Abstract
The bacterial RNA polymerase (RNAP) holoenzyme consists of a catalytic core enzyme (α2ββ′ω) in complex with a σ factor that is essential for promoter recognition and transcription initiation. During early elongation, the stability of interactions between σ and the remainder of the transcription complex decreases. Nevertheless, there is no mechanistic requirement for release of σ upon the transition to elongation. Furthermore, σ can remain associated with RNAP during transcription elongation and influence regulatory events that occur during transcription elongation. Here we demonstrate that promoter-like DNA sequence elements within the initial transcribed region that are known to induce early elongation pausing through sequence-specific interactions with σ also function to increase the σ content of downstream elongation complexes. Our findings establish σ-dependent pausing as a mechanism by which initial transcribed region sequences can influence the composition and functional properties of the transcription elongation complex over distances of at least 700 base pairs.
Keywords: RNA polymerase, σ factor, transcription initiation, promoter-proximal pausing, transcription elongation complex
The σ subunit of bacterial RNA polymerase (RNAP) is required for promoter-specific transcription initiation (Gross et al. 1998). When complexed with the RNAP core enzyme (subunit structure α2ββ′ω), different σ factors specify the recognition of different classes of promoters (Gruber and Gross 2003). The primary σ factor in Escherichia coli, σ70, typically directs transcription initiation from promoters defined by two conserved hexameric DNA sequence elements, termed the −10 and −35 elements for their relationship to the transcription start site (position +1). During the transition from transcription initiation to transcription elongation, the growth of the nascent RNA destabilizes the interaction between σ70 and the RNAP core enzyme (Mekler et al. 2002; Murakami et al. 2002; Vassylyev et al. 2002; Nickels et al. 2005), but the complete release of σ70 is not required for entry into the elongation phase of transcription (Ring et al. 1996; Bar-Nahum and Nudler 2001; Mukhopadhyay et al. 2001; Mooney et al. 2005). Thus, although historically defined as an initiation factor, σ70 can also remain associated with the transcription elongation complex and influence the transcription process during elongation.
A functional role for σ70 during elongation was first established in the context of bacteriophage λ late gene transcription (for review, see Roberts et al. 1998). Specifically, the expression of the phage late genes under the control of the λQ anti-terminator protein depends on a σ70-dependent pause that occurs during early elongation, shortly after the σ70-containing RNAP holoenzyme has escaped the late promoter PR′. Formation of this paused early elongation complex, which contains a stably bound nascent RNA that is 16 or 17 nucleotides (nt) in length, depends on an interaction between σ70 and a DNA sequence element in the initial transcribed region positioned from +1 to +6 that resembles a promoter −10 element (Ring et al. 1996). Furthermore, the same module of σ70 that contacts the promoter −10 element during transcription initiation (σ70 region 2) contacts the −10-like element in the initial transcribed region during early elongation pausing (Ring et al. 1996). Similar −10-like initial transcribed region DNA sequence elements have been shown to induce σ70-dependent pausing in the context of the late promoters of other lambdoid phages (Ring et al. 1996) and in the context of several E. coli promoters, including the lac, tnaA, and cspD promoters (Brodolin et al. 2004; Nickels et al. 2004; Hatoum and Roberts 2008). Furthermore, both bioinformatics analysis and a candidate-based approach suggest that ∼20% of all E. coli promoters may carry σ70-dependent pause-inducing sequence elements in their initial transcribed regions (Nickels et al. 2004; Hatoum and Roberts 2008).
Here we establish a new function for initial transcribed region σ70-dependent pause elements: to modify the functional properties of downstream elongation complexes by increasing their σ70 content. In particular, we show that an elongation complex that encounters a σ70-dependent pause element in the initial transcribed region is more likely to retain σ70, and thus is more likely to pause at a second σ70-dependent pause element positioned downstream. We further demonstrate that the effect of a promoter-proximal σ70-dependent pause element on the σ70 content of downstream elongation complexes can be detected over distances of at least 700 base pairs (bp). Our findings establish σ70-dependent pausing as a mechanism whereby initial transcribed region sequences can influence the composition and functional properties of the elongation complex throughout a transcription unit. Unlike cis-acting sequences that function to recruit trans-acting factors to the elongation complex, these promoter-like initial transcribed region sequences functionally alter the transcription complex by inhibiting loss of an RNAP subunit. We propose that engagement of a promoter-proximal σ70-dependent pause element increases the σ70 content of downstream elongation complexes by stabilizing the association of σ70 with the early elongation complex during a series of critical nucleotide addition steps when the probability of σ70 release is relatively high.
Results
Preliminary considerations
Biophysical characterization of artificially stalled transcription complexes indicates that the association of σ70 with the early elongation complex is stabilized by interactions between σ70 and a pause-inducing sequence element in the initial transcribed region. Specifically, ensemble and single-molecule fluorescence resonance energy transfer (FRET) measurements of the σ70 content of elongation complexes halted during early elongation indicate that a pause-inducing sequence element positioned from +1 to +6 (like that at λPR′) functions to stabilize the association of σ70 with stalled elongation complexes containing nascent RNAs 11–15 nt in length (Nickels et al. 2004; Kapanidis et al. 2005). However, conflicting results have been obtained regarding whether the presence of a σ70-dependent pause-inducing sequence element in the initial transcribed region increases the proportion of elongation complexes that contain σ70 at positions downstream from the pause site. Ensemble FRET results suggest that a σ70-dependent pause-inducing sequence element in the initial transcribed region increases the σ70 content of elongation complexes halted at position +50 (Mukhopadhyay et al. 2001; Nickels et al. 2004), but single-molecule FRET results do not (Kapanidis et al. 2005). Given the conflicting evidence obtained from these prior biophysical studies of halted elongation complexes, we sought to investigate whether the presence of a σ70-dependent pause-inducing sequence element in the initial transcribed region influences the σ70 content of downstream elongation complexes using approaches that would enable us to monitor the σ70 content of actively transcribing complexes.
Presence of σ70-dependent pause element in the initial transcribed region increases the σ70 content of downstream elongation complexes in vitro
To determine whether the presence of a pause-inducing promoter −10-like element in the initial transcribed region increases the proportion of elongation complexes that contain σ70 at downstream positions, we took advantage of prior in vitro observations indicating that, in contrast with promoter-proximal −10-like elements (i.e., those that extend no further than approximately +15 relative to the transcription start site), promoter-distal −10-like elements can induce efficient pausing only when high concentrations (1 μM) of free σ70 are present (Mooney and Landick 2003; Mooney et al. 2005; Sevostyanova et al. 2008). These findings suggest that efficient engagement of promoter-distal σ70-dependent pause elements requires that free σ70 rebind the elongation complex, likely because the majority of complexes lose σ70 during the earliest phase of elongation (Mooney and Landick 2003). We therefore sought to determine whether the presence of a promoter −10-like element in the initial transcribed region could inhibit σ70 loss during the earliest phase of elongation and enable σ70-dependent pausing at a promoter-distal site in the absence of high concentrations of free σ70.
