMed] [Google Scholar]