Abstract
In bacteria that divide by binary fission, cell division starts with the polymerization of the tubulin homolog FtsZ at mid-cell to form a cell division scaffold (the Z ring), followed by recruitment of the other divisome components. The current view of bacterial cell division control starts from the principle of negative checkpoints that prevent incorrect Z-ring positioning. Here we provide evidence of positive control of cell division during sporulation of Streptomyces, via the direct recruitment of FtsZ by the membrane-associated divisome component SsgB. In vitro studies demonstrated that SsgB promotes the polymerization of FtsZ. The interactions are shown in vivo by time-lapse imaging and Förster resonance energy transfer and fluorescence lifetime imaging microscopy (FRET-FLIM), and are corroborated independently via two-hybrid studies. As determined by fluorescence recovery after photobleaching (FRAP), the turnover of FtsZ protofilaments increased strongly at the time of Z-ring formation. The surprising positive control of Z-ring formation by SsgB implies the evolution of an entirely new way of Z-ring control, which may be explained by the absence of a mid-cell reference point in the long multinucleoid hyphae. In turn, the localization of SsgB is mediated through the orthologous SsgA, and premature expression of the latter is sufficient to directly activate multiple Z-ring formation and hyperdivision at early stages of the Streptomyces cell cycle.
Keywords: divisome, cytokinesis, development, peptidoglycan, live imaging, FLIM
Most bacteria grow and divide by binary fission. The process starts with the increase of the cell size, followed by genome replication and segregation. Once completed, the cell is split to generate two daughter cells, each typically containing a single copy of the chromosome. The prokaryotic cell division scaffold consists of overlapping protofilaments of the tubulin homolog FtsZ (Bi and Lutkenhaus 1991), which, in all prokaryotes studied so far, is the first protein of the cell division machinery or divisome. Several proteins are known to assist in septum site localization and stabilizing the Z ring, including FtsA and ZipA (Hale and de Boer 1997; RayChaudhuri 1999; Pichoff and Lutkenhaus 2002), ZapA (Gueiros-Filho and Losick 2002), and SepF (Hamoen et al. 2006). The Z ring then mediates the recruitment of the cell division machinery or divisome to the mid-cell position (for review, see Goehring and Beckwith 2005; Adams and Errington 2009). Recent evidence suggests that this is a two-step mechanism, with a significant lag in Bacillus between the formation of the Z ring associated with FtsA, ZapA, and EzrA and the recruitment of the other components of the cytokinetic machinery, such as FtsL, FtsW, DivIB, and DivIVA (Gamba et al. 2009). The process of Z-ring (dis)assembly during division is actively controlled (for review, see Romberg and Levin 2003).
In most bacteria, the positioning and timing of septum formation involves the action of negative control systems such as Min, which prevents Z-ring assembly at the cell poles (Raskin and de Boer 1997; Marston et al. 1998), and nucleoid occlusion, which prevents formation of the Z ring over nonsegregated chromosomes (Wu and Errington 2004). While most studies so far support the commonly accepted principle that, like mitosis in eukaryotes, the initiation of prokaryotic cell division is negatively controlled, the question is whether this is an absolute prerequisite. One candidate system where perhaps cell division may be positively controlled is the filamentous soil bacterium Streptomyces, since the enhanced expression of a single protein designated SsgA triggers cell division in a direct or indirect manner (van Wezel et al. 2000b, 2006). Streptomycetes are Gram-positive soil bacteria that have a complex life cycle similar to that of filamentous fungi, and they produce >50% of all known antibiotics and many other bioactive natural products (Hopwood 2007). The vegetative mycelium consists of syncytial cells separated by septa (cross-walls) spaced at 5–10 μm (Wildermuth and Hopwood 1970). During the reproductive phase, streptomycetes produce long chains of spores in aerial hyphae, following a complex cell division event whereby ladders of up to a hundred Z rings are produced in a short time span (Chater 2001; Flardh and Buttner 2009). While the Streptomyces cell division machinery resembles that of unicellular bacteria, there are some rather remarkable differences. For one, streptomycetes are the only known organisms where cell division is not essential for growth (McCormick et al. 1994; McCormick and Losick 1996). Second, the canonical control systems like Min, Noc, and SulA are missing. Two MinD homologs are present in Streptomyces coelicolor, but these do not function in septum site localization (J McCormick and GP van Wezel, unpubl.), and MinC is absent, while DivIVA functions in driving tip growth (Flardh 2003). Hence, cell division control likely requires actinomycete-specific proteins, with SsgA as an obvious candidate. SsgA is a member of the family of SsgA-like proteins (SALPs), with seven orthologs in S. coelicolor (Noens et al. 2005; Traag and van Wezel 2008). Null mutants of ssgA and ssgB are blocked at a stage preceding the onset of sporulation-specific cell division, suggesting they may play a role in the control of this process (van Wezel et al. 2000a; Keijser et al. 2003). Interestingly, while SsgA has a major impact on morphogenesis, its expression is independent of the early whi genes, which regulate the different stages of aerial development and sporulation (Chater 2001; Traag et al. 2004). In this study, we demonstrate positive control of septum site localization and Z-ring formation as a third major difference with cell division in most other bacteria. We show that SsgA orchestrates division by facilitating the correct localization of SsgB, which then directly recruits FtsZ to the future septum sites, and tethers the Z ring to the cytoplasmic membrane. Protein interactions are assessed quantitatively in vivo using fluorescence lifetime imaging microscopy (FLIM), and the sequence of localization of the proteins involved was followed live via time-lapse imaging.