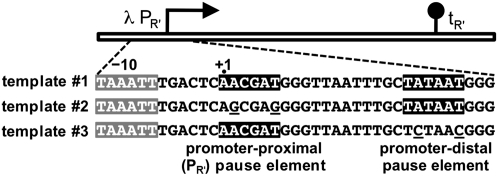
We performed a series of in vitro transcription assays using three templates (Fig. 1). The first template (template #1) carried both a promoter-proximal σ70-dependent pause-inducing sequence element and a promoter-distal σ70-dependent pause-inducing sequence element. In particular, template #1 carried the well-characterized pause element associated with the λ late promoter (λPR′) positioned between +1 and +6, as well as a consensus promoter −10 element positioned between +19 and +24. The second template (template #2) carried two base pair substitutions in the promoter-proximal pause element (A+2G/T+6G) that destabilize interactions with σ70 and eliminate σ70-dependent pausing (Ring and Roberts 1994). The third template (template #3) carried two base pair substitutions in the promoter-distal pause element (A+20C/T+24C); these substitutions were predicted to destabilize interactions with σ70 and eliminate σ70-dependent pausing at the promoter-distal site.
Figure 1.
Schematic depiction and relevant sequence of the in vitro transcription templates. The −10 element of the λPR′ promoter is highlighted in gray. The promoter-proximal λPR′ −10-like pause-inducing element (positioned between +1 and +6) and the promoter-distal pause-inducing element (consensus −10 hexamer positioned between +19 and +24) are highlighted in black. The distal pause element also includes a TG dinucleotide upstream of the −10 hexamer, and thus is a consensus extended −10 element. The mutations that disrupt the promoter-proximal pause element on template #2 and the mutations that disrupt the promoter-distal pause element on template #3 are underlined. Terminator tR′ (black lollipop) is positioned to terminate transcription at ∼116 nt.
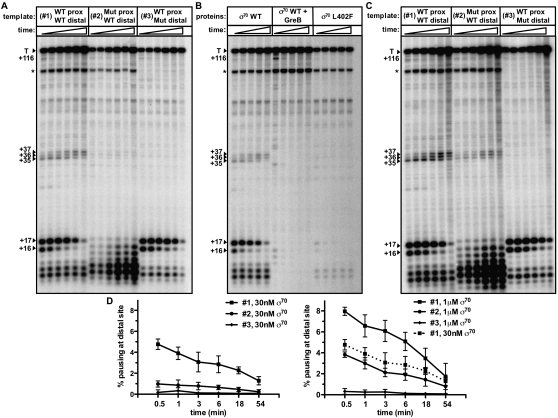
When single-round transcription time-course reactions were performed with low concentrations of σ70 (30 nM, a twofold molar excess over RNAP core) using template #1, the expected 16- and 17-nt λPR′ pause species, which appeared early in the time course and decayed over time, were observed (Fig. 2A). In addition to these 16- and 17-nt RNA transcripts, less-prominent 35- to 37-nt RNA transcripts were observed throughout the time course (Fig. 2A). Strikingly, when the transcription reactions were performed using template #2, which carries mutations in the promoter-proximal pause element, these 35- to 37-nt RNA transcripts were not detected, consistent with the idea that the presence of a σ70-dependent pause element in the initial transcribed region can facilitate σ70-dependent pausing at promoter-distal sites. Several lines of evidence confirmed that the 35- to 37-nt RNA transcripts are, in fact, the result of σ70-dependent pausing at the promoter-distal site. First, the promoter-distal pause element is displaced by 18 bp relative to the position of the promoter-proximal pause element (register +19 compared with register +1), and the 35- to 37-nt RNA transcripts are ∼18 nt longer than the paused species that result from engagement of the promoter-proximal pause element. Second, the 35- to 37-nt RNA species are absent when reactions are performed using template #3, which carries mutations in the promoter-distal pause element (Fig. 2A). Third, production of the 35- to 37-nt RNA species (like production of the 16- and 17-nt RNA species) is reduced when reactions are done using template #1 in the presence of RNAP holoenzyme reconstituted with a well-characterized σ70 mutant (σ70 L402F) that is deficient in σ70-dependent pausing (Fig. 2B; Ko et al. 1998). Fourth, the 35- to 37-nt RNA species are sensitive to the transcript cleavage factor GreB (Fig. 2B), a hallmark of previously characterized σ70-dependent pause species (Marr and Roberts 2000). We therefore conclude that the presence of a promoter-proximal σ70-dependent pause element increases the likelihood that σ70-dependent pausing will occur at a promoter-distal site in vitro.
Figure 2.
A promoter −10-like element in the initial transcribed region inhibits σ70 loss during early elongation and enables σ70-dependent pausing at a promoter-distal site in vitro. (A) Single-round in vitro transcription reactions performed with RNAP holoenzyme (15 nM core reconstituted with 30 nM σ70) and three different templates (#1, #2, and #3, as depicted in Fig. 1) bearing the indicated wild-type (WT) or mutant (Mut) promoter-proximal and promoter-distal pause elements. The reactions were performed as a time course with samples taken 0.5, 1, 3, 6, 18, and 54 min after transcription was initiated. RNA species that reflect σ70-dependent pausing at the promoter-proximal element (+16 and +17) or the promoter-distal element (+35, +36, and +37) are indicated. The 35- to 37-nt RNA species, which were annotated by comparison with a reaction done in the presence of a chain-terminating nucleotide, were also detected when the RNA was end-labeled with [γ-32P]ATP, indicating that they are not cleavage products derived from an internal portion of the RNA (Supplemental Fig. S1). We note the accumulation of abortive RNA products in reactions performed with template #2, which we attribute to the two base pair substitutions in the initial transcribed region; initial transcribed region alterations can have dramatic effects on abortive transcript production (Hsu et al. 2003; Chander et al. 2007). The ∼116-nt terminated transcript (T) is indicated, and the asterisk (*) indicates a shorter terminated transcript that is the result of transcription initiating under the control of the promoter-distal extended −10 element. (B) Single-round in vitro transcription reactions performed using template #1 (wild-type promoter-proximal and promoter-distal pause elements) with RNAP holoenzyme (15 nM core reconstituted with either 30 nM σ70 wild type or 30 nM σ70 L402F, as indicated). The assay was performed and the gel annotated as in A, except that samples were taken 0.5, 1, 6, 18, and 54 min after transcription was initiated. Where indicated, GreB was included in the reactions at 100 nM. (C) As in A, except the assay was performed with RNAP holoenzyme that was reconstituted with 1 μM σ70. (D) Graph showing the percentage of σ70-dependent pausing detected at the promoter-distal site [distal pause signal/(distal pause signal + terminated signal)] for reactions performed in the presence of 30 nM σ70 (left graph) as in A, or 1 μM σ70 (right graph) as in C. Plotted on the graphs are the mean and SD of three independent experiments. In the right graph, we show that the percent of pausing detected at the distal site on template #2 in the presence of 1 μM σ70 was approximately the same as that detected at the distal site on template #1 in the presence of low concentrations (30 nM) of σ70 (dotted line).
