Abstract
Transcription of non-protein-coding DNA (ncDNA) and its noncoding RNA (ncRNA) products are beginning to emerge as key regulators of gene expression. We previously identified a regulatory system in Saccharomyces cerevisiae whereby transcription of intergenic ncDNA (SRG1) represses transcription of an adjacent protein-coding gene (SER3) through transcription interference. We now provide evidence that SRG1 transcription causes repression of SER3 by directing a high level of nucleosomes over SRG1, which overlaps the SER3 promoter. Repression by SRG1 transcription is dependent on the Spt6 and Spt16 transcription elongation factors. Significantly, spt6 and spt16 mutations reduce nucleosome levels over the SER3 promoter without reducing intergenic SRG1 transcription, strongly suggesting that nucleosome levels, not transcription levels, cause SER3 repression. Finally, we show that spt6 and spt16 mutations allow transcription factor access to the SER3 promoter. Our results raise the possibility that transcription of ncDNA may contribute to nucleosome positioning on a genome-wide scale where, in some cases, it negatively impacts protein–DNA interactions.
Keywords: ncDNA, intergenic transcription, chromatin, repression
Over the past decade, genome-wide expression studies in eukaryotes have revealed that transcription is not limited to protein-coding DNA, but rather occurs throughout entire genomes, often involving both DNA strands (Kapranov et al. 2007; Pheasant and Mattick 2007; Berretta and Morillon 2009; Jacquier 2009). Although the extent of transcription of non-protein-coding DNA (ncDNA) has been questioned recently (van Bakel et al. 2010), it is clear that eukaryotes produce many RNA molecules that do not encode proteins (noncoding RNAs [ncRNAs]) (Goodrich and Kugel 2009; Harrison et al. 2009; Mercer et al. 2009; Costa 2010). ncRNAs have diverse properties, ranging in size from short (microRNAs [miRNAs]) to long (long RNAs [lnRNAs]) and ranging in stability from stable to unstable. With the exception of several families of well-studied ncRNAs—including rRNAs, tRNAs, snRNAs, snoRNAs, and miRNAs—the biological functions of these ncRNAs are only beginning to be understood.
Although it is likely that some ncRNAs may represent transcriptional noise (Struhl 2007; Seila et al. 2009), it has become increasingly clear that transcription of noncoding regions of eukaryotic genomes plays important biological functions, primarily in regulating gene expression (Goodrich and Kugel 2009; Harrison et al. 2009; Mercer et al. 2009). Examples of this include the Xist/Tsix RNAs involved in mammalian X inactivation (Lee 2009), the roX1 and roX2 RNAs involved in dosage compensation in Drosophila (Gelbart and Kuroda 2009), the human HOTAIR involved in the regulation of developmental genes (Rinn et al. 2007), the mouse Air and Kcnq1ot1 RNAs involved in establishing genomic imprinting (Royo and Cavaille 2008), and the mouse VL30 RNA and human PSF-binding ncRNAs that regulate cell proliferation and tumorigenesis (Li et al. 2009; Wang et al. 2009).
Significant advances have been made in understanding widely diverse mechanisms by which transcription of ncDNAs regulate gene expression. In some cases, it is the ncRNA product that regulates gene expression. ncRNAs have been shown to recruit complexes that modify chromatin, interact with activator and coactivator proteins and modulate their function, and interact with RNA polymerase II (Pol II) and other basal transcription factors to control their activity (Goodrich and Kugel 2009; Harrison et al. 2009; Mercer et al. 2009). Alternatively, the act of transcribing ncDNA has also been shown to both positively and negatively regulate gene expression. In most of these cases, a transcription interference mechanism has been proposed. Examples include mouse and human globin genes (Ashe et al. 1997; Gribnau et al. 2000); the Drosophila Hox genes (Schmitt et al. 2005; Mazo et al. 2007); and Saccharomyces cerevisiae SER3 (Martens et al. 2004), ADH1/ADH3 (Bird et al. 2006), IME4 (Hongay et al. 2006), and FLO11 (Bumgarner et al. 2009) genes. Although several mechanisms of transcription interference have been described, most involving RNA Pol II directly, experiments that distinguish between these mechanisms at specific genes have not been performed.
Interestingly, several studies in yeast have implied that transcription of ncDNA may contribute to gene regulation by altering chromatin structure. Transcription of a series of ncRNAs 5′ of the Schizosaccharomyces pombe fbp1+ gene was found to facilitate an open chromatin conformation, allowing transcription factors access to the fbp1+ promoter during glucose induction (Hirota et al. 2008). Antisense transcription has been shown to silence the expression of PHO84 by a mechanism that requires Hda1/2/3-dependent deacetylation of histones located at the PHO84 promoter (Camblong et al. 2007, 2009). Finally, two recent studies provide evidence that transcription of DNA antisense to the GAL10 gene alters post-translational modifications of histones that facilitate repression of the divergently transcribed GAL10 and GAL1 genes (Houseley et al. 2008; Pinskaya et al. 2009).
Previously, we showed that serine-dependent transcription of ncDNA (SRG1) in S. cerevisiae represses expression of the adjacent SER3 gene (Martens et al. 2004, 2005). In the presence of serine, transcription of SRG1 extends across the promoter of the adjacent SER3 gene, yielding two short transcripts that terminate 75 base pairs (bp) 5′ and 25 bp 3′ of the SER3 translational start (Thompson and Parker 2007), and a minor SRG1–SER3 readthrough transcript that extends to the end of SER3 (Martens et al. 2004; Thompson and Parker 2007). We provided evidence that it is the act of transcribing SRG1 across the SER3 promoter, rather than the SRG1 RNA products, that represses SER3 (Martens et al. 2004). In this study, we elucidate the mechanism whereby serine-dependent transcription of ncDNA (SRG1) in S. cerevisiae represses expression of the adjacent SER3 gene. We show that SER3 repression correlates with a broad region of strong micrococcal nuclease (MNase) protection spanning the entire SRG1 transcription unit, suggesting that nucleosomes are loosely positioned across this region. Surprisingly, conditions that reduce SRG1 transcription result in dramatically reduced MNase protection at the SER3 promoter, indicating a loss of nucleosome occupancy. By analyzing mutations in SPT6 and SPT16, two genes that encode subunits of the Spt6/Spn1(Iws1) and FACT elongation complexes, we provide evidence that it is the nucleosomes assembled at the SER3 promoter by intergenic SRG1 transcription, not RNA Pol II itself, that interfere with the binding of transcription factors to the SER3 promoter. Our data are consistent with a general model in which transcription of ncDNA can assemble nucleosomes that occlude DNA from binding by sequence-specific DNA-binding proteins.
Results
Evidence that nucleosomes occupy the SER3 promoter in repressing conditions
Previously, we showed that transcription of intergenic SRG1 DNA is required for SER3 repression (Martens et al. 2004). Several pieces of data suggest that chromatin structure also plays an important role in SER3 repression. First, we identified histones and two activators of histone gene expression, Spt10 and Spt21 (Dollard et al. 1994; Hess et al. 2004; Eriksson et al. 2005), in a genetic screen for repressors of SER3 expression (J Pruneski, unpubl.). Second, DNA microarray experiments revealed that depletion of histone H4 resulted in strong SER3 derepression (Wyrick et al. 1999). Third, a mutation in SPT6, a gene that encodes a protein required to maintain proper chromatin structure over genes during transcription (Kaplan et al. 2003; Cheung et al. 2008), also results in SER3 derepression (Kaplan et al. 2003).