Results
Hierarchical order of SsgA, SsgB, and FtsZ localization
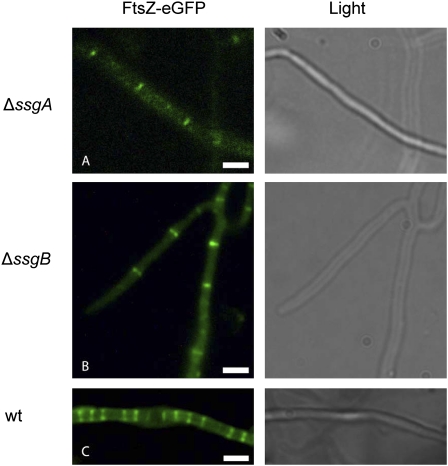
As an initial step in uncovering the possible role of SsgA and SsgB in cell division control, their temporal and spatial localization was determined in relation to that of FtsZ. During sporulation-specific cell division, FtsZ initially forms long filaments in the aerial hyphae, followed by localization in a focal pattern during the onset of cell division, and finally by Z-ring formation, visible as spectacular FtsZ ladders that are typical of streptomycetes (Fig. 1; Schwedock et al. 1997; Grantcharova et al. 2005; Willemse and van Wezel 2009). Interestingly, SsgB also localizes to septum sites and forms the same ladder-like patterns as FtsZ, highlighting SsgB as a likely divisome component (Fig. 1, bottom, corroborated in vivo by immuno-FM; Supplemental Fig. S1), which localizes prior to FtsZ (Fig. 1, top row). To see whether SsgA and SsgB are able to localize to septum sites independently of FtsZ, we made use of the unique feature of streptomycetes that ftsZ-null mutants are viable despite their inability to divide (McCormick et al. 1994). Interestingly, foci of SsgA-eGFP as well as of SsgB-eGFP accumulated in similar fashion as in wild-type cells in the ftsZ-null mutant S. coelicolor HU133 (Fig. 2). SsgB foci accumulated with a spacing (1.25 ± 0.12 foci per micron) very similar to that of Z ladders, clearly indicating that SsgB can find future septum sites independently of FtsZ. Therefore, SsgB may well be the first example of a divisome component that localizes to the septum site prior to and independently of FtsZ (Fig. 2). However, SsgB cannot form rings in the absence of FtsZ, nor are any septa produced, illustrating that FtsZ is required for the ring-like localization of SsgB. Expectedly, TRITC-WGA staining of the cell wall did not identify any accumulation of peptidoglycan in the absence of FtsZ (Fig. 2). Conversely, the formation of Z ladders depends on SsgA and SsgB, with only occasional and widely spaced septa formed in the ssgA- and ssgB-null mutants, with a frequency similar to that of cross-wall formation in vegetative hyphae (Fig. 3). Again, like in vegetative hyphae, these cross-walls do not lead to physical separation of the adjacent compartments, as no constrictions were observed at the exterior of the hyphae with light or electron microscopy. The failure of FtsZ to form ladders of septa was further supported by time-lapse imaging, again identifying occasional and nonconstricting cross-walls (Supplemental Fig. S2).
Figure 1.
SsgB and FtsZ colocalize during sporulation-specific cell division in S. coelicolor. (From left to right) SsgB, FtsZ, an overlay of the two, and the corresponding bright-field image. Developmental stages are shown as follows (for explanation, see the model in Fig. 9): early aerial hyphae (stage IIA; 0.6–0.7 μm in diameter) (top row); predivision foci in aerial hyphae (stage IIC; diameter, 0.8 μm) (middle row); Z rings (stage III; diameter, 0.9 μm) (bottom row). SsgB localizes when ftsZ is still diffusely distributed, while, at later stages, including Z-ring formation, they fully colocalize. See also Figure 4. The approximate developmental stages were determined on the basis of their position in the mycelium (proximity to the vegetative hyphae) and the width of the hyphae (ranging from ∼600 nm for emerging aerial hyphae to ∼900 nm [i.e., spore width] for sporulating aerial hyphae). Note that, for all stages, images were obtained for both combinations of SsgB-eGFP + FtsZ-mCherry (S. coelicolor JSC2) and SsgB-mCherry + FtsZ-eGFP (S. coelicolor JSC3), with the top row presenting JSC2, and the middle and bottom rows presenting JSC3. Most representative images are shown (mCherry fusions are shown in red, and GFP fusions are in green). Bar, 1 μm.
Figure 2.
Localization of SsgA and SsgB and dependence on FtsZ. (Left) Fluorescence micrographs. (Middle) Corresponding TRITC-WGA cell wall stains. (Right) Light images. (Top to bottom) (Row 1) Localization of SsgA-GFP in the ftsZ-null mutant Hu133 (strain JSC15). (Row 2) Localization of SsgA in wild-type cells (strain JSC5; cells in stage prior to septum formation). (Row 3) SsgB localization in the ftsZ-null mutant Hu133, showing normal localization of foci at future septum sites, but failure to produce rings due to the absence of FtsZ (strain JSC10). (Row 4) SsgB localization in the ssgA-null mutant, showing that proper SsgB localization depends on SsgA (strain JSC8). (Row 5) SsgB localization in wild-type cells (strain JSC6). The lectin TRITC-WGA stains peptidoglycan subunits. Bar, 1 μm.
Figure 3.
FtsZ localization in ssgA- and ssgB-null mutants. Fluorescence micrographs showing FtsZ-eGFP (left panel) and corresponding light image (right panel) of aerial hyphae of the ssgA-null mutant (strain JSC12) (top row) and the ssgB-null mutant (strain JSC11) (middle row). In these genetic backgrounds, FtsZ-eGFP forms septa in a pattern similar to that observed in vegetative hyphae. This conforms well to the notion that SsgA and SsgB are required per se for the formation of Z ladders typical of sporulation-specific cell division, but not for individual septa such as cross-walls in vegetative hyphae (see the text). The hyphal width of all strains shown is ∼900 nm, indicative of the normal ring stage in septum formation. (Bottom row) Wild-type localization of FtsZ is shown as a reference (strain K202). Bar, 1 μm.
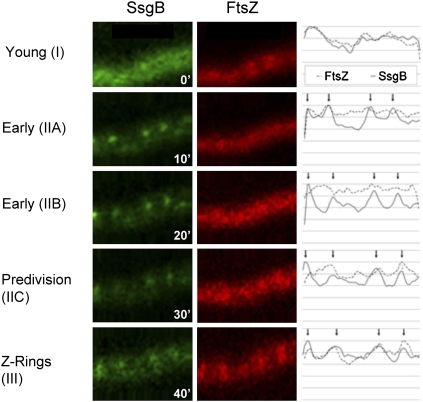
To visualize the localization hierarchy of SsgB-eGFP and FtsZ-mCherry live during the initiation of sporulation-specific cell division, we further developed the technique of time-lapse imaging (Jyothikumar et al. 2008) to image proteins in aerial hyphae. This unequivocally supported the fluorescence microscopy data, with SsgB localizing to cell division sites first, followed some minutes later by FtsZ (Fig. 4; Supplemental Movie). The time between the emergence of SsgB foci and the formation of Z ladders was ∼30 min. The intensity profiles of the still images corroborate the colocalization of SsgB and FtsZ (Fig. 4).
Figure 4.
Still images from the time-lapse Supplemental Movie, which provides a live recording of the order of recruitment of SsgB and FtsZ. The fluorescence intensity profiles (right column) highlight the overlap between the foci of SsgB-eGFP (left column) and FtsZ-mCherry (middle column), up to and including the stage of Z-ring formation. The strain that was used for imaging is S. coelicolor JSC2 (Supplemental Table S3). The time line was from 0 min (top image) to 45 min (bottom image). The low resolution of the fluorescence images is an inevitable consequence of the strong magnification from the time-lapse series. Arrows refer to the peaks in fluorescence intensity of the SsgB profile. For an explanation of stages (roman numerals between brackets), refer to the model in Figure 9. The intensity profiles were prepared using ImageJ.