To test the hypothesis that the presence of a promoter-proximal pause element enables promoter-distal pausing by inhibiting the loss of σ70 from the early elongation complex, we asked whether the addition of excess σ70 could restore promoter-distal σ70-dependent pausing on template #2. Specifically, we performed the transcription reactions in the presence of 1 μM σ70 (a 67-fold molar excess over RNAP core) and found that the abundance of the 35- to 37-nt RNA species generated using template #2 increased over their abundance when the reactions were performed with low concentrations of σ70 (Fig. 2C,D); thus, the abundance of the 35- to 37-nt RNAs produced from template #2 at high concentrations of σ70 and from template #1 at low concentrations of σ70 were approximately the same (Fig. 2D, right). Furthermore, the addition of high concentrations of σ70 also increased the abundance of the 35- to 37-nt RNA species generated in reactions performed with template #1, but did not result in the appearance of these species with template #3 (Fig. 2C,D). We conclude that the presence of a σ70-dependent pause element in the initial transcribed region facilitates σ70-dependent pausing at the promoter-distal site in vitro by inhibiting loss of σ70 during the earliest phase of elongation.
Presence of σ70-dependent pause element in the initial transcribed region increases the σ70 content of downstream elongation complexes in vivo
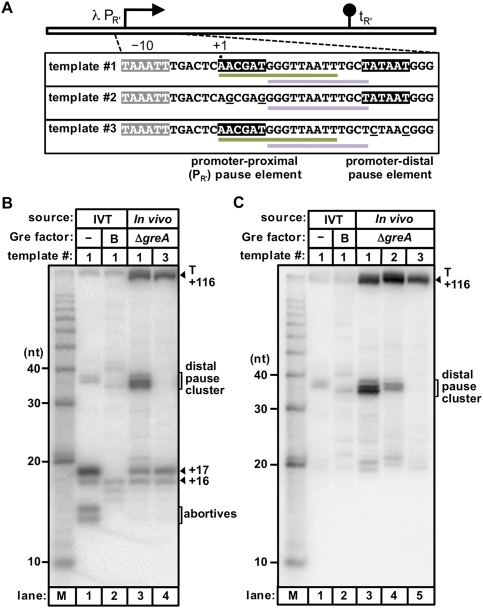
Next, we sought to determine whether the presence of a promoter −10-like element in the initial transcribed region could inhibit σ70 loss during the earliest phase of elongation and enable σ70-dependent pausing at a promoter-distal site in vivo. To do this, we introduced a multicopy plasmid vector carrying template #1, #2, or #3 into E. coli cells. We then isolated total RNA from the cells and detected RNA transcripts generated in vivo from these templates using hybridization with radiolabeled locked nucleic acid (LNA) probes (Válóczi et al. 2004). We identified transcripts associated with σ70-dependent pausing in vivo based on a comparison with transcripts generated during in vitro transcription reactions.
We first sought to identify RNA transcripts produced from templates #1 and #3 in vivo using an LNA probe complementary in sequence to nucleotides +1 to +15 (Fig. 3A), which enabled the detection of RNA transcripts associated with promoter-proximal σ70-dependent pausing (the 16- and 17-nt species) and RNA transcripts associated with promoter-distal σ70-dependent pausing (the 35- to 37-nt species) (Fig. 3B, lane 1). (Note that the +1 to +15 probe is not fully complementary in sequence to transcripts produced from template #2, which bears mutations at positions +2 and +6, within the promoter-proximal pause element.) With the +1 to +15 probe, we detected RNA transcripts associated with promoter-proximal σ70-dependent pausing in RNA samples isolated from cells carrying either template #1 or template #3 (Fig. 3B). In contrast, RNA transcripts associated with promoter-distal σ70-dependent pausing were detected only in RNA samples isolated from cells carrying template #1, which carries an intact promoter-distal pause element (Fig. 3B).
Figure 3.
A promoter −10-like element in the initial transcribed region facilitates σ70-dependent pausing at a promoter-distal site in vivo. (A) Schematic of templates and LNA probes. Templates #1, #2, and #3 are depicted as in Figure 1. Sequences complementary to the +1 to +15 probe (green line) or the +7 to +19 probe (lavender line) are indicated. (B,C) Detection of RNA transcripts with the +1 to +15 probe (B) or the +7 to +19 probe (C). The in vivo generated RNA samples isolated from a ΔgreA strain to facilitate the detection of pause species (Marr and Roberts 2000) were loaded alongside transcripts generated in nonradioactive reactions performed in vitro (IVT) using template #1 in the absence (labeled −) or presence (labeled B) of GreB; a 10-nt RNA ladder is included (lane M). We note that in vivo generated promoter-distal pause products (distal pause cluster) include a faster-migrating species that is likely due to the action of GreB, as this species is also present when GreB is included in the in vitro transcription reaction (see also Fig. 2B). A control experiment excluded the possibility that the apparent effect of the promoter-proximal pause element on the abundance of the promoter-distal pause products is due to an effect of the PR′ pause mutations on the efficiency of probe hybridization (Supplemental Fig. S2).
We next determined the effect of mutating the promoter-proximal pause element on promoter-distal σ70-dependent pausing in vivo. Thus, we compared the abundance of transcripts associated with promoter-distal pausing produced from template #1 (which carries an intact promoter-proximal pause element) with the abundance of transcripts associated with promoter-distal pausing produced from template #2 (which carries an inactivated promoter-proximal pause element). Because of the mutations present on template #2 (at positions +2 and +6), we used an LNA probe complementary in sequence to nucleotides +7 to +19 for this analysis (Fig. 3A). In contrast to the +1 to +15 probe, which enabled detection of transcripts associated with promoter-proximal and promoter-distal pausing, the +7 to +19 probe enabled detection only of transcripts associated with promoter-distal σ70-dependent pausing (the 35- to 37-nt species) (Fig. 3C). With the +7 to +19 probe, we again detected the promoter-distal 35- to 37-nt pause species in RNA samples isolated from cells carrying template #1 and did not detect these 35- to 37-nt species when the promoter-distal pause element was mutationally inactivated (template #3) (Fig. 3C). Furthermore, the abundance of the 35- to 37-nt transcripts associated with promoter-distal pausing was reduced significantly (approximately fivefold) when the PR′ pause element was mutationally inactivated (template #2) (Fig. 3C). We conclude that the presence of a σ70-dependent pause element in the initial transcribed region increases the likelihood that σ70-dependent pausing will occur at a promoter-distal site in vivo, presumably by inhibiting loss of σ70 during the earliest phase of elongation, as was revealed by our in vitro analysis (see Fig. 2).