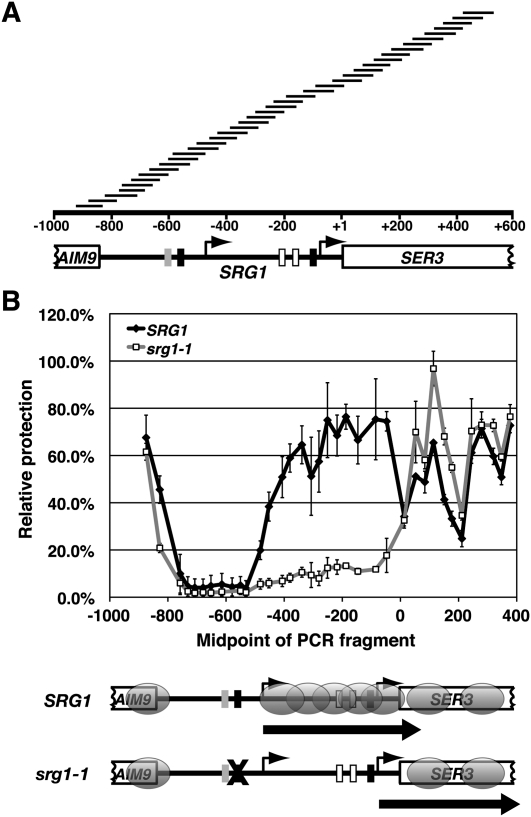
To investigate a possible role for chromatin structure in SER3 repression, we first determined the positions of nucleosomes across the SER3 locus in wild-type cells grown in SER3-repressing conditions (YPD) by a nucleosome scanning assay (Sekinger et al. 2005; Lee et al. 2007). Briefly, cells are treated with formaldehyde, spheroplasted, and then incubated with increasing amounts of MNase to digest nonnucleosomal DNA (see the Materials and Methods for details). As described previously (Brickner et al. 2007), we monitored MNase digestion of two sequences located in the GAL1–10 promoter—one within a well-positioned nucleosome (GAL1 NB), and one within an adjacent MNase-sensitive region (GAL1 NUB)—by quantitative PCR (qPCR) (Supplemental Fig. S1). DNA isolated from the MNase concentration where we observed significant protection of GAL1 NB relative to GAL1 NUB was then used to assess MNase protection across SRG1–SER3. We performed qPCR with 38 unique primer pairs to amplify overlapping SRG1–SER3 sequences (Fig. 1A) from both MNase-digested and undigested DNA. MNase protection for each of these sequences was quantified as the ratio of template present in MNase-digested DNA over undigested DNA that was then normalized to the amount of MNase-protected GAL1 NB template. Using this method, we identified peaks of MNase protection, indicating the presence of a positioned nucleosome at the 3′ end of AIM9 (the gene adjacent to SRG1) and two at the 5′ end of the SER3 ORF (Fig. 1B). We also found a 200-bp MNase-sensitive region (from −750 to −550 with respect to the SER3 ATG) corresponding to the SRG1 promoter, indicating a nucleosome-depleted region that is a hallmark of many yeast promoters (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007). In addition, we identified a broad region of MNase protection that begins at the SRG1 transcription start site (−475) and extends across the SER3 promoter to the SER3 translational start site, a region that defines the SRG1 transcription unit. This pattern of strong MNase protection implies the presence of nucleosomes that are positioned randomly across the SRG1 transcription unit. Therefore, the SER3 promoter lacks the typical nucleosome-depleted region (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007). These results are consistent with our previously reported indirect-labeling experiments (Martens and Winston 2002) and with genome-wide nucleosome positioning experiments (Lee et al. 2007).
Figure 1.
Nucleosome positions and relative occupancy at SER3 in the presence and absence of SRG1 transcription. (A) Schematic of SER3 locus, including the 3′ 161 bp of AIM9 (−1000 to −839 relative to SER3 ATG) and the 5′ 600 bp of the SER3 ORF. The arrows at −475 and −75 indicate the transcription start sites of SRG1 and SER3, respectively. Blocks of intergenic sequence identity between S. cerevisiae and four related yeast strains are marked, including the SRG1 and SER3 TATAs (black boxes), sequences required for SER3 activation (white boxes), and a Cha4-binding site (gray box). The scale represents the distance from the SER3 translation start (+1). The tiled black bars above the scale indicate the DNA fragments amplified by qPCR to quantify nucleosome position and relative occupancy (see Supplemental Table S2 for details). (B) Nucleosome scanning assay was performed on wild-type (FY4, FY2097, and FY1350) and srg1-1 (YJ582, FY2250, and YJ585) cells that were grown in YPD medium (SER3 repressed) at 30°C. Using qPCR, the relative MNase protection of each SER3 template was calculated as a ratio to the control GAL1 NB template found within a well-positioned nucleosome in the GAL1–10 promoter (see Supplemental Fig. S1). Each point on the graph shows the mean ± SEM from three independent experiments that are plotted at the midpoint of each PCR product. Results for amplicons SER3-5 to SER3-41 are shown. Below the graph, a diagram of the SER3 locus indicates the positions of nucleosomes (gray ovals) extrapolated from the MNase protection data. The block arrows indicate the transcription activity of SRG1 and SER3, respectively. srg1-1 strains have a mutated TATA sequence (marked by an X) that inhibits SRG1 transcription, causing SER3 derepression.
To determine if SRG1 transcription affects the chromatin structure at SER3, we repeated the nucleosome scanning assay using srg1-1 strains, which carry a mutation of the SRG1 TATA sequence. This mutation severely reduces SRG1 transcription, resulting in strong derepression of SER3 (Martens et al. 2004). In the srg1-1 cells, MNase protection was reduced specifically over the SRG1 transcription unit as compared with wild-type cells, indicating a dramatic loss of nucleosome occupancy (Fig. 1B). Our results reveal a positive correlation between SRG1 transcription and nucleosome occupancy across SRG1, an unexpected finding given the negative correlation between transcription and nucleosome occupancy generally observed for protein-coding genes (Lee et al. 2004; Schwabish and Struhl 2004).