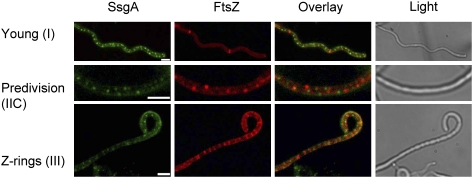
In turn, SsgA forms a distinct focal pattern in young aerial hyphae at a time when SsgB is still diffusely distributed (Fig. 5, top row), while SsgA and FtsZ do not colocalize at any time (Fig. 6). However, there is significant overlap between SsgA and SsgB foci at a specific stage just prior to septum formation in sporogenic aerial hyphae (Fig. 5, middle row), but this was seen only in ∼3% of the sporogenic aerial hyphae (with >200 hyphae analyzed in still images), indicating that the interaction is most likely transient. SsgA foci do not remain localized at the division site during divisome assembly, and the protein is relocated to a position adjacent to the septa (Fig. 5, bottom row), which is in accordance with earlier observations (Noens et al. 2007). Thus, the localization studies strongly suggest a hierarchical localization order, SsgA–SsgB–FtsZ, whereby FtsZ is dispensable for the initial division site localization of SsgB. Expectedly, SsgB fails to accumulate properly in ssgA-null mutants (Fig. 2). Most ssgA mutant hyphae no longer accumulate SsgB, while the number of SsgB foci was strongly reduced in others (average of 0.53 ± 0.07 foci per micron, as compared with 1.25 ± 0.14 per micron in the parent). In precious few aerial hyphae of ssgA-null mutants, SsgB localized in a similar pattern as in the parental strain, but the foci contained around threefold less SsgB, as derived from the fluorescence intensities (37 ± 2 and 12 ± 1 arbitrary units for foci in M145 and the ssgA-null mutant, respectively).
Figure 5.
Localization of SsgA and SsgB during different stages of development in S. coelicolor. (Left to right) SsgA-eGFP (green), SsgB-mCherry (red), an overlay, and a bright-field image. The strain that was used is S. coelicolor JSC4 (Supplemental Table S3). SsgA localizes prior to SsgB in young aerial hyphae (0.6 μm wide) (top row), and the two proteins colocalize only briefly during an early stage of cell division initiation in sporogenic aerial hyphae (diameter, 0.7 μm) (middle row). (Bottom row) At the time of Z-ring formation, SsgA and SsgB do not colocalize (diameter of aerial hyphae, 0.8 μm). For an explanation of stages (roman numerals between brackets), refer to the model in Figure 9. Bar, 1 μm.
Figure 6.
SsgA and FtsZ localize differentially in S. coelicolor. (Left to right) SsgA-eGFP (green), FtsZ-mCherry (red), an overlay of the two, and the corresponding bright-field image. The strain that was used is S. coelicolor JSC1. The activation of cell division by SsgA is governed via the recruitment of SsgB (Fig. 5), and SsgA does not colocalize with FtsZ. Developmental stages are shown as follows: young aerial hyphae (note that the exposure time for FtsZ was halved to prevent overexposure of the cross-walls) (top row); aerial hyphae producing predivision foci (middle row); sporulating aerial hyphae (note constrictions) (bottom row). For an explanation of stages (roman numerals between brackets), refer to the model in Figure 9. Bar, 1 μm.
In contrast, overexpression of SsgA stimulates the accumulation of foci and rings of SsgB-eGFP and FtsZ-mCherry in vegetative hyphae, while they are distributed uniformly in wild-type cells (Supplemental Fig. S3); this underlines the role of SsgA as an activator of cell division, most likely by allowing the correct positioning of SsgB, which then recruits FtsZ (see below). In line with the notion that the activation of cell division by SsgA should be mediated via SsgB and FtsZ, enhanced expression of SsgA fails to stimulate septum formation in either the ssgB-null mutant GSB1 or the ftsZ mutant Hu133 (Supplemental Fig. S4).
Protein dynamics and interactions
To analyze sporulation stage-specific dynamics of SsgA, SsgB, and FtsZ in Streptomyces, fluorescence recovery after photobleaching (FRAP) was applied (Supplemental Fig. S5; Supplemental Table S1), which monitors the redistribution of bleached molecules. Prior to septation, SsgA recovered to bleached foci in just over half a minute (T1/2 of 37 ± 4 sec), approximately twice as fast as SsgB and FtsZ (T1/2 of 62 ± 11 sec and 71 ± 4 sec, respectively). Expectedly, unassembled FtsZ recovered very rapidly (T1/2 of 1 sec). Interestingly, at the time of Z-ring formation, the dynamics of FtsZ and SsgB increased strongly (T1/2 of 5 ± 2 sec and 8 ± 3 sec, respectively), with FtsZ protofilament turnover now at least as dynamic as in Bacillus subtilis (Anderson et al. 2004).
SsgB and FtsZ interact strongly in vivo
The data presented above paint a picture of SsgAB as a positive control system for Z-ring formation during sporulation in the aerial hyphae of Streptomyces. To quantitatively assess the in vivo proximity and direct interactions between SsgA, SsgB, and FtsZ, Förster resonance energy transfer (FRET) combined with FLIM was applied (Cremazy et al. 2005). Its application was successfully applied recently to study the interaction between members of the Escherichia coli divisome (Alexeeva et al. 2010). FRET is the capacity of a low-wavelength fluorophore to transfer its excited state energy to a longer-wavelength counterpart given a sufficient overlap of the corresponding emission and excitation spectra, if the proximity between the two fluorescent proteins is between 2 and 10 nm. FLIM imaging allows determining the FRET efficiency, and this efficiency acts as a molecular ruler that measures the intermolecular distances between the donor and acceptor molecules (e.g., one protein translationally fused to eGFP and one fused to mCherry). In line with literature, the fluorescence lifetime of eGFP fusion proteins alone was 2.6 ± 0.1 nsec (Fig. 7A,B). The same lifetime (i.e., no interaction) was found when SsgA-eGFP and SsgB-mCherry were imaged together during early stage aerial growth (Table 1), but, when SsgA and SsgB colocalized in presporulation foci, the fluorescence lifetime of SsgA-eGFP was reduced to 1.03 ± 0.3 nsec (Fig. 7C,D), corresponding to an intermolecular distance of 5.1 ± 0.3 nm. To provide perspective for the interaction strength, the lifetime reductions are in the same order as for histone H2B interacting with DNA (1.6 nsec) (Cremazy et al. 2005). This corroborates an order of recruitment of SsgA > SsgB > FtsZ, in line with the localization studies. Similarly, the lifetime of FtsZ-eGFP/mCherry decreased strongly on colocalization with SsgB-mCherry/eGFP (1.3 and 1.5 ± 0.3 nsec, respectively) (Fig. 7E,F), with an intermolecular distance of ∼5.6 ± 0.3 nm, thus corroborating the temporal order of recruitment of SsgA–SsgB–FtsZ suggested by the localization studies. As a reference for the later divisome components, we also determined the intermolecular distance of SsgB and FtsZ to FtsI, a transpeptidase involved in septal peptidoglycan synthesis (Pogliano et al. 1997). The fluorescence lifetime for the SsgB–FtsI interaction was 1.67 ± 0.29 nsec and 1.90 ± 0.15 nsec with SsgB and FtsI as donors, respectively (6.3 ± 0.3-nm distance), and thus SsgB is closer to FtsZ than to FtsI. Expectedly, FtsZ interacts very closely with FtsI, with a very short fluorescence lifetime of 0.55 ± 0.08 nsec, corresponding to a 4.3 ± 0.1 nm intermolecular distance (Table 1).