Presence of σ70-dependent pause element in the initial transcribed region increases the σ70 content of downstream elongation complexes over distances of at least 700 bp
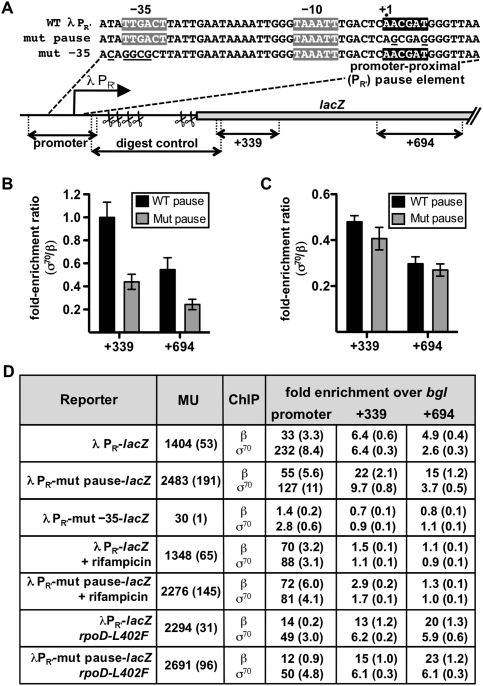
Our results obtained in vitro and in vivo suggest that the presence of a σ70-dependent promoter-proximal pause element facilitates recognition of a more downstream pause element because it enhances the σ70 content of the early elongation complex. That is, transcription elongation complexes that arrive at the promoter-distal pause element are more likely to contain σ70 if they have already engaged a promoter-proximal pause element. We next used chromatin immunoprecipitation (ChIP) to determine whether the effect of a promoter-proximal σ70-dependent pause element on the σ70 content of downstream elongation complexes could be detected throughout a transcription unit. Using cells containing a single-copy PR′-lacZ fusion with either the wild-type pause element or a mutationally inactivated pause element (A+2G/T+6G), we assessed the occupancy of both the β subunit and the σ70 subunit of RNAP across each transcription unit. To do this, we determined the fold enrichment of the promoter/pause region (−92 to +45) and two downstream regions (centered at positions +339 and +694) in both the β- and σ70-immunoprecipitated material relative to a nontranscribed region of the DNA (within the bgl operon). To ensure that promoter-bound transcription complexes did not contribute to the immunoprecipitation of downstream DNA (due to the difficulty of shearing the DNA to a sufficiently small average fragment length), we separated promoter DNA from downstream DNA using restriction enzymes (Fig. 4A; see also the Materials and Methods).
Figure 4.
Effect of the λPR′ promoter-proximal σ70-dependent pause element on the σ70 content of elongation complexes on a λPR′-lacZ fusion. (A) Schematic depiction and relevant sequence of the single-copy λPR′-lacZ fusions used in the ChIP assay. The −35 and −10 elements of the λPR′ promoter are highlighted in gray. The −10-like pause-inducing element (positioned between +1 and +6) is highlighted in black. The mutations that disrupt the promoter-proximal pause element (on the template designated “mut pause”) and the mutations within and upstream of the −35 element that disrupt the λPR′ promoter (on the template designated “mut −35”) are underlined. The black two-headed arrows in the schematic represent the target regions (promoter, +339, +694, and digest control) that were amplified by real-time PCR in the ChIP assay. The scissor symbols represent the locations of the restriction enzyme recognition sites (all of which are within the digest control region and outside the other target regions) that were used to separate the β-immunoprecipitated (or σ70-immunoprecipitated) promoter region from the downstream regions. The efficiency of digestion at these sites was monitored by quantitative amplification of the digest control region. (B,C) Graphs showing the ChIP fold enrichment ratios (σ70/β) from cells containing a λPR′-lacZ fusion with either the wild-type pause element (WT pause) or an inactivated pause element (Mut pause), as indicated. The σ70/β ratios were obtained by dividing the fold enrichment over background (bgl) of a given DNA region (+339 or +694) in the σ70-immunoprecipitated material by the fold enrichment of the same region in the β-immunoprecipitated material (see the Materials and Methods). We note that these fold enrichment ratios do not represent the absolute molar ratios of σ70 and β in the elongation complex (see the Discussion). In B, the cells contained wild-type σ70, whereas in C, the cells contained chromosomally encoded σ70 L402F. The means and SDs of three independent experiments are graphed. (D) Fold enrichments over bgl for both the β- and σ70-immunoprecipitated DNA from cells containing the templates shown in A and either wild-type σ70 or chromosomally encoded σ70 L402F, as indicated. The fold enrichments over bgl were determined as described in the Materials and Methods and represent the means of three independent experiments, with SDs in parentheses. These fold enrichment values were used to calculate the fold enrichment ratios plotted in B and C. The cell cultures that were analyzed by ChIP were also assayed for β-galactosidase (before the addition of formaldehyde or rifampicin), and the results are expressed in Miller units (MU) with the SDs in parentheses.
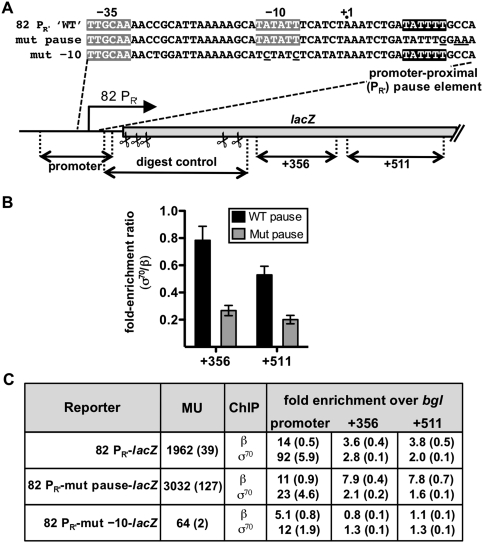
To compare the σ70 content of elongation complexes on the two fusion constructs (PR′-lacZ containing the wild-type or mutated pause element), we calculated fold enrichment ratios (σ70/β) by dividing the fold enrichment of a given DNA region in the σ70-immunoprecipitated material by the fold enrichment of the same region in the β-immunoprecipitated material. Although we observed a general decrease in the σ70 content of elongation complexes with increasing distance from the promoter on both templates (Fig. 4B), we found that the σ70 content of elongation complexes in both the +339 and +694 regions was approximately twofold higher when the PR′ pause element was intact (see Fig. 4B,D). In contrast, when the ChIP assays were done in strains containing chromosomally encoded σ70 L402F, which is deficient in σ70-dependent pausing (see Fig. 2B), the fold enrichment (σ70/β) ratios were the same within error for each downstream region of the DNA regardless of whether the template carried a wild-type or mutated pause element (Fig. 4C,D). We conclude, therefore, that the presence of a functional σ70-dependent pause element in the initial transcribed region enhances the wild-type σ70 content of downstream transcription elongation complexes, and that this effect can be detected over at least 700 bp.