Serine-dependent transcription of SRG1 intergenic DNA controls nucleosome occupancy of the SER3 promoter
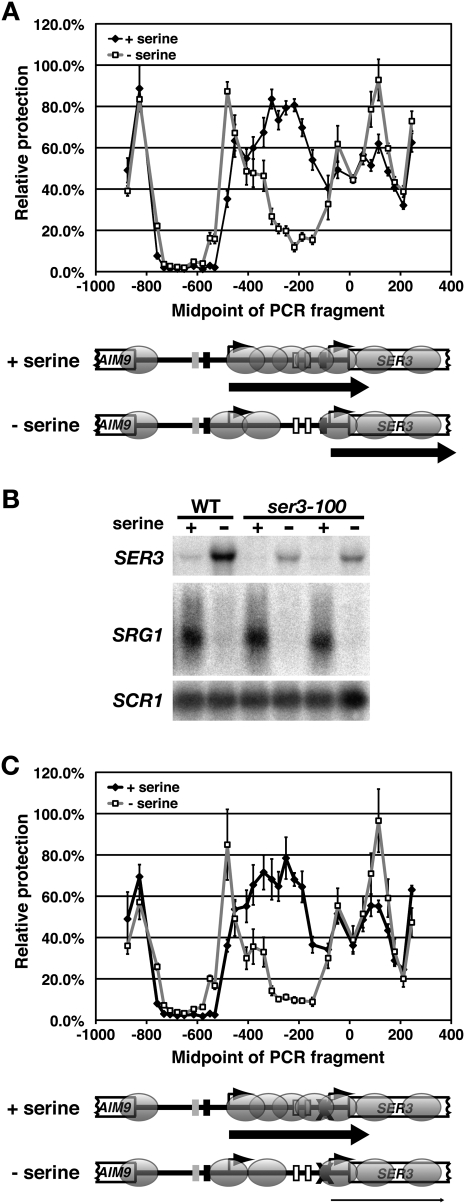
We showed previously that SER3 expression is tightly controlled by the serine-dependent regulation of SRG1 transcription (Martens et al. 2005). Therefore, we also measured MNase accessibility at SER3 in wild-type strains that were grown in synthetic complete (SC) + serine (SRG1 induced; SER3 repressed) and then shifted to SC − serine (SRG1 repressed, SER3 induced) for 25 min. Since the extent of the MNase digestion of the GAL1 NB region was identical in these different growth conditions (Supplemental Fig. S2), we again normalized all SER3 data to this region. As expected for cells grown in serine-rich media, the relative MNase protection across SRG1–SER3 is nearly identical to that observed for cells grown in YPD (cf. wild-type strains in Figs. 1B, 2A). When cells were shifted to media lacking serine, we measured a significant decrease in MNase protection over the SRG1 transcribed region. However, rather than extending across the entire SRG1 transcription unit, as was observed for srg1-1, the reduced MNase protection was restricted to a 200-bp region that included sequences that had been determined previously to be required for SER3 activation (Martens et al. 2004). An MNase-protected region of ∼350 bp, consistent with two closely associated nucleosomes or possibly one nucleosome that adopts multiple positions, remains near the 5′ end of SRG1. This MNase-protected region begins at a more 5′ position, including the SRG1 transcription start site and possibly the SRG1 TATA, as compared with the beginning of the broad peak of MNase protection that was measured for cells grown in serine-rich media. Thus, in contrast to the complete loss of nucleosomes across SRG1 that occurs in the srg1-1 strains, serine starvation depletes nucleosomes specifically over sequences required for SER3 activation. Therefore, in response to serine starvation, the SER3 promoter adopts the typical promoter architecture, with +1 and −1 nucleosomes flanking a nucleosome-depleted UAS (Albert et al. 2007; Lee et al. 2007).
Figure 2.
Effect of serine on nucleosome positions and relative occupancy at SER3. (A) Nucleosome scanning assay was performed on wild-type cells (FY2097 and FY4) that were grown at 30°C in SC + serine media (+ serine) and then shifted to SC − serine media (− serine) for 25 min as described in Figure 1. Each point on the graph shows the mean relative MNase protection ± SEM from four independent experiments (two for each strain) plotted at the midpoint of each PCR product. Results for amplicons SER3-7 to SER3-41 are shown. (B) Northern analysis of SER3 and SRG1 was performed on a wild-type (FY2097) and two ser3-100 strains (YJ275 and FY2099) that have a mutated SER3 TATA. Cells were grown at 30°C in SC + serine media (+ serine) and then shifted to SC − serine media (− serine) for 25 min. SCR1 serves as a loading control. (C) Nucleosome scanning assay was performed on ser3-100 strains (YJ275 and FY2099) as described in A.
To determine if the loss of nucleosome occupancy at the SER3 promoter is caused by a loss of SRG1 transcription and is not simply an effect of the resulting increase in SER3 transcription, we repeated the nucleosome scanning assay using strains that contain a mutation in the SER3 TATA sequence (ser3-100). Although the ser3-100 mutation strongly inhibits SER3 activation when cells are shifted from serine-rich to serine starvation media (10-fold decrease in SER3 mRNA levels) (Fig. 2B), the changes in MNase protection between these growth conditions were identical to those observed for a wild type (Fig. 2, cf. A and C). Therefore, reduced nucleosome occupancy over the SER3 promoter is not a consequence of increased SER3 expression.
FACT and Spt6/Spn1(Iws1) are required to repress SER3
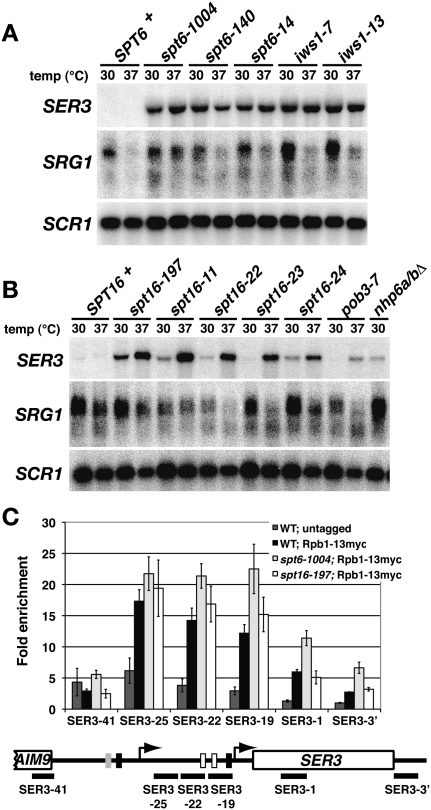
Our results thus far are consistent with two possible mechanisms for transcription interference at SER3. In the first possibility, similar to the conventional transcription interference mechanism (Greger et al. 2000), RNA Pol II elongating across SRG1 competes with transcription factors for binding to the SER3 promoter. In the second possibility, the nucleosomes maintained over the SER3 promoter by SRG1 transcription compete with transcription factor access to the SER3 promoter. If the latter possibility is true, we reasoned that disrupting nucleosome reassembly during transcription might cause SER3 derepression. Several studies have implicated the essential, highly conserved FACT and Spt6/Spn1(Iws1) transcription elongation complexes in transcription-dependent chromatin reassembly (Belotserkovskaya et al. 2003; Kaplan et al. 2003; Mason and Struhl 2003; Cheung et al. 2008; Jamai et al. 2009). Northern analyses were performed on several temperature-sensitive mutants of the Spt6/Spn1(Iws1) and FACT complexes that were grown in YPD at permissive (30°C) and nonpermissive (37°C) temperatures. Large increases in SER3 mRNA levels were detected in multiple spt6 and spn1(iws1) mutants at both 30°C and 37°C (Fig. 3A). While increases were more modest and variable in the FACT mutants (spt16, pob3, and nhp6), we did find that, in at least one mutant, spt16-197, a significant increase in SER3 mRNA levels occurred at 30°C (Fig. 3B). Importantly, SRG1 RNA levels were not significantly reduced in most of the mutant strains as compared with a wild type at 30°C.
Figure 3.