Figure 7.
Molecular interactions between SsgA, SsgB, FtsZ, and the membrane studied by FLIM. Fluorescence intensity images (A,C,E,G,I) and corresponding pseudocolored fluorescence lifetime images ranging from 0 nsec (red) to 3 nsec (blue) (B,D,F,H,J) are presented for SsgA alone (strain JSC5) (A,B) and the interaction between SsgA and SsgB (strain JSC4) (C,D), SsgB and FtsZ (strain JSC2) (E,F), SsgB and the FM5-95-stained membrane (strain JSC6) (G,H), and FtsZ and the membrane (strain JSC7) (I,J).
Table 1.
FLIM measurements of molecular interactions
The data reveal direct interactions between SsgA and SsgB in aerial hyphae; between SsgB, FtsI, and FtsZ; and between SsgB/FtsI/FtsZ and the cell membrane.
(ND) Not determined.
SsgB and FtsI both interact strongly with the cytoplasmic membrane (stained with FM 5-95), as shown by the extraordinary short lifetime of 0.65 ± 0.01 nsec (4.5 ± 0.1-nm distance) for SsgB (Fig. 7G,H) and 0.39 ± 0.25 nsec (4.1 ± 0.4 nm) for FtsI. In contrast, FtsZ is relatively far away from the cell membrane (lifetime decreased to 2.20 ± 0.01 nsec, or 7.6 ± 0.1-nm distance) (Fig 7I,J). Based on these distances and the three-dimensional structures of SsgB (which forms trimers) (Xu et al. 2009) and FtsZ (Lowe and Amos 1998), it is likely that the SsgB ring lies between (and is concentric with) the cell membrane and the Z ring.
To independently corroborate the molecular interactions established in vivo, we tested the ability of SsgA, SsgB, and FtsZ to establish pair-wise interactions using the bacterial two-hybrid system (BACTH) (Karimova et al. 1998). Since the S. coelicolor SsgB protein fails to form multimers in E. coli due to folding problems, we used the SsgB ortholog from the thermophilic actinomycete Thermobifida fusca (SsgBTfus), which was used for structure elucidation and whose functionality was demonstrated by its ability to complement ssgB-null mutants of S. coelicolor (Xu et al. 2009). DNA fragments encoding the selected proteins were cloned into pKT25 and/or pUT18 vectors to generate recombinant plasmids expressing hybrid proteins fused C-terminally to the T25 or T18 fragment of Bordetella pertussis adenylate cyclase, respectively. The two-hybrid studies corroborated the direct interaction between SsgA and SsgBTfus as well as that between SsgBTfus and the extremely well-conserved N-terminal domain of FtsZ (amino acids 1–195) (Supplemental Fig. S6). However, SsgBtfus did not show interaction with the less well-conserved C-terminal domain of FtsZ (amino acids 196–399), while SsgA did not interact with either part of FtsZ or with the full-length protein. These data fully support our in vivo interaction studies performed with FRET-FLIM.
Quantification and stoichiometry of SsgA, ssgB, FtsI, and FtsZ
To obtain insight into the dynamics as well as to establish whether SsgB may form complete rings during division, we determined the absolute number and stoichiometry of the different fusion proteins during different stages of Z-ring formation. For this, we used 80-nm rotavirus particles containing exactly 120 eGFP molecules as an internal standard (Dundr et al. 2002), which were confined in a diffraction-limited spot. In predivision foci, SsgA, SsgB, and FtsI accumulated in similar amounts (∼300 molecules), while FtsZ was 1.5-fold to twofold more abundant (SsgA:SsgB:FtsI:FtsZ ratio of 1:1.2:1:1.7) (Supplemental Table S2). At the divisome—where SsgA does not accumulate—in particular, SsgB and FtsZ were much more abundant, with, on average, ∼700 SsgB, ∼350 FtsI, and ∼1500 FtsZ molecules per Z ring (average ratio of 2:1:4) (Supplemental Table S2). Notably, the amount of FtsZ was highly variable in Z rings, varying from ∼600 molecules at Z-ring initiation (corresponding to the number of molecules in predivision foci) to a maximum of ∼6000 molecules on the Z ring in dividing cells. This conforms well to recent computer modeling studies, which predict that the Z ring should undergo a transition from a low-density to a high-density state (Lan et al. 2009; see the Discussion).
In vitro polymerization of FtsZ
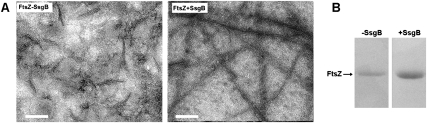
To determine whether SsgB has a positive influence on the polymerization of FtsZ, we applied in vitro polymerization assays. FtsZ-His6 from T. fusca was allowed to polymerize in the presence of GTP and with either SsgB-His6 or BSA (negative control). MgCl2 concentrations were such as to favor protofilament formation of FtsZ (Low et al. 2004). Electron microscopy of negatively stained samples was applied to characterize the FtsZ polymers formed in the presence of SsgB. This revealed a strongly positive effect on filament formation, whereby length had increased significantly in comparison with the filaments formed in the control samples, demonstrating that SsgB is able to stimulate FtsZ polymerization (Fig 8A). Average filament length was 435 ± 36 nm (P < 0.0001; n = 56) for FtsZ with SsgB, and 110 ± 6 nm (P < 0.0001; n = 100) in the absence of SsgB.