Two control experiments confirmed that the association of σ70 (and β) with downstream DNA was dependent on transcription initiating from promoter PR′. First, neither downstream DNA region was enriched above background in either the β- or the σ70-immunoprecipitated material when PR′ was inactivated (as confirmed by the results of parallel β-galactosidase assays) on the template with the intact pause element (Fig. 4D). Second, the fold enrichment values for the promoter region were high, whereas the fold enrichment values for the downstream DNA regions dropped to background or near background in both the β- and the σ70-immunoprecipitated samples when the cells were pretreated with rifampicin, which traps RNAP at the promoter (Fig. 4D; Herring et al. 2005).
To explore the generality of these observations, we tested the effect of another promoter-proximal σ70-dependent pause element, which associated with the late promoter of phage 82. Displaced by 8 bp relative to the position of the λPR′ pause element, the 82 pause element induces pausing at a nascent RNA length of ∼25 nt (Goliger and Roberts 1989), as compared with 16–17 nt in the case of the λ pause element. Using a pair of 82 PR′-lacZ fusion constructs (bearing the wild-type or a mutated +25 pause element), we again used ChIP to examine the β and σ70 occupancies within the promoter/pause region and two downstream regions (centered at positions +356 and +511) (Fig. 5A). We found that the σ70 content of elongation complexes was between 2.5-fold and threefold higher in both the +356 and +511 regions when the 82 PR′ pause element was intact (Fig. 5B,C). As we did in the case of the λ PR′ construct, we inactivated the 82 PR′ promoter and confirmed that the association of σ70 (and β) with downstream DNA was dependent on transcription initiating from this promoter (Fig. 5C). We conclude that a promoter-proximal σ70-dependent pause element can enhance the σ70 content of downstream transcription elongation complexes when located in at least two distinct positions within the initial transcribed region.
Figure 5.
Effect of the 82 PR′ promoter-proximal σ70-dependent pause element on the σ70 content of elongation complexes on an 82 PR′-lacZ fusion. (A) Schematic depiction and relevant sequence (see also the legend for Supplemental Fig. S3) of the single-copy 82 PR′-lacZ fusions used in the ChIP assay. The −35 and −10 elements of the 82 PR′ promoter are highlighted in gray. The +25 pause element consists of a −10-like hexamer (highlighted in black and positioned from +9 to +14) as well as an upstream TG dinucleotide, and thus is an extended −10-like element. The mutations that disrupt the +25 pause element (on the template designated “mut pause”) and the mutations within the −10 element that disrupt the 82 PR′ promoter (on the template designated “mut −10”) are underlined. An in vitro transcription assay confirmed the elimination of σ70-dependent pausing on the mut pause template (Supplemental Fig. S3). The black two-headed arrows in the schematic represent the target regions (promoter, +356, +511, and digest control) that were amplified by real-time PCR in the ChIP assay. The scissor symbols represent the locations of the restriction enzyme recognition sites (all of which are within the digest control region and outside the other target regions) that were used to separate the β-immunoprecipitated (or σ70-immunoprecipitated) promoter region from the downstream regions. The efficiency of digestion at these sites was monitored by quantitative amplification of the digest control region. (B) Graph showing the ChIP fold enrichment ratios (σ70/β) from cells containing a single-copy 82 PR′-lacZ fusion with either the +25 pause element (WT pause) or a mutationally inactivated pause element (Mut pause), as indicated. The σ70/β ratios were obtained by dividing the fold enrichment over background (bgl) of a given DNA region (+356 or +511) in the σ70-immunoprecipitated material by the fold enrichment of the same region in the β-immunoprecipitated material (see the Materials and Methods). The means and SDs of three independent experiments are graphed. (C) Fold enrichments over bgl for both the β- and σ70-immunoprecipitated DNA from cells containing the templates shown in A. The fold enrichments over bgl were determined as described in the Materials and Methods, and represent the means of three independent experiments with SDs in parentheses. These fold enrichment values were used to calculate the fold enrichment ratios plotted in B. The cell cultures that were analyzed by ChIP were also assayed for β-galactosidase (before the addition of formaldehyde), and the results are expressed in Miller units (MU) with the SDs in parentheses.
Discussion
Here we identify σ70-dependent pausing as a mechanism by which the sequence of the initial transcribed region can influence both the composition and functional properties of downstream elongation complexes. In particular, our results demonstrate that the presence of a σ70-dependent pause element in the initial transcribed region inhibits σ70 loss during the earliest stage of elongation, increasing the σ70 content of elongation complexes at least 700 bp into the transcription unit. Moreover, the analysis of actively transcribing complexes in vitro and in vivo indicates that σ70 is retained in a functionally competent state that permits it to engage downstream pause-inducing elements.
A model for how the presence of a promoter-proximal σ70-dependent pause element influences the σ70 content of downstream elongation complexes
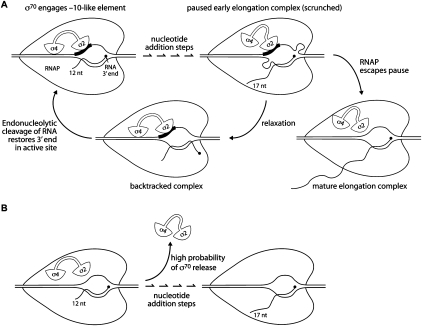
Before providing a mechanistic explanation for the effect of a promoter-proximal σ70-dependent pause element on the σ70 content of downstream elongation complexes, we consider how the paused early elongation complex at λPR′ is formed. The position of the λPR′ −10-like pause element relative to the promoter −10 element (displaced downstream by 12 bp) suggests that σ70 might initially engage this element at a nascent RNA length of ∼12 nt. Thus, it was proposed that formation of the paused elongation complex at λPR′ involves the establishment of sequence-specific interactions between σ70 (region 2 in particular) and the −10-like element at a nascent RNA length of ∼12 nt, followed by several nucleotide addition steps during which σ70 maintains contact with the pause element, until the transcript reaches a critical length (16 or 17 nt) at which pausing is detected (Marr and Roberts 2000). Consistent with the suggestion that σ70 can maintain contact with the pause element through several successive nucleotide addition steps, the results of ensemble FRET analysis indicate that a σ70-dependent pause element positioned from +1 to +6 can stabilize the association of σ70 with elongation complexes halted after the synthesis of RNA transcripts ranging in length from 11 nt to at least 15 nt (Nickels et al. 2004). According to the model, the nucleotide addition steps that occur while σ70 maintains contact with the pause element require expansion of the transcription bubble and concomitant DNA scrunching (see Fig. 6A; Marr and Roberts 2000), as has been shown to occur in the context of promoter-bound initial transcribing complexes during the process of promoter escape/abortive initiation (Kapanidis et al. 2006; Revyakin et al. 2006). In the case of initial transcription, DNA scrunching generates a “stressed intermediate” that can be relieved in two ways: Either σ70 relinquishes its contacts with the promoter DNA and RNAP escapes the promoter, or the expanded bubble collapses and an abortive transcript is released (Kapanidis et al. 2006; Revyakin et al. 2006). In the case of early elongation pausing, the strain generated by DNA scrunching can similarly be resolved in two ways: Either σ70 relinquishes its contacts with the pause element and RNAP escapes the pause, or the expanded bubble collapses, in this case without release of the nascent RNA, generating a backtracked complex in which the catalytic center of the enzyme is no longer in register with the 3′ end of the nascent transcript (Fig. 6A; Marr and Roberts 2000; Perdue and Roberts 2010).