Repression of SER3 is dependent on Spt6/Spn1(Iws1) and the FACT complex. (A) Northern analysis of SER3, SRG1, and SCR1 (loading control) was performed on wild-type (FY4), spt6-1004 (FY2425), spt6-140 (FY111), spt6-14 (FY1221), iws1-7 (GHY1199), and iws1-13 (GHY1200) strains. Cells were grown in YPD at 30°C to mid-log and then shifted for 60 min to 37°C. (B) Northern analysis of SER3, SRG1, and SCR1 (loading control) was performed on wild-type (FY4), spt16-197 (FY346), spt16-11 (TF8030-1), spt16-22 (YJ832), spt16-23 (YJ833), spt16-24 (TF7783-24), pob3-7 (TF8031-1), and nhp6aΔ∷URA3 nhp6bΔ∷URA3 (FY1411) strains that were grown in YPD. (C) ChIP analysis was performed on chromatin isolated from wild-type (YJ877, YJ878, YJ879, and YJ884), spt6-1004 (YJ886, YJ887, YJ888, and YJ892), and spt16-197 (YJ841, YJ842, and YJ843) strains expressing Rpb1-C13myc and untagged control strains (FY4, FY5, and YJ586). Rpb1-C13myc was immunoprecipitated with α-myc A14 antibody from chromatin prepared from cells that were grown in YPD at 30°C. The amount of immunoprecipitated DNA was determined by qPCR as a percentage of the input material and is expressed as the fold enrichment over a control region of chromosome V that lacks ORFs (Supplemental Table S2, No ORF). Each bar represents the mean ± SEM from at least three independent experiments. Below the graph is a schematic of SER3 with black bars corresponding to the regions amplified by qPCR (see Supplemental Table S2 for details).
We also performed chromatin immunoprecipitation (ChIP) experiments to measure RNA Pol II occupancy across the SRG1/SER3 locus in a wild-type strain and two of these mutants (spt6-1004 and spt16-197) that express either untagged Rpb1 (control) or a myc-tagged version of Rpb1 (Rpb1-13myc). The spt6-1004 and spt16-197 mutants have both been well characterized and share similar phenotypes characteristic of transcription defects, including sensitivity to the nucleotide analog 6-azauracil, suppression of Ty insertions, and cryptic intragenic transcription (Kaplan et al. 2003; Mason and Struhl 2003). Consistent with our Northern data, RNA Pol II strongly associates with the SRG1 transcription unit (Fig. 3C) to similar levels in wild-type, spt6-1004, and spt16-197 cells. Taken together, these results show that SER3 repression is strongly dependent on both Spt6/Spn1(Iws1) and FACT. When these factors are mutated, SER3 is derepressed without affecting RNA Pol II levels at SRG1. This result argues against a model in which it is the level of active transcription that confers transcription interference.
Beyond the primary sites of SRG1 transcription termination, we found a twofold increase in RNA Pol II occupancy in the spt6-1004 cells as compared with wild-type cells, which is consistent with our Northern data (Fig. 3C). However, we did not detect an increase in RNA Pol II in the spt16-197 cells. Although surprising given the increase in SER3 mRNA levels in this mutant, this result may be reconciled if we consider that SRG1 transcription does not always terminate properly, resulting in the production of a minor readthrough that extends to the end of SER3 (Martens et al. 2004; Thompson and Parker 2007). Importantly, we found that the level of SRG1–SER3 readthrough product is reduced in both spt6-1004 and spt16-197 mutants (S Hainer, unpubl.), which is likely due to increased initiation at the SER3 promoter. Therefore, increased RNA Pol II occupancy in these mutant strains that would better reflect the observed increases in SER3 transcription are likely masked by the RNA Pol II that occupies SER3 as a result of the synthesis of an SRG1–SER3 readthrough product.
Nucleosome occupancy of the SER3 promoter is reduced in spt6-1004 and spt16-197 mutants at the permissive temperature
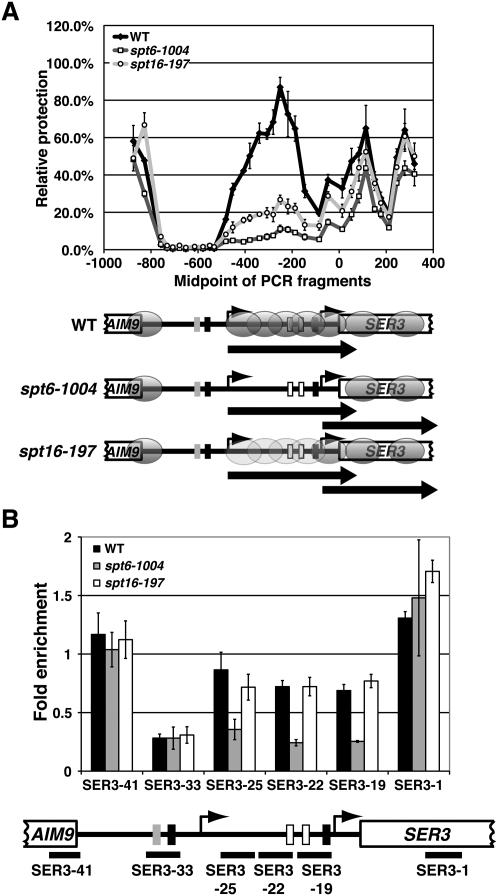
To test whether the level of nucleosomes over SRG1 affects SER3 repression, we next performed nucleosome scanning assays to compare MNase accessibility across SRG1 in wild-type, spt6-1004, and spt16-197 cells that were grown in YPD at 30°C. We again normalized MNase protection of each SRG1–SER3 region to the GAL1 NB region, as the MNase accessibility of the GAL1 control regions was indistinguishable between these strains (Supplemental Fig. S3A). Compared with wild-type cells, we measured a significant reduction of MNase protection specifically across the SRG1 transcribed unit in spt6-1004 cells (fourfold decrease) and to a slightly lesser extent in spt16-197 cells (threefold decrease) (Fig. 4A), indicating nucleosome depletion across SRG1. These results are strikingly similar to the nucleosome scanning results we obtained for the srg1-1 mutant (Fig. 1B). However, while SRG1 transcription was greatly reduced in srg1-1 strains, it remained at wild-type levels in the spt6-1004 and spt16-197 mutants.
Figure 4.
Nucleosome positions and relative occupancy at SER3 in spt6-1004 and spt16-197 mutants. (A) Nucleosome scanning assay was performed on wild-type (FY2134, YJ864, and YJ847), spt6-1004 (FY2180, YJ855, YJ862), and spt16-197 (FY346, YJ859, and YJ916) strains that were grown in YPD at 30°C as described in Figure 1. The light-gray ovals over the SRG1 transcription unit in the spt16-197 strain reflect that this region is slightly more protected from MNase digestion as compared with the spt6-1004 strain. (B) Histone H3 ChIP was performed on chromatin isolated from wild-type (FY4, FY5, and YJ586), spt6-1004 (YJ886, YJ887, and YJ888), and spt16-197 (YJ844, YJ845, and YJ846) cells that were grown in YPD. The amount of immunoprecipitated DNA was determined by qPCR as a percentage of the input material and is expressed as the fold enrichment over GAL1 NB (see Supplemental Fig. S1). Each bar represents the mean ± SEM of at least three independent experiments. Below the graph is a schematic of SER3 with black bars corresponding to the regions amplified by qPCR (see Supplemental Table S2 for details).