Figure 8.
Effect of SsgB on FtsZ polymerization in vitro. (A) Electron microscopy analysis of filament formation in vitro. (Left) Typical FtsZ filaments formed in the absence of SsgB (BSA was used as control). (Right) FtsZ filaments formed in the presence of SsgB. Note the strong increase in filament formation as induced by the presence of SsgB. For these experiments, we used purified FtsZ from T. fusca (FtsZTfus) in combination with either purified SsgBTfus or BSA, and in the presence of 0.2 mM GTP. Each protein in the reactions was present at 2 μM. Bar, 50 nm. (B) Complementary pelleting assay. The protein samples used were FtsZTFus pelleted without SsgBTfus (left) and with SsgBTfus (right). In total, 3.8 ± 0.4 times more FtsZ protein was pelleted with ultracentrifugation in the presence of SsgBTfus as compared with the control without SsgBTfus. For details, see the Materials and Methods.
The polymerized FtsZ fraction was then pelleted by ultracentrifugation at 165,000g and analyzed via SDS-PAGE, followed by quantification of the relative amounts of FtsZ. The amount of polymerized FtsZ increased with a factor 3.8 ± 0.4 in the presence of SsgB, whereas a control experiment with BSA revealed no change in the amount of polymerized FtsZ (Fig. 8B). These data demonstrate that SsgB has a strong stimulatory effect on FtsZ polymerization and filament length.
Discussion
In this study, we demonstrate that FtsZ is recruited to the septum site by the novel divisome component SsgB during sporulation in Streptomyces. In turn, the temperospatial localization of SsgB is controlled by the paralogous SsgA, and the enhanced expression of SsgA alone suffices to activate the accumulation of SsgB, followed by Z-ring formation and active cell division. The evolution of a new way of Z-ring control again underlines that, in terms of cell division, streptomycetes are the odd ones out; in these organisms, cell division is dispensable for growth—with viable ftsQ- and ftsZ-null mutants (McCormick et al. 1994; McCormick and Losick 1996)—and multiple cell division occurs during sporulation, initiated by forming spectacular ladders of Z rings (Schwedock et al. 1997). A model for the control of cell division during sporulation of Streptomyces is presented (Fig. 9).
Figure 9.
Model for the control of Z-ring formation in Streptomyces. (Left) In young aerial hyphae (stage I), SsgA forms foci, while SsgB and FtsZ are still diffuse. Then, SsgA and SsgB colocalize briefly at one and typically an alternating side of the aerial hyphae (stage IIA). Prior to sporulation-specific cell division, FtsZ is often seen to form long spiral-like filaments through the hyphae (Grantcharova et al. 2005; Willemse and van Wezel 2009; our unpublished results), and we hypothesize that these attach at alternating sides to the now assembled SsgB foci (stage IIB). From this point onward, FtsZ and SsgB continue to colocalize, whereby SsgB strongly interacts with the membrane and FtsZ to tether FtsZ (stage IIC). Z rings are then formed at the sporulation stage, followed by chromosome condensation and segregation and formation of the sporulation septa (stage III). During spore maturation, SsgA localizes at either side of the septa, and finally marks the two sites where spores germinate (Noens et al. 2007).
Following the recruitment of FtsZ, SsgB remains associated with it at the divisome. Hence, in contrast to the principle that FtsZ is typically the first divisome component to arrive at division sites followed by the sequential recruitment of the cytokinetic machinery (Goehring and Beckwith 2005; Dajkovic and Lutkenhaus 2006; Gamba et al. 2009), in Streptomyces, SsgB arrives prior to FtsZ at the septum site. This was further demonstrated by time-lapse imaging, and was corroborated by the observation that FtsZ ladder formation depends on SsgA and SsgB, while SsgB localizes at septum sites independently of FtsZ. A few septa are formed in the absence of SsgA or SsgB, with a large spacing very similar to that found in vegetative hyphae, and, like in vegetative hyphae, these only lead to the formation of compartments, without cytokinesis. This underlines an important issue; namely, that the formation of cross-walls is controlled in a different way than sporulation-specific cell division.
The Streptomyces divisome is highly dynamic, with turnover of FtsZ filaments increasing sharply during cytokinesis. In B. subtilis, the half-life of FtsZ protofilaments at the septum is close to 10 sec, a turnover rate that is in line with that of mitotic tubulin (Anderson et al. 2004). The turnover rate of microtubules is ∼10 times higher in mitotic metaphase cells than in (nondividing) interphase cells (Salmon et al. 1984). FRAP imaging of Streptomyces cell division proteins showed dynamics similar to those of microtubules, with FtsZ and SsgB forming relatively static assemblies prior to active cell division, which then changes dramatically during active division, with FtsZ and SsgB half-life reduced from ∼1 min to ∼5 and 8 sec, respectively. Hence, the Streptomyces divisome is at least as dynamic as that of Bacillus. Recent computer modeling studies predicted that the Z ring should undergo a transition from a low-density to a high-density state, required in, e.g., E. coli to generate sufficient force to drive the contraction during cytokinesis (Lan et al. 2009). Indeed, where the number of SsgB proteins doubles from ∼350–700 molecules per septum during division, the amount of FtsZ increases from ∼600 molecules at Z-ring initiation (corresponding to the number of molecules in predivision foci) to a maximum of ∼6000 molecules in dividing cells. In contrast, the amount of the peptidoglycan-synthesizing FtsI remains more or less constant. To form a continuous ring inside hyphae with a diameter of ∼800 nm, at least twice as much SsgB is required, as observed in our experiments, strongly suggesting that not all FtsZ molecules in the Z ring interact directly with an SsgB protein. This is supported by the FLIM data, where we used either SsgB or FtsZ as the donor (and the other as acceptor). The significant increase in lifetime when FtsZ was the donor relative to when SsgB was the donor shows that, while all SsgBs were immediately adjacent to FtsZ, a significant proportion of the FtsZ proteins in the Z ring is not immediately adjacent to an SsgB protein. This is in accordance with the notion that the number of FtsZ molecules strongly increases to form a complete Z ring, while SsgB does not.
FRET-FLIM imaging further demonstrated that SsgB is closely attached to both the cytoplasmic membrane and FtsZ, while FtsZ interacts with the membrane but is positioned significantly farther from it than SsgB. Like SsgB, the N-terminal half of FtsZ is highly conserved in streptomycetes, and two-hybrid studies strongly suggest that this is precisely the domain that SsgB interacts with. These data support a model whereby FtsZ protofilaments are tethered to the cell membrane by SsgB trimers. Taken together with the observed stimulation of FtsZ filament length in vitro and the continued colocalization of the proteins throughout the whole process of cell division (live-imaging data), this suggests that SsgB remains associated with the FtsZ filaments, perhaps protecting the FtsZ filaments from dissociating.