Figure 6.
Model to explain how a promoter-proximal σ70-dependent pause element increases the σ70 content of downstream elongation complexes. (A) Formation of paused early elongation complex at λPR′ (see also Fig. 8 in Perdue and Roberts 2010). (Top left) Domain 2 of σ70 (σ2) is shown engaging the promoter-proximal −10-like pause element (black rectangle) at a nascent RNA length of 12 nt. (Top right) The nascent RNA is extended to 17 nt via a series of nucleotide addition steps that occur while σ2 maintains contact with the pause element, requiring DNA scrunching (depicted as ssDNA loops in the transcription bubble). The scrunched complex can undergo two alternative fates: Either RNAP escapes the pause (bottom right), requiring that σ2 relinquish contact with the pause element, or the expanded transcription bubble collapses (bottom left), generating a backtracked complex in which the catalytic center of the enzyme is no longer in register with the 3′ end of the RNA transcript. Endonucleolytic cleavage of the transcript (stimulated by Gre factor) (Marr and Roberts 2000) can regenerate a catalytically proficient complex with a correctly positioned RNA 3′ end, allowing the cycle to repeat. (B) Critical window of nucleotide addition steps during which σ70 release is “fast.” In the absence of a promoter-proximal pause element that allows σ70 to maintain stabilizing interactions with the DNA, the transcription complex releases σ70 with high probability during a critical window of early nucleotide addition steps (here when the nascent RNA is extended from 12 to 17 nt).
To explain how a promoter-proximal σ70-dependent pause element increases the σ70 content of downstream elongation complexes, we propose that the sequence-specific interactions between σ70 and the pause element stabilize the association of σ70 with the early elongation complex during a “critical window” of nucleotide addition steps when the probability of σ70 release is relatively high. Support for the notion of such a critical window comes from the results of both ensemble and single-molecule FRET measurements of the σ70 content of halted elongation complexes (Nickels et al. 2004; Kapanidis et al. 2005), which suggest that release of σ70 is biphasic, with an initial “fast” phase that occurs after synthesis of an RNA transcript ∼12 nt in length and a subsequent “slow” phase. Thus, according to our model, the interaction between σ70 and an early elongation pause element stabilizes the association of σ70 with the RNAP core enzyme during critical nucleotide addition steps when σ70 release is “fast” (due, perhaps, to clashes between the nascent RNA and specific portions of σ70) (for review, see Mooney et al. 2005) and a significant fraction of the transcription complexes would otherwise release σ70 (Fig. 6B). Our finding that the effect of an early elongation pause element on the σ70 content of downstream elongation complexes was manifest when the pause element was positioned from +1 to +6 (λ PR′) or from +9 to +14 (82 PR′) suggests that the critical window of fast σ70 release might occur at different template positions, depending on the sequence context. Subsequently, as the elongation complexes leave the critical region, they appear to enter a phase where release of σ70 is “slow,” based on the gradual decrease in the σ70 content of mature elongation complexes with increasing distance from the promoter observed in both prior studies (Raffaelle et al. 2005; Mooney et al. 2009a) and the results presented here (Figs. 4, 5).
Prevalence of promoter-proximal σ70-dependent pause elements in vivo
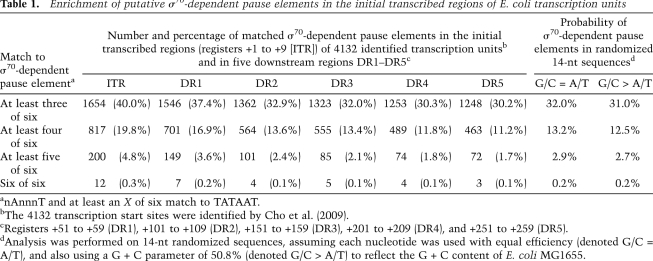
Previous bioinformatics analysis of ∼650 promoters suggested that σ70-dependent promoter-proximal pause elements are associated with ∼20% of all promoters in E. coli (Nickels et al. 2004). We took advantage of a more comprehensive set of 4132 transcription start sites defined from high-throughput sequencing of the 5′ ends of primary RNA transcripts isolated from E. coli (Cho et al. 2009) to re-examine this estimate (Supplemental Table S1). The λ pause element consists of a three of six match to the −10 element consensus sequence (TATAAT) in register +1 (where register is defined by the first base of the −10-like hexamer), including the highly conserved A and T nucleotides at positions 2 and 6 of the hexamer (McClure 1985), whereas the 82 pause element consists of a four of six match to the −10 element consensus sequence in register +9, again including the highly conserved A and T nucleotides at positions 2 and 6 of the hexamer. For our analysis, we identified transcription units containing a three of six, four of six, five of six, or six of six match to the −10 element consensus, with an A and a T at positions 2 and 6 of the hexamer, respectively, in registers +1 to +9; 40% (1654) of the transcription units contained at least a three of six match, 20% (817) contained at least a four of six match, ∼5% (200) contained at least a five of six match, and ∼0.3% (12) contained a six of six match (Table 1). Although we do not know what fraction of the 4132 transcription start sites used for this analysis are associated with a σ70-dependent promoter, it is likely to be the major fraction, because most of the transcripts were present in RNA samples harvested from mid-exponential-phase cells (Cho et al. 2009).
Table 1.
Enrichment of putative σ70-dependent pause elements in the initial transcribed regions of E. coli transcription units
anAnnnT and at least an X of six match to TATAAT.
bThe 4132 transcription start sites were identified by Cho et al. (2009).
cRegisters +51 to +59 (DR1), +101 to +109 (DR2), +151 to +159 (DR3), +201 to +209 (DR4), and +251 to +259 (DR5).
dAnalysis was performed on 14-nt randomized sequences, assuming each nucleotide was used with equal efficiency (denoted G/C = A/T), and also using a G + C parameter of 50.8% (denoted G/C > A/T) to reflect the G + C content of E. coli MG1655.
To determine whether the initial transcribed regions of transcription units (i.e., registers +1 to +9) are enriched for putative σ70-dependent pause elements, we computed the probabilities of finding the target sequence motifs by chance within random sequences (Table 1). Based on this analysis, we would have expected 31% of the transcription units to contain at least a three of six match, 12.5% to contain at least a four of six match, 2.7% to contain at least a five of six match, and 0.2% to contain a six of six match. This analysis therefore suggests that initial transcribed regions are enriched in σ70-dependent pause elements. Consistent with this inference, we found that, when we searched for the target sequence motifs at five distinct promoter-distal positions (registers +51 to +59, +101 to +109, +151 to +159, +201 to +209, and +251 to +259), the percentages of transcription units containing matches in each of these regions approached the percentages that were calculated based on the probabilities of finding the target sequence motifs by chance (Table 1).