To complement our MNase experiments, we performed histone H3 ChIP assays in these same strains grown under the same conditions (Fig. 4B). In wild-type cells, we detected significant histone H3 occupancy over the SER3 promoter as compared with the SRG1 promoter, which is consistent with nucleosomes occupying the SER3 promoter. Moreover, at least for spt6-1004 cells, there is a twofold to threefold decrease in histone H3 occupancy specifically over the SER3 promoter that parallels the increase in MNase sensitivity over this region. Curiously, we did not observe a similar decrease in histone H3 occupancy over the SER3 promoter in spt16-197 cells. Since the loss of MNase protection is less pronounced in the spt16-197 mutants as compared with the spt6-1004 mutants, it is possible that histone H3 ChIP is not sensitive enough to detect a change in histone occupancy between wild-type and spt16-197 strains. Alternatively, nucleosomes may only partially reassemble in the spt16-197 mutant in a manner that makes them more accessible to MNase without altering histone H3 occupancy. Based on previous studies (Belotserkovskaya et al. 2003; Xin et al. 2009), an intriguing possibility is that reassembly of the H2A/H2B dimers at the SER3 promoter may be specifically reduced by the spt16-197 mutation. Taken together, these data support a model whereby FACT and Spt6/Spn1(Iws1) are required for SRG1 transcription-dependent assembly of nucleosomes that repress SER3.
spt6-1004 and spt16-197 mutants are defective for transcription interference at SER3
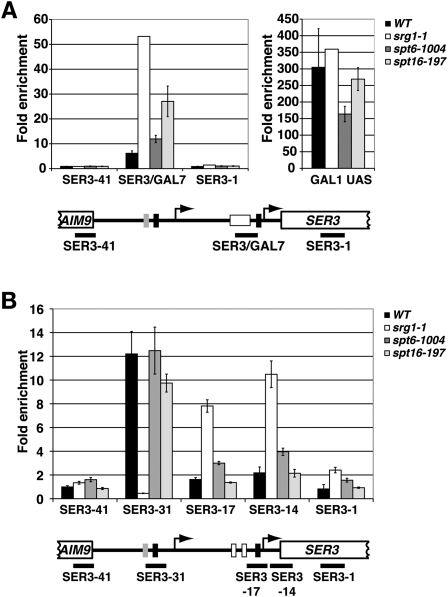
To test whether SRG1 transcription-dependent nucleosomes interfere with transcription factor binding to the SER3 promoter, we performed ChIP experiments in spt6-1004 and spt16-197 mutants. Because sequence-specific activators of SER3 remain unknown, we first used a previously described ser3∷GAL7UAS allele in which the putative SER3 UAS is replaced with two binding sites for the Gal4 transcription activator (Martens et al. 2004). We then measured Gal4 occupancy by ChIP in wild-type, srg1-1, spt6-1004, and spt16-197 strains that all contain the ser3∷GAL7UAS allele and were grown in YPgal (Fig. 5A). Consistent with our previous data (Martens et al. 2004), Gal4 occupancy at the SER3 promoter increases eightfold in the srg1-1 control strain where SRG1 is no longer transcribed and the SER3 promoter is depleted of nucleosomes. In the spt6-1004 and spt16-197 strains where SRG1 is transcribed at wild-type levels but nucleosome occupancy at the SER3 promoter is reduced, Gal4 occupancy at the SER3 promoter was also increased twofold and fourfold, respectively (Fig. 5A, left panel). Based on our SER3 expression and nucleosome occupancy data (Figs. 3A, 4A), the twofold increase in Gal4 occupancy at the SER3 promoter in the spt6-1004 strains was lower than expected. However, this result is likely related to the fact that we also found reduced Gal4 occupancy at the control GAL1 promoter in spt6-1004 cells as compared with wild-type, srg1-1, and spt16-197 cells (Fig. 5A, right panel).
Figure 5.
spt6-1004 and spt16-197 mutants are defective for transcription interference at SER3. (A) Gal4 ChIP was performed on wild-type (YJ871, YJ872, and YJ873), spt6-1004 (YJ875, YJ876, and YJ850), spt16-197 (YJ867, YJ868, and YJ869), and positive control srg1-1 (FY2260) cells that all contain the ser3∷GAL7UAS allele. Chromatin was prepared from cells grown at 30°C in YPraf to 0.8 × 107 cells per milliliter, and then for an additional 4 h at 30°C after the addition of 2% galactose. Gal4 ChIP signals were determined by qPCR at the three SER3 locations (left histogram), and at GAL1 as a positive control (right histogram). All values were normalized to a control region located near the telomere of chromosome VI (TELVI) (Supplemental Table S2) and represent the mean ± SEM. Below the graph is a diagram of the ser3∷GAL7UAS allele in which the putative SER3 UAS region was replaced with the GAL7 UAS region containing two Gal4-binding sites (white box). The black bars indicate the regions of SER3 amplified by qPCR. (B) TBP ChIP was performed on chromatin isolated from wild-type (FY4, FY5, YJ586, and KY719), spt6-1004 (YJ886, YJ887, YJ888, and YJ892), spt16-197 (YJ841, YJ842, YJ843, and YJ844), and positive control srg1-1 (FY2471, YJ582, YJ583, and YJ585) strains that were grown in YPD at 30°C as described in Figure 3C.
We also compared TBP occupancy by ChIP at the SRG1 and SER3 TATA sequences in wild-type, srg1-1, spt6-1004, and spt16-197 strains that contain the endogenous SRG1–SER3 locus (Fig. 5B). The SRG1 and SER3 TATA sequences are both conserved among related yeast strains, bind TBP, and are required for SRG1 and SER3 transcription, respectively (Martens and Winston 2002; Martens et al. 2004). At the SRG1 TATA, there is little difference in TBP occupancy in the spt6-1004 and spt16-197 mutants as compared with the wild-type strains, which agrees with our Northern and RNA Pol II ChIP data (see Fig. 3). At the SER3 TATA, TBP occupancy increased twofold in spt6-1004 cells as compared with a fourfold increase in srg1-1 control cells, suggesting that the loss of nucleosomes over the SER3 promoter in the spt6-1004 mutants either increases TBP binding directly or possibly indirectly by allowing an unknown SER3 activator protein better access to the SER3 promoter. Interestingly, we did not observe a significant difference in TBP occupancy in the spt16-197 mutant. This result may not be surprising, considering the increase in SER3 expression is significantly lower in this mutant as compared with the spt6-1004 mutant (Fig. 3; see Supplemental Fig. S4B for a direct comparison). Therefore, this assay may lack the sensitivity to detect a significant difference in TBP occupancy between wild-type and spt16-197 cells.
From these data, we conclude that transcription interference at SER3 is dependent, at least in part, on Spt6 and Spt16. Taken together with results described earlier, our findings suggest that transcription interference of SER3 is partially mediated by nucleosomes that occupy the SER3 promoter as a consequence of SRG1 transcription from intergenic DNA.