Several proteins that stimulate filament formation with different modes of action have been described in the literature. For example, ZipA stimulates the formation of filament networks (RayChaudhuri 1999), while ZapA promotes bundling by forming lateral connections between FtsZ filaments (Gueiros-Filho and Losick 2002; Low et al. 2004). The activity of SsgB is more similar to that of ZipA than ZapA, with, on average, around fourfold increase of FtsZ filament length in comparison with control samples, and lack of extensive bundling. In eukaryotes, specific protein complexes act as nucleation complexes for filament formation, such as the ARP2/3 complex and formins for actin filaments and the microtubule organizing center (MTOC) microtubules (Luders and Stearns 2007; Chesarone and Goode 2009). Interestingly, evidence for a complex involved in filament capping and polymerization in bacteria has been provided for the actin-like ParM filament involved in plasmid segregation, which is stabilized by the small ParR protein in complex with a centromere-like parC DNA fragment (Salje and Lowe 2008). Whether SsgB functions in an analogous fashion on FtsZ protofilaments remains to be elucidated.
Where SsgB acts to recruit FtsZ and stimulate filament formation, the mode of action of SsgA is less clear. We demonstrated previously that SsgA marks the sites where the cell wall needs to be remodeled; namely, during apical growth, branching, germination, and cell division (Noens et al. 2007). High-level overexpression of SsgA induces submerged sporulation in vegetative hyphae of S. coelicolor (which normally forms only dense mycelial clumps) (van Wezel et al. 2000a) and affects hypergermination, with up to eight germ tubes protruding from a single spore (Noens et al. 2007). It is rather unlikely that SsgA acts in all of these cases via interaction with (and localization of) SsgB. One possibility is that other members of the family of SALPs interact with SsgA to control the localization of cell wall remodeling enzymes—such as PBPs (peptidoglycan-binding proteins)—during germination, branching, and apical growth, as the different ssg mutants all have specific defects in peptidoglycan synthesis or autolysis. Major candidates are, in particular, SsgD and SsgG. Null mutants of ssgD have major defects in lateral wall integrity, while ssgG mutants occasionally skip septa in the Z ladders, thus resulting in many large and multinucleoid spores (Noens et al. 2005). It will be interesting to see whether S. coelicolor mutants lacking multiple or even all seven SALPs may also lack cross-walls.
The decisive checkpoints in nature are typically mediated through negative control systems, including the mitotic checkpoints during eukaryotic cell division (Oliferenko et al. 2009) and mid-cell positioning of FtsZ in most bacteria (Marston et al. 1998). This is deemed necessary to avoid premature cell division, as incorrect or premature septum positioning is generally devastating, resulting in minicells and/or incorrectly segregated or damaged chromosomes. There is, perhaps, an obvious biological explanation for the fact that, in streptomycetes, positive control of cell division is possible, owing to the different way cell division is organized. If, occasionally, a septum is misplaced during sporulation-specific cell division, this is without too much consequence, as many viable spores will still be produced. Similarly, during vegetative growth, only occasional cross-walls are formed, with highly variable spacing and multinucleoid compartments as a result, and, again, a misplaced septum generally will not be problematic. The positive control of Z-ring localization is, perhaps, a necessary step in the evolution of a system to produce chains of spores. A Min-type control system is probably less feasible in Streptomyces, as the aerial hyphae are up to 100 times as long as E. coli cells and lack a mid-cell reference point or two relatively adjacent poles to oscillate between. A major issue to address is how the perfect spacing of so many Z rings is controlled during sporulation-specific cell division. The size of the nucleoid candidates is a natural ruler to set the minimum boundaries for the spore compartments during sporulation. The first step will be to determine what controls the localization of the cell division activator SsgA, and whether this is also positively controlled. With this, some very important questions lie ahead of us, and their solution should shed more light on the unique features of positive cell division control in filamentous bacteria.
Materials and methods
Bacterial strains and constructs
Bacterial strains and growth conditions
All strains and plasmids used in this study are presented in Supplemental Table S3, and the oligonucleotides are presented in Supplemental Table S4. S. coelicolor M145 was obtained from the John Innes Centre strain collection, and the S. coelicolor ftsZ mutant HU133 (McCormick et al. 1994) was kindly provided by Joe McCormick; the ssgA and ssgB mutants of S. coelicolor were published previously (van Wezel et al. 2000a; Keijser et al. 2003). FM145 is a spontaneous low-autofluorescent derivative of M145 (Willemse and van Wezel 2009). BACTH complementation assays were carried out with the nonreverting adenylate cyclase-deficient (cya) E. coli strain BTH101 (Karimova et al. 1998).
Fluorescent fusion constructs
Constructs were created expressing SsgA, SsgB, or FtsZ translationally fused to either eGFP or mCherry, with all fusion genes expressed from the natural promoters of the respective genes. The colonies were phenotypically and microscopically very similar to the parental strain S. coelicolor FM145, and functionality of the ssgA (Noens et al. 2007), ssgB (normal phenotype of the genomic integrant), and ftsZ fusion constructs was demonstrated by the full complementation of the respective mutants. Fusion genes expressing SsgB-eGFP or SsgB-mCherry were introduced into the genome by gene replacement, so as to replace the wild-type ssgB. For this, we cloned egfp directly behind the coding region of ssgB in the vector pIJ786, and gene replacement was carried out using the Redirect method developed by Gust et al. (2004) (for details, see Gust et al. 2004 and references therein). An mCherry variant of pIJ786 was constructed by replacing the gene for eGFP with that for mCherry; for this, we amplified the gene for mCherry with oligonucleotides pIJ786-mCherry-Fw and pIJ786-mCherry-Rev and inserted the fragment into pIJ786 as an NdeI–HindIII fragment so as to replace egfp, thus generating pGWS510. Subsequently, SsgB-786Fw and SsgB-786Rev were used to amplify the entire insert of either pIJ786 or pGWS510, generating DNA fragments encompassing a cassette of either egfp or mcherry followed by the apramycin resistance cassette aacC4, which is inserted between the last seven codons of ssgB and the first 20 nucleotides (nt) of the ssgB downstream region. Using high-efficiency recombination achieved in a λ red background (Gust et al. 2004 and references therein) derivatives of cosmid L2, which contains an ∼40-kb section of the S. coelicolor chromosome including ssgB, were obtained with either egfp or mcherry fused immediately behind ssgB. Using apramycin resistance for selection of the desired recombinants, the wild-type ssgB gene was replaced with the respective fusion genes according to routine methods. The correct recombinants were verified by PCR amplification and DNA sequencing using oligonucleotides SsgB-forward and eGFP-reverse. KF41 (Grantcharova et al. 2005), a kind gift from Klas Flärdh, was used to obtain construct pGWS523 expressing ftsZ-mCherry. For this, ftsZ-egfp was replaced with ftsZ-mcherry. To achieve this, mcherry was amplified with oligonucleotides pKF41-mCherry-Fw and pKF41-mCherry-Rev, digested with BamHI and NotI, and inserted into KF41 digested with the same enzymes.