Functional roles for σ70 during transcription elongation
The results of prior ChIP analyses reveal unexplained differences in the σ70 content of transcription elongation complexes on different transcription units (Wade and Struhl 2004; Grainger et al. 2005; Reppas et al. 2006; Mooney et al. 2009a). Our finding that the presence of a promoter-proximal σ70-dependent pause element influences the σ70 content of transcription complexes up to ∼700 bp downstream from the transcription start site suggests that the sequence of the initial transcribed region may be a key determinant of at least some of these differences. We note that our ChIP data do not permit an estimate of the actual fractional occupancy of σ70 in the elongation complex at any particular position within a transcription unit because we do not know how the cross-linking efficiencies of σ70 and RNAP in a promoter-bound or paused early elongation complex compare with those in an actively transcribing elongation complex.
Using an artificial construct, we demonstrated that the presence of a promoter-proximal σ70-dependent pause element facilitated σ70-dependent pausing at a promoter-distal pause site both in vitro and in vivo. Interestingly, promoter-distal σ70-dependent pausing has been proposed to occur at a cluster of sites extending as far as ∼100 bp downstream from the transcription start site of the E. coli tnaA promoter, which also carries a σ70-dependent pause element in its initial transcribed region (Hatoum and Roberts 2008). Our findings suggest that the observed promoter-distal pausing in the tnaA transcription unit may be influenced by the presence of the σ70-dependent pause element in the initial transcribed region.
σ70-dependent pausing has been shown to affect the production of full-length transcripts, presumably by modulating RNAP density within the transcription unit (see also Fig. 4D; Hatoum and Roberts 2008). Such effects on RNAP density within a transcription unit could, in principle, affect the overall elongation rate and modulate the responsiveness of RNAP to termination signals by influencing the extent of RNAP “cooperation” (Epshtein and Nudler 2003). As a component of the elongation complex, σ70 could also influence transcription through mechanisms independent of its ability to induce pausing. For example, σ70 could influence the accessibility of the transcription elongation complex to regulatory factors. In this regard, the β′ coiled-coil, which binds σ70 region 2, has also been identified as a binding site for at least two elongation factors, including the highly conserved general elongation factor NusG and the NusG paralog RfaH (Belogurov et al. 2007; Mooney et al. 2009b; Nickels 2009). Essential in E. coli, NusG influences the rate of transcription elongation and functions as both a termination and an anti-termination factor (for review, see Roberts et al. 2008). As NusG is expected to compete with σ70 for access to the elongation complex (Sevostyanova et al. 2008), the presence of a promoter-proximal σ70-dependent pause element could potentially influence gene expression by decreasing the NusG content of downstream elongation complexes. Finally, σ70 might exert a direct effect (either positive or negative) on the process of transcription termination (for example, see Arndt and Chamberlin 1988).
Materials and methods
Strains and plasmids
A list of strains and plasmids is provided in Supplemental Table S2.
In vitro transcription
His-tagged wild-type σ70 and σ70 L402F were purified as described (Nickels et al. 2004). E. coli RNAP core enzyme was purchased from Epicentre and GreB was a gift from J. Gelles (Brandeis University). Open complexes were formed by incubating 15 nM RNAP (preincubated with either 30 nM wild-type σ70, 30 nM σ70 L402F, or 1 μM wild-type σ70) with 20 nM linear DNA template for 5 min at 37°C in transcription buffer (20 mM Tris-HCl at pH 8.0, 0.1 mM EDTA, 50 mM KCl, 100 μg/mL BSA, 10 mM DTT) plus 200 μM GTP, ATP, and CTP, and 50 μM UTP (supplemented with 1 μCi/μL [α-32P]-UTP). A single round of transcription was initiated by adding 4 mM MgCl2 and 10 μg/mL rifampicin. Reactions were stopped and processed at the indicated times (procedure as described in Nickels et al. 2005), and RNA transcripts were electrophoresed on 12% polyacrylamide sequencing gels. Bands were visualized by PhosphorImager, and the data were analyzed using ImageQuant software.
Detection of RNA transcripts by hybridization
ML176 cells containing plasmid pFW11tet-PR′_+19, pFW11tet-mut PR′_+19, or pFW11tet-PR′_mut+19 were grown in LB supplemented with tetracycline (10 μg/mL) to an OD600 of 0.5. Cell suspensions (5 mL) were mixed with RNAlater (15 mL; Ambion), collected by centrifugation (20 min at 17,000g), and resuspended in TRI Reagent (1 mL; Molecular Research Center), and RNA species <200 nt in size were isolated using mirVana miRNA Isolation kit (Ambion). Transcripts were detected as described in Goldman et al. (2009).
β-Galactosidase assays
Reporter strain cells (in triplicate) were harvested from cultures grown at 37°C to mid-exponential phase (OD600 = 0.5) in LB broth supplemented with tetracycline (10 μg/mL). β-Galactosidase assays were performed as described (Nickels 2009) using microtiter plates and a microtiter plate reader. Miller units were calculated as described (Nickels 2009).
ChIP assay
Cultures were grown as described for the β-galactosidase assays. Formaldehyde-induced cross-linking, cell lysis, and DNA shearing were preformed as described (Deighan and Hochschild 2007). When present, rifampicin was added to 150 μg/mL and cells were incubated for 20 min prior to addition of formaldehyde. Protein/DNA complexes were immunoprecipitated in immunoprecipitation buffer (50 mM HEPES at pH 7, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM PMSF) using a monoclonal antibody reactive against the β subunit of RNAP (clone NT63, Neoclone) or a monoclonal antibody reactive against σ70 (clone 2G10, Neoclone) and 20 μL of protein-A/G magnetic beads (New England Biolabs). Immunoprecipitated complexes were washed once in restriction enzyme buffer 2 (New England Biolabs), and then incubated in the same buffer containing a restriction enzyme cocktail for 1 h at 37°C. The restriction enzymes cleave within the “digest control region” (a region nested between the promoter region and the first downstream region) (Figs. 4A, 5A) and serve to increase the resolution of the assay by separating promoter-associated β (or σ70)/DNA complexes from downstream regions. For ChIP experiments with the λPR′-lacZ fusion, the enzymes used were AciI, AhdI, HindIII, XmnI, and NdeI. For ChIP experiments with the 82 PR′-lacZ fusion, the enzymes used were BglI, HindIII, NdeI, PstI, and PvuII. Complexes were then washed four times with 1× immunoprecipitation buffer and once with TE buffer (10 mM Tris at pH 8, 1 mM EDTA), and were resuspended in elution buffer (TE buffer containing 1% SDS). The cross-links were reversed for 7 h at 65°C, and the β- and σ70-immunoprecipitated DNAs were purified using Zymo Research spin columns.