Histone modifications that suppress cryptic intragenic transcription are not required for SER3 repression
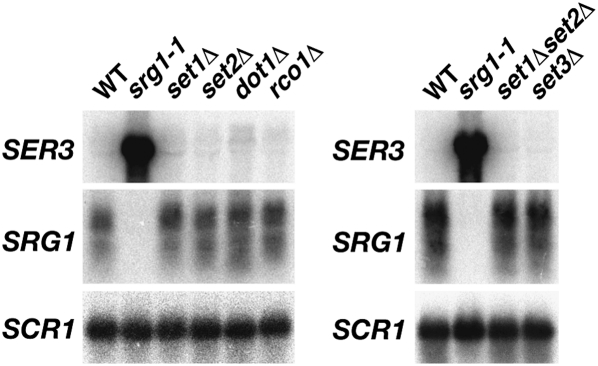
Spt6 and Spt16 have been shown previously to suppress transcription initiation from cryptic promoters that are located within protein-coding regions (Mason and Struhl 2003; Kaplan et al. 2009). Cryptic intragenic transcription is also suppressed by a cascade of transcription-dependent post-translational histone modifications (Lee and Shilatifard 2007; Li et al. 2007a). During transcription, Set2 methylates Lys 36 of histone H3, thereby marking nucleosomes associated with recently transcribed DNA (Pokholok et al. 2005; Rao et al. 2005). Dimethylated H3K36 acts as a binding site for the Rpd3S histone deacetylase complex (Youdell et al. 2008). Upon recruitment, Rpd3S deacetylates the reassembled nucleosomes on the N-terminal tails of histones H3 and H4, which suppresses cryptic intragenic transcription, presumably by occluding transcription factor access (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). Recently, Set1-dependent methylation of H3K4 has also been implicated as a signal for transcription-dependent histone deacetylation by Rpd3S (Pinskaya et al. 2009) and the Set3 complex (Kim and Buratowski 2009). Because of these observations, a likely hypothesis is that Set1 and Set2 may contribute to SER3 repression by regulating similar histone modifications over the SER3 promoter in response to SRG1 transcription. To test this possibility, we performed a Northern analysis to measure the effect of deleting the genes encoding the Set1, Set2, and Dot1 histone methyltransferases; the Rco1 subunit of Rpd3S; and the Set3 subunit of the Set3 complex on SER3 and SRG1 expression. Deletions of any one of these genes or a set1Δset2Δ double deletion has no effect on SER3 or SRG1 mRNA levels (Fig. 6). Moreover, mutations of histone H3 Lys 4 (methylated by Set1), Lys 36 (methylated by Set2), or Lys 79 (methylated by Dot1) also has little to no effect on SER3 repression (S Hainer, unpubl.). Therefore, our results suggest that the relative contribution of these histone reassembly mechanisms may vary at different loci throughout the genome.
Figure 6.
Repression of SER3 does not require histone methyltransferases or the Rpd3S and Set3C histone deacetylase complexes. Northern analysis of SER3, SRG1, and SCR1 (loading control) was performed on wild-type (YJ586), srg1-1 (FY2471), set1Δ (KY938), set2Δ (KY912), dot1Δ (KY934), rco1Δ (KY1235), set1Δset2Δ (KY1822), and set3Δ (KY1806) strains that were grown in YPD at 30°C.
Discussion
In this study, we provide evidence that intergenic transcription represses adjacent gene transcription by assembling a repressive chromatin structure, rather than by the act of transcription. First, we showed that SRG1 intergenic transcription is required not only for repression of the adjacent SER3 gene, but also to maintain MNase protection of the SER3 promoter. Second, we determined that changes in the MNase protection of the SER3 promoter are caused by changes in SRG1 transcription and are not an effect of the changes to SER3 transcription. Third, we found that cells expressing mutant versions of the Spt6 and Spt16 elongation factors derepress SER3 and reduce MNase protection across the SER3 promoter without altering SRG1 RNA levels or RNA Pol II occupancy across SRG1. These results clearly implicate the nucleosomes assembled on the SER3 promoter as the key factor in SER3 repression. Finally, we found that Spt6 and Spt16 are required to inhibit transcription factor binding to the SER3 promoter, which suggests that the nucleosomes assembled at the SER3 promoter by these factors interfere with the binding of transcription factors to their sites on DNA.
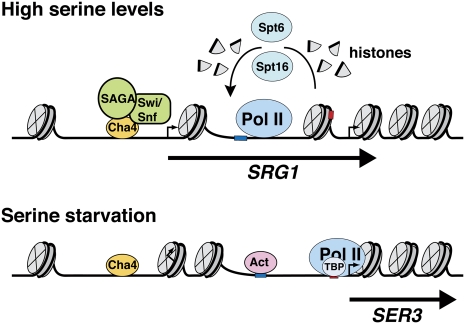
Taken together with our previous studies (Martens and Winston 2002; Martens et al. 2004, 2005), we propose the following model for SER3 regulation (Fig. 7). When cells are grown in serine-rich medium, the Cha4 DNA-binding protein recruits the Swi/Snf and SAGA complexes, resulting in the induction of SRG1 transcription. RNA Pol II transcribes SRG1 across the SER3 promoter, disassembling nucleosomes in its path and then reassembling them in its wake by a mechanism that involves both Spt6 and Spt16. SRG1 transcription is thus required to maintain nucleosomes across the SER3 promoter, interfering with transcription factor binding. When cells are then transferred to serine starvation conditions, Cha4 no longer recruits Swi/Snf and SAGA, resulting in decreased SRG1 transcription. Without intergenic transcription to maintain them, nucleosomes are depleted over the SER3 UAS, allowing transcription factors—either an as yet unknown site-specific DNA-binding activator or possibly TBP and RNA Pol II—to bind and activate SER3. Two positioned nucleosomes remain at the 5′ end of SRG1, where they are likely to inhibit SRG1 transcription.
Figure 7.
A model for SER3 regulation by SRG1 intergenic transcription. When serine is available to the cells, DNA-bound Cha4 recruits SAGA and Swi/Snf to initiate SRG1 transcription, possibly by remodeling the two nucleosomes located at the 5′ end of SRG1 to expose the SRG1 transcription start site. RNA Pol II transcribes SRG1 and, through Spt6 and Spt16, disassembles nucleosomes in its path and then reassembles them in its wake. As a result, nucleosomes continuously occupy the SER3 UAS where they repress SER3 by occluding the SER3 promoter from transcription factor binding. In the absence of serine, SRG1 transcription is repressed, possibly due to the presence of two nucleosomes at its 5′ end that encompass its transcription start site. In the absence of SRG1 transcription, the SER3 UAS is depleted of nucleosomes, allowing an as yet unknown activator (Act) and/or TBP and RNA Pol II to bind and activate SER3 transcription.
In addition to its role in nucleosome assembly during transcription, Spt6 has also been reported to reassemble nucleosomes at the promoters of PHO5 and several other yeast genes during repression (Adkins and Tyler 2006). Therefore, an alternative model for SER3 repression is that Spt6 and, possibly, Spt16 reassemble nucleosomes over the SER3 promoter independently of SRG1 transcription. Thus, mutations in these factors may bypass the normal role for SRG1 transcription, which is to interfere with the recruitment of chromatin remodeling factors needed to displace the repressive nucleosomes at the SER3 promoter. A prediction of this model is that the increased levels of SER3 expression observed in the spt6-1004 and spt16-197 mutants would no longer be dependent on sequence-specific activators to recruit chromatin remodeling factors, analogous to what has been observed for PHO5 (Adkins and Tyler 2006). To test this alternative model, we first identified a 37-bp sequence within the SER3 promoter (from −192 to −228; SER3 ATG = +1) that is required for SER3 activation in response to serine starvation (Supplemental Fig. S4A). When this sequence was deleted in the spt6-1004 and spt16-197 strains, SER3 mRNA levels were reduced as compared with similar strains expressing wild-type SER3 (Supplemental Fig. S4B). Therefore, spt6-1004 and spt16-197 mutations do not bypass the requirement of the SER3 UAS for SER3 activation, which argues against this alternative model.
Although MNase accessibility has been used extensively to predict nucleosome occupancy in eukaryotic organisms (for examples, see Yuan et al. 2005; Lee et al. 2007), we cannot rule out the possibility that DNA-binding proteins may contribute to the protection of the SER3 promoter from MNase digestion in serine-rich conditions. However, our observation that MNase protection over the SER3 promoter was reduced in spt6-1004 and spt16-197 mutants without affecting RNA Pol II occupancy suggests that at least RNA Pol II and its associated factors do not affect MNase digestion.