Constructs for BACTH screening
For construction of the recombinant plasmids for BACTH assays, the genes of interest were PCR-amplified using Pfu polymerase (Stratagene) and oligonucleotide pairs as described in Supplemental Table S4. PCR products were cloned as XbaI/XmaI-digested (for ssgBTfus, and N-terminal-coding and C-terminal-coding parts of ftsZ) or XbaI/KpnI-digested (for ssgASc) DNA fragments in frame with either the T18 or the T25 fragment of the catalytic domain of B. pertussis adenylate cyclase (cyaA) into pUT18 and/or pKT25 vectors (BACTH System Kit, Euromedex), which were digested with the same enzymes. This resulted in pUT18 derivatives pBTH069 (ssgASc) and pBTH121 (for ssgBTfus), and pKT25 derivatives pBTH123 (ssgBTfus), pBTH147 (FtsZ [N-term; amino acids 1–195]), and pBTH151 (FtsZ [C-term]; amino acids 196–399) (Supplemental Table S3).
BACTH complementation assays
For BACTH complementation assays, recombinant pKT25 and pUT18 carrying the genes of interest were used in various combinations to cotransform E. coli BTH101 cells. The transformants were plated onto LB-X-Gal-IPTG medium with ampicillin at 100 μg/mL and kanamycin at 50 μg/mL, and were incubated for 24–36 h at 30°C. For each combination, 10 independent colonies were restreaked on M63 minimal medium supplemented with maltose as the sole carbon source, which allows growth only when the two proteins expressed by the transformants interact.
Microscopy
Fluorescence microscopy
Fluorescence and corresponding light micrographs were obtained with either an Olympus bh-2 or a Zeiss Axioscope A1 upright fluorescence microscope (with an Axiocam Mrc5 camera at a resolution of 37.5 nm/pixel), with, for the green channel, 470- to 490-nm excitation and 515 long-pass detection; and for the red channel, 530- to 550-nm excitation and 590 long-pass detection. The green fluorescent images were created using 470/40-nm bandpass excitation and 525/50 bandpass detection; for the red channel, 550/25-nm bandpass excitation and 605/70 bandpass detection were used. For staining of the cell wall (peptidoglycan), we used TRITC-WGA; for membrane staining, we used FM5-95 (both obtained from Molecular Probes). All images were background-corrected, setting the signal outside the hyphae to 0 to obtain a sufficiently dark background. These corrections were made using Adobe Photoshop CS4
Time-lapsed (live) imaging
Uncoated μ-dishes (Ibidi GmbH) were perforated at the side while closed tightly, and subsequently were semifilled with SFM medium. These dishes were inoculated, turning the lid so it was supported on the vents, allowing gas exchange, and were sealed of by two layers of Parafilm to prevent drying of the medium. Samples were incubated at 30°C and imaged with a Zeiss Observer A1 microscope with a Hamamatsu EM-CCD C9100-02 camera. Images were taken with 10-sec intervals for 50 min. In order to minimize focal drift, the microscope stage and imaging chamber were allowed to equilibrate for 60 min before imaging.
FRAP
FRAP measurements were performed on a Zeiss LSM 510 using a 40× plan neofluor, 1.3 NA lens with 15× digital zoom. Monitoring was done with a 25-mW Ar laser with 25% power output at 1%–2% intensity. Photobleaching was performed by scanning two to 10 times using 50%–75% laser intensity.
FRET-FLIM
FRET is the possibility of a fluorescent donor molecule transfering its excitation energy to an acceptor molecule. For FRET to occur, conditions must be such that the emission spectrum of the donor molecule (e.g., eGFP) overlaps the excitation spectrum of the acceptor (e.g., mCherry), and the distance between the proteins must be between 2 nm and 10 nm. All FLIM measurements were performed on a Bio-Rad Radiance mp 2100 system, with a Nikon TE 300 inverted microscope. Two-photon excitation pulses were generated by a Ti:Sapphire laser (Coherent Mira) pumped by a 5-W Coherent Verdi laser. Pulse trains of 76 MHz (150-fsec pulse duration, 860-nm center wavelength) were produced. The excitation light was coupled directly into the microscope and focused into the sample using a CFI Plan Apochromat 63 water immersion objective lens (numerical aperture, 1.2). For the FLIM measurements, the Hamamatsu R3809U MCP PMT was used. The frame size of the acquired images is 64 × 64 pixels. Cell membrane was stained using a 0.2 μg/mL solution of FM 5-95 (N-[3-trimethylammoniumpropyl]-4-[6-{4-(diethylamino) phenyl} hexatrienyl]pyridinium-dibromide; Molecular Probes) prepared in Hanks' Balanced Salt Solution (HBSS). All FRET-FLIM measurements were performed on aerial hyphae.
From the images, complete fluorescence lifetime decays were calculated per pixel, and were fitted using a double-exponential decay model as well as a single exponential decay selecting the best fit based on χ2 of the fitted curve. For double-exponential fitting, the fluorescence lifetime of one component was fixed to the value found for eGFP fused to the same gene (2.5–2.7 nsec). For each two-component fit, a combination of both fluorescence lifetimes revealed the same fluorescence lifetime as determined by a single-component fit. Based on several characteristics of donor and acceptor dyes, the R0 (the distance at which half of the energy is transferred from the donor to the acceptor) can be calculated (Stryer 1978). The efficiency of energy transfer between the donor and acceptor is a measure for the intermolecular average distance. For eGFP and mCherry, the R0 is 5.4 nm (Goedhart et al. 2007); a similar R0 is calculated for eGFP and FM 5-95.
Fluorescence quantification
By making use of virus-like particles with the coat protein-fused to eGFP (Charpilienne et al. 2001), internal quantification of protein expression can be achieved, since the particles contain exactly 120 eGFP fusions (Dundr et al. 2002). By comparing the relative intensity of the viral particles in the image to the intensity of the fluorescently tagged proteins of interest, the absolute number of proteins can be derived as (I × 120)/IVLP, where I is the intensity of the signal of interest, and IVLP is the intensity of the virus-like particles.
Protein isolation
C-terminally His6-tagged FtsZ and SsgB from T. fusca were purified from E. coli BL21(DE3) harboring the pET22b-based expression constructs pGWS806 and pGWS807, respectively. The procedure was generally as described previously (Mahr et al. 2000), except that cells were disrupted via osmotic shock and protein was purified from the supernatants following centrifugation at 37,000 rpm for 45 min. The protein was purified on a HisPur Cobalt Resin column (Pierce Biotechnology), as recommended by the supplier. Protein-containing fractions were combined and dialyzed against a buffer containing 50 mM HEPES/NaOH (pH 7.2) and 50 mM KCl.
In vitro polymerization of FtsZ and pelleting assay
For polymerization assays, 2 μM FtsZTfus with or without equimolar ratios of SsgBTfus or BSA (negative control) was prewarmed at 30°C. Subsequently, polymerization and staining were carried out as published (Mohammadi et al. 2009) in the presence of 5 mM MgCl2 and 0.2 mM GTP. After 10 min of incubation at 30°C, samples were applied to a microscope grid, stained with uranyl acetate, and examined with transmission electron microscopy. Samples were imaged with a Philips EM 410 transmission electron microscope. Filament lengths were determined using ImageJ.
The FtsZ filament pelleting assay was carried out on as described previously (Small et al. 2007) except that the initial volume was 100 μL. Samples contained FtsZTfus together with either SsgBTfus or (as negative control) BSA. Centrifugation was carried out in an Airfuge A-100 miniultracentrifuge (Beckman-Coulter, Inc.) at 165,000g for 20 min. Pellets were dissolved in 100 μL of sample buffer, heated for 10 min at 100°C, and analyzed on a 10% SDS-PAGE gel. The relative intensities of the Coomassie brilliant blue-stained protein bands were determined via ImageJ.
Acknowledgments
We are grateful to Richard Losick, Lotte Søgaard-Andersen, and Bjørn Traag for discussions; to Joe Mc Cormick for providing the ftsZ-null mutant HU133; to Klas Flärdh for providing KF41; and to Didier Poncet for providing the rotavirus particles. Roman Koning and Ronald Limpens are thanked for help with transmission electron microscopy. The work was supported by an ECHO from the Netherlands Society for Scientific Research (NWO), and a VICI grant from the Dutch Applied Research Council (STW).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.600211.
Supplemental material is available for this article.
References
- Adams DW, Errington J 2009. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 [DOI] [PubMed] [Google Scholar]
- Alexeeva S, Gadella TW Jr, Verheul J, Verhoeven GS, den Blaauwen T 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol 77: 384–398 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol 186: 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Charpilienne A, Nejmeddine M, Berois M, Parez N, Neumann E, Hewat E, Trugnan G, Cohen J 2001. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J Biol Chem 276: 29361–29367 [DOI] [PubMed] [Google Scholar]
- Chater KF 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): A checkpoint multiplex? Curr Opin Microbiol 4: 667–673 [DOI] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL 2009. Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol 21: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremazy FG, Manders EM, Bastiaens PI, Kramer G, Hager GL, van Munster EB, Verschure PJ, Gadella TJ Jr, van Driel R 2005. Imaging in situ protein-DNA interactions in the cell nucleus using FRET-FLIM. Exp Cell Res 309: 390–396 [DOI] [PubMed] [Google Scholar]
- Dajkovic A, Lutkenhaus J 2006. Z ring as executor of bacterial cell division. J Mol Microbiol Biotechnol 11: 140–151 [DOI] [PubMed] [Google Scholar]
- Dundr M, McNally JG, Cohen J, Misteli T 2002. Quantitation of GFP-fusion proteins in single living cells. J Struct Biol 140: 92–99 [DOI] [PubMed] [Google Scholar]
- Flardh K 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49: 1523–1536 [DOI] [PubMed] [Google Scholar]
- Flardh K, Buttner MJ 2009. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7: 36–49 [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol 191: 4186–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW Jr 2007. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS One 2: e1011 doi: 10.1371/journal.pone.0001011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J 2005. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr Biol 15: R514–R526 doi: 10.1016/j.cub.2005.06.038 [DOI] [PubMed] [Google Scholar]
- Grantcharova N, Lustig U, Flardh K 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J Bacteriol 187: 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF 2004. λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54: 107–128 [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PA 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88: 175–185 [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol 59: 989–999 [DOI] [PubMed] [Google Scholar]
- Hopwood DA 2007. Streptomyces in nature and medicine: The antibiotic makers. Oxford University Press, New York [Google Scholar]
- Jyothikumar V, Tilley EJ, Wali R, Herron PR 2008. Time-lapse microscopy of Streptomyces coelicolor growth and sporulation. Appl Environ Microbiol 74: 6774–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci 95: 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser BJ, Noens EE, Kraal B, Koerten HK, van Wezel GP 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol Lett 225: 59–67 [DOI] [PubMed] [Google Scholar]
- Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX 2009. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci 106: 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Moncrieffe MC, Lowe J 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol 341: 839–852 [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206 [DOI] [PubMed] [Google Scholar]
- Luders J, Stearns T 2007. Microtubule-organizing centres: A re-evaluation. Nat Rev Mol Cell Biol 8: 161–167 [DOI] [PubMed] [Google Scholar]
- Mahr K, van Wezel GP, Svensson C, Krengel U, Bibb MJ, Titgemeyer F 2000. Glucose kinase of Streptomyces coelicolor A3(2): Large-scale purification and biochemical analysis. Antonie van Leeuwenhoek 78: 253–261 [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12: 3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Losick R 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J Bacteriol 178: 5295–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Su EP, Driks A, Losick R 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol 14: 243–254 [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, Martos A, Alfonso C, van Marle J, Rivas G, den Blaauwen T 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48: 11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noens EE, Mersinias V, Traag BA, Smith CP, Koerten HK, van Wezel GP 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol Microbiol 58: 929–944 [DOI] [PubMed] [Google Scholar]
- Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol 64: 1244–1259 [DOI] [PubMed] [Google Scholar]
- Oliferenko S, Chew TG, Balasubramanian MK 2009. Positioning cytokinesis. Genes Dev 23: 660–674 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci 94: 559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA 1997. The MinE ring: An FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91: 685–694 [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J 18: 2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Levin PA 2003. Assembly dynamics of the bacterial cell division protein FTSZ: Poised at the edge of stability. Annu Rev Microbiol 57: 125–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Lowe