ChIP assay quantitation by real-time PCR
For each ChIP experiment, there were five target regions of interest (Figs. 4A, 5A): the promoter region, two downstream regions within the fused lacZ gene, a digest control region, and, as a background control, a region of the bglF gene within the bgl operon. The chromosome-encoded bgl operon is transcriptionally silent in wild-type E. coli; therefore, the analysis of bgl product amplification provided an internal standard for DNA that was nonspecifically immunoprecipitated by the β and σ70 antibodies or by the protein-A/G magnetic beads. All primer pairs (purchased from IDT) (Supplemental Table S3) permitted the amplification of a single product of the predicted size with ∼1.93-fold amplification per cycle using 10-μL reactions containing iTaq SYBR Green Supermix with ROX (Bio-Rad) and a StepOnePlus Real-Time PCR System (Applied Biosystems). Quantitative PCRs were run in triplicate for each immunoprecipitated DNA sample, and the averaged threshold cycle (Ct) of the amplification products was determined at a specific ΔRn [(fluorescence signal of reporter dye/fluorescence signal of passive ROX dye) − baseline]. For each β- and σ70-immunoprecipitated sample, the fold enrichment of target DNA regions over background (bgl) was calculated using the formula 1.93Ct(bgl) − Ct(target region). All fold enrichment values represent the average of three biological replicates, and standard deviations are presented in parentheses (Figs. 4D, 5C). For the digest control regions, the fold enrichments over bgl were in the range of 0.03–0.38, indicating that promoter-bound transcription complexes did not contribute to the immunoprecipitation of downstream DNA.
Acknowledgments
We thank J. Gelles and J.W. Roberts for helpful discussion, J. Gelles for purified GreB, and S. Dove for comments on the manuscript. We thank Pinal Kanabar (Waksman Genomics Core Facility) for analysis of initial transcribed region sequences, Peter Hochschild for computing the probabilities that random sequences contain the target sequence motifs, and Jonathan Livny for assistance with generating Supplemental Table S1. Work was supported by NIH GM44025 to A.H., and a Pew Scholars Award to B.E.N.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1991811.
Supplemental material is available for this article.
References
- Arndt KM, Chamberlin MJ 1988. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J Mol Biol 202: 271–285 [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G, Nudler E 2001. Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell 106: 443–451 [DOI] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I 2007. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell 26: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodolin K, Zenkin N, Mustaev A, Mamaeva D, Heumann H 2004. The σ70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat Struct Mol Biol 11: 551–557 [DOI] [PubMed] [Google Scholar]
- Chander M, Austin KM, Aye-Han N-N, Sircar P, Hsu LM 2007. An alternate mechanism of abortive release marked by the formation of very long abortive transcripts. Biochemistry 46: 12687–12699 [DOI] [PubMed] [Google Scholar]
- Cho B-K, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 11: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighan P, Hochschild A 2007. The bacteriophage λQ anti-terminator protein regulates late gene expression as a stable component of the transcription elongation complex. Mol Microbiol 63: 911–920 [DOI] [PubMed] [Google Scholar]
- Epshtein V, Nudler E 2003. Cooperation between RNA polymerase molecules in transcription elongation. Science 300: 801–805 [DOI] [PubMed] [Google Scholar]
- Goldman SR, Ebright RH, Nickels BE 2009. Direct detection of abortive RNA transcripts in vivo. Science 324: 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliger JA, Roberts JW 1989. Sequences required for antitermination by phage 82 Q protein. J Mol Biol 210: 461–471 [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrson M, Holdstock J, Busby SJ 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci 102: 17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B 1998. The functional and regulatory roles of σ factors in transcription. Cold Spring Harb Symp Quant Biol 63: 141–155 [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA 2003. Multiple σ subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466 [DOI] [PubMed] [Google Scholar]
- Hatoum A, Roberts JW 2008. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol 68: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO 2005. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J Bacteriol 187: 6166–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LM, Vo NV, Kane CM, Chamberlin MJ 2003. In vitro studies of transcript initiation by Escherichia coli RNA Polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry 42: 3777–3786 [DOI] [PubMed] [Google Scholar]
- Kapanidis A, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S 2005. Retention of transcription initiation factor σ70 in transcription elongation: Single-molecule analysis. Mol Cell 20: 347–356 [DOI] [PubMed] [Google Scholar]
- Kapanidis A, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH 2006. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314: 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, Marr MT, Guo TS, Roberts JW 1998. A surface of Escherichia coli σ70 required for promoter function and antitermination by phage λ Q protein. Genes Dev 12: 3276–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, Roberts JW 2000. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell 6: 1275–1285 [DOI] [PubMed] [Google Scholar]
- McClure WR 1985. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54: 171–204 [DOI] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH 2002. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108: 599–614 [DOI] [PubMed] [Google Scholar]
- Mooney RA, Landick R 2003. Tethering a σ70 to RNA polymerase reveals high in vivo activity of σ factors and σ70-dependent pausing at promoter-distal locations. Genes Dev 17: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R 2005. σ and RNA polymerase: An on-again, off-again relationship? Mol Cell 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R 2009a. Regulator trafficking on bacterial transcription units in vivo. Mol Cell 33: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R 2009b. Two structurally independent domains of E. coli NusG create a regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391: 341–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH 2001. Translocation of σ70 with RNA polymerase during transcription: Fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106: 453–463 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA 2002. Structural basis of transcription initiation: T. aquaticus RNA polymerase holoenzyme at 4 Å resolution. Science 296: 1280–1284 [DOI] [PubMed] [Google Scholar]
- Nickels BE 2009. Genetic assays to define and characterize protein–protein interactions involved in gene regulation. Methods 47: 53–62 [DOI] [PubMed] [Google Scholar]
- Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A 2004. The σ70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat Struct Mol Biol 11: 544–550 [DOI] [PubMed] [Google Scholar]
- Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A 2005. The interaction between σ70 and the β-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci 102: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue SA, Roberts JW 2010. A backtrack-inducing sequence is an essential component of E. coli σ70-dependent promoter-proximal pausing. Mol Microbiol 78: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ 2005. Holoenzyme switching and stochastic release of σ factors from RNA polymerase in vivo. Mol Cell 20: 357–366 [DOI] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K 2006. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell 24: 747–757 [DOI] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR 2006. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314: 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring BZ, Roberts JW 1994. Function of a nontranscribed DNA strand site in transcription elongation. Cell 78: 317–324 [DOI] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW 1996. Function of E. coli RNA polymerase σ factor σ70 in promoter-proximal pausing. Cell 86: 485–493 [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW 1998. Antitermination by bacteriophage λ Q protein. Cold Spring Harb Symp Quant Biol 63: 319–325 [DOI] [PubMed] [Google Scholar]
- Roberts JW, Shankar S, Filter JJ 2008. RNA polymerase elongation factors. Annu Rev Microbiol 62: 211–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I 2008. The elongation factor RfaH and the initiation factor σ bind to the same site on the transcription elongation complex. Proc Natl Acad Sci 105: 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z 2004. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32: e175 doi: 10.1093/nar/gnh171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Wade JT, Struhl K 2004. Association of RNA polymerase with transcribed regions in Escherichia coli. Proc Natl Acad Sci 101: 17777–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]