If SRG1 transcription from intergenic DNA is required to maintain nucleosomes over the SER3 UAS, then from where might these nucleosomes originate? An intriguing source of these nucleosomes would be those positioned over the SRG1 transcription start site and TATA (Fig. 7) that likely inhibit SRG1 transcription in the absence of serine. Based on this study and our previous work (Martens and Winston 2002; Martens et al. 2004, 2005), Swi/Snf, when recruited to the SRG1 promoter in response to serine, may slide these nucleosomes toward SER3 to facilitate preinitiation complex assembly and SRG1 transcription. Once RNA Pol II begins to transcribe SRG1, the nucleosomes originally moved by Swi/Snf are disassembled to allow passage of RNA Pol II, and then are reassembled behind RNA Pol II by Spt6 and Spt16. Therefore, the activities of Swi/Snf, Spt6/Spn1, and FACT may combine to establish and maintain nucleosomes over the SER3 promoter that interfere with transcription factor binding to this region. This scenario would also explain the difference in nucleosome occupancy at the 5′ end of SRG1 observed for wild-type cells grown in the serine starvation media as compared with srg1-1 cells grown in serine-rich media, two conditions in which SER3 is strongly derepressed (Figs. 1, 2A). In contrast to wild-type cells grown in serine starvation medium, where it is no longer recruited, Swi/Snf is presumably still recruited by Cha4 in the srg1-1 (SRG1 TATA mutant) cells that are grown in serine-rich media. Thus, Swi/Snf can remodel the nucleosomes at the 5′ end of SRG1; however, these nucleosomes cannot be maintained in the absence of SRG1 transcription.
In addition to the nucleosome reassembly activity of Spt6/Spn1 and FACT, it has been well documented that a cascade of transcription-dependent post-translational modifications of histones found within nucleosomes over protein-coding genes contributes to the repression of intragenic transcription initiation (Lee and Shilatifard 2007; Li et al. 2007a). However, our studies show that SER3 repression appears to be independent of at least some of these marks, including Set1-mediated methylation of histone H3 K4, Set2-mediated methylation of K36, and the removal of histone H3 and H4 acetylation by the Rpd3S and Set3C histone deacetylase complexes. Although we cannot rule out the possibility that other post-translational histone modifications may be involved, our results indicate a difference in the requirement of transcription-dependent post-translational histone modifications between SER3 repression by SRG1 transcription and repression of cryptic intragenic transcription. This difference may be related to the fact that SRG1 is a relatively short transcription unit (∼400 bp) that is highly transcribed. It has been reported recently that cryptic intragenic transcription preferentially occurs at lowly transcribed genes (Li et al. 2007b; Cheung et al. 2008; Lickwar et al. 2009). Therefore, it is possible that highly transcribed SRG1 may not be dependent on H3K36 methylation and subsequent histone deacetylation for protection from intragenic transcription, because of the frequent passage of RNA Pol II. Alternatively, short, highly transcribed genes may never establish this histone mark, since histone H3K36 methylation predominates toward the 3′ ends of transcribed genes (Pokholok et al. 2005). In support of this possibility, genome-wide analyses of K36 methylation indicate little K36 trimethylation at SRG1 (Pokholok et al. 2005).
In contrast to the characteristic transcription-dependent depletion of nucleosomes seen at protein-coding genes (Yuan et al. 2005; Lee et al. 2007), we show transcription-dependent assembly of nucleosomes across intergenic SRG1. How does one account for this apparent contradiction between nucleosome occupancy and transcription? Several recent studies have indicated that DNA sequences can either favor or refract nucleosome formation, thereby influencing genome-wide nucleosome positioning (Yuan et al. 2005; Ioshikhes et al. 2006; Segal et al. 2006; Peckham et al. 2007; Field et al. 2008; Kaplan et al. 2009). As has been proposed for yeast genes containing nucleosome-depleted promoter regions (Segal and Widom 2009), one possibility is that the underlying DNA sequence of the SER3 promoter may normally disfavor nucleosome formation to facilitate transcription factor binding. Therefore, by reassembling nucleosomes after each passage of RNA Pol II, SRG1 transcription effectively maintains nucleosomes over DNA that is normally refractory to nucleosomes. Several observations support this possibility. First, the SER3 UAS region that is nucleosome-depleted in the absence of SRG1 transcription contains poly(dA:dT) tracts, a sequence motif that resists bending and thus disfavors nucleosome formation (Segal and Widom 2009). Second, the SER3 UAS sequence is predicted to have a low nucleosome-forming potential by an algorithm developed using comparative genomics (Ioshikhes et al. 2006). Finally, the SER3 UAS sequence failed to form a stable nucleosome in a genome-wide in vitro nucleosome reconstitution assay (Kaplan et al. 2009).
In S. cerevisiae, cells respond to changes in serine availability by rapidly inducing or repressing transcription of SER3. This response involves a dynamic competition between nucleosomes and transcription factors that is controlled by the transcription of SRG1 from intergenic ncDNA. Our findings raise the intriguing possibility that widespread transcription of ncDNA may impact genome-wide chromatin architecture. In doing so, transcription of ncDNA may influence not only gene expression, but also other cellular processes that are dependent on protein–DNA interactions.
Materials and methods
S. cerevisiae strains and media
All S. cerevisiae strains used in this study (Supplemental Table S1) are isogenic with a GAL2+ derivative of S288C (Winston et al. 1995). Strains were constructed using standard genetic crosses or by transformation (Ausubel et al. 1991). The C termini of RPB1 and SPT16 were tagged with 13 copies of the c-Myc epitope by PCR-mediated transformation of diploid strains using pFA6a-13myc-KanMX and pFA6a-13myc-HIS3MX, respectively (Longtine et al. 1998). The spt16-22 and spt16-23 alleles (Formosa et al. 2001) were integrated into a diploid strain by two-step gene replacement using SnaBI-digested pTF142-23 and pTF142-22 plasmids (kindly provided by T. Formosa, University of Utah, Salt Lake City, UT). The ser3ΔUAS mutation was constructed by replacing 37 bp of SER3 promoter sequence (from −228 to −198; SER3 ATG = +1) with an AvrII restriction site by QuikChange mutagenesis (Agilent Technologies) to yield pRM08 plasmid. The ser3ΔUAS allele was then integrated into a diploid strain by two-step gene replacement using AfeI-digested pRM08. Several strains contain a KanMX-marked deletion of the SER33 gene, which is a paralog of SER3. Based on previous studies (Martens and Winston 2002; Martens et al. 2004) and the results presented in this study, the deletion of SER33 does not affect SER3 regulation. Strains were grown in the following media as indicated in the figure legends: YPD (1% yeast extract, 2% peptone, 2% glucose), YPgal (1% yeast extract, 2% peptone, 2% galactose), YPraf (1% yeast extract, 2% peptone, 2% raffinose), and SC with 1 mM serine (SC + serine) or without serine (SC − serine) (Rose et al. 1990).
Nucleosome scanning assay
Nucleosome scanning experiments were performed using a method adapted from those described previously (Whitehouse and Tsukiyama 2006; Brickner et al. 2007; Lee et al. 2007). Cells were grown to 2 × 107 to 3 × 107 cells per milliliter and were treated with formaldehyde (2% final concentration) for 30 min at 30°C and then glycine (125 mM final concentration) for 10 min at room temperature. Formaldehyde-treated cells (1.2 × 109) were harvested by centrifugation, washed with Tris-buffered saline, and then incubated in ZDB buffer (50 mM Tris Cl at pH 7.5, 1 M sorbitol, 10 mM β-mercaptoethanol) containing 1.5 mg of zymolase 20T for 30 min at 30°C on a rocker platform. Spheroplasts were pelleted by low-speed centrifugation, gently washed with NP buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris Cl at pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 0.075% NP-40, 1 mM β-mercaptoethanol, 500 μM spermidine), and resuspended in 1.8 mL of NP buffer. Samples were divided into six 300-μL aliquots that were then digested with 0, 1, 2.5, 5, 10, and 20 U of MNase (Nuclease S7 from Roche) for 45 min at 37°C. Digestions were stopped with 75 μL of Stop buffer (5% SDS, 50 mM EDTA) and were treated with 100 μg of proteinase K for 12–16 h at 65°C. DNA was extracted by phenol/chloroform using PLG-H tubes (5 Prime), and was incubated with 50 μg of RNase A for 1 h at 37°C. DNA was re-extracted with phenol/chloroform, precipitated with an equal volume of isopropanol, washed with 80% ethanol, and resuspended in 100 μL of TE. MNase digestions were evaluated by two methods. First, one-fifth of digested DNA was separated by gel electrophoresis. Second, previously characterized GAL1 promoter sequences (Lohr 1984; Brickner et al. 2007; Floer et al. 2010)—one within a positioned nucleosome (GAL1 NB), and a second adjacent region (GAL1 NUB) that is rapidly digested by MNase—were amplified by qPCR from MNase-treated and untreated samples. The MNase concentration that resulted in mostly mononucleosome-sized DNA (see Supplemental Fig. S1) with a GAL1 NUB/NB ratio of <15% was subjected to further qPCR using tiled SER3 primer pairs (SER3-1 to SER3-41) (Supplemental Table S2). For each SER3 primer set, the amount of protected template was calculated as a ratio between MNase-digested and undigested samples and then normalized to the amount of protected GAL1 NB template. All nucleosome scanning assays were done in triplicate using at least two independent strains as indicated in the figure legends.
Northern analysis
Northern analysis was performed as described previously (Ausubel et al. 1991) on 20 μg of total RNA isolated from cells grown to 1 × 107 to 2 × 107 cells per milliliter. DNA probes were generated by random prime-labeling PCR fragments for SER3 (ChrV: 324059–324307), SRG1 (ChrV: 322258–322559), and SCR1 (ChrV: 441741–442266). SCR1 serves as a loading control, since its RNA levels are unaffected by the mutations and growth conditions used in this study.
ChIP analysis
For histone H3, TBP, and Rpb1-C13myc ChIPs, cells were grown in YPD at 30°C to 1 × 107 to 2 × 107 cells per milliliter. For Gal4 ChIPs, cells were grown in YPraf at 30°C to 0.8 × 107 cells per milliliter, and then an additional 4 h at 30°C after addition of 2% galactose. Chromatin preparation and treatment were preformed as described previously (Shirra et al. 2005). Gal4, histone H3, TBP, and Rpb1-13myc were immunoprecipitated by incubating sonicated chromatin overnight at 4°C with 1 μL of anti-GAL4 DBD antibody (sc-577, Santa Cruz Biotechnology), 5 μL of anti-histone H3 antibody (ab1791, Abcam), 2 μL of anti-TBP antibody (kind gift from G. Prelich, Albert Einstein College of Medicine), and 4 μL of anti-c-myc A-14 antibody (sc-789, Santa Cruz Biotechnology), respectively. Dilutions of input and immunoprecipitated DNA were subjected to qPCR. All ChIP signals were normalized to a control: either GAL1 NB template (histone H3 ChIP), TELVI template located within a telomeric region on chromosome VI (Gal4 ChIP), or “No ORF” template located within a region of chromosome V that lacks ORFs (Rpb1-C13myc and TBP ChIPs). Details regarding the primers used for qPCR in each ChIP experiment are listed in Supplemental Table S2.
qPCR
All qPCR data were obtained using an ABI 7300 or StepOnePlus Real-Time PCR system, SYBR green reagents (Fermentas), and the primer sets listed in Supplemental Table S2. All calculations were performed using Pfaffl methodology for relative quantitation of real-time PCR (Pfaffl 2001).
Acknowledgments
We thank Karen Arndt, Andrea Duina, Fred Winston, and Travis Mavrich for critical reading of this manuscript prior to submission. We are grateful to Karen Arndt, Tim Formosa, Grant Hartzog, Greg Prelich, and Fred Winston for providing us with antibodies, strains, and plasmids used in this work. This work was supported by NIH grant GM080470, and by an award from Pittsburgh Life Sciences Greenhouse to J.A.M.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1975011.
Supplemental material is available for this article.
References
- Adkins MW, Tyler JK 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev 11: 2494–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl KE 1991. Current protocols in molecular biology. John Wiley and Sons, New York [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Berretta J, Morillon A 2009. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 10: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ, Winge DR 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J 25: 5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81 doi: 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F 2009. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277 doi: 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF 2010. Non-coding RNAs: Meet thy masters. Bioessays 32: 599–608 [DOI] [PubMed] [Google Scholar]
- Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol 14: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson PR, Mendiratta G, McLaughlin NB, Wolfsberg TG, Marino-Ramirez L, Pompa TA, Jainerin M, Landsman D, Shen CH, Clark DJ 2005. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol Cell Biol 25: 9127–9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ 2001. Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 20: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: A complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF 2009. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol 44: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Aranda A, Proudfoot N 2000. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 8415–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Harrison BR, Yazgan O, Krebs JE 2009. Life without RNAi: Noncoding RNAs and their functions in Saccharomyces cerevisiae. Biochem Cell Biol 87: 767–779 [DOI] [PubMed] [Google Scholar]
- Hess D, Liu B, Roan NR, Sternglanz R, Winston F 2004. Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21. Mol Cell Biol 24: 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134 [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF 2006. Nucleosome positions predicted through comparative genomics. Nat Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol Cell 35: 377–383 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT 2009. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev 23: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A 2007. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res 618: 130–134 [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007a. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL 2007b. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Feng T, Lian Y, Zhang G, Garen A, Song X 2009. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci 106: 12956–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD 2009. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS ONE 4: e4886 doi: 10.1371/journal.pone.0004886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D 1984. Organization of the GAL1–GAL10 intergenic control region chromatin. Nucleic Acids Res 12: 8457–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev 16: 2231–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martens JA, Wu PY, Winston F 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 19: 2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23: 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW 2007. Transcriptional interference: An unexpected layer of complexity in gene regulation. J Cell Sci 120: 2755–2761 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z 2007. Nucleosome positioning signals in genomic DNA. Genome Res 17: 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS 2007. Raising the estimate of functional human sequences. Genome Res 17: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 28: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD 2005. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 25: 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Winston F, Hieter P 1990. Methods in yeast genetics; A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Royo H, Cavaille J 2008. Non-coding RNAs in imprinted gene clusters. Biol Cell 100: 149–166 [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R 2005. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 19: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. What controls nucleosome positions? Trends Genet 25: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA 2009. Divergent transcription: A new feature of active promoters. Cell Cycle 8: 2557–2564 [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K 2005. Intrinsic histone–DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Shirra MK, Rogers SE, Alexander DE, Arndt KM 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169: 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Cui Y, Zhang G, Garen A, Song X 2009. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci 106: 16794–16798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T 2006. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol 13: 633–640 [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T 2009. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol Cell 35: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol 28: 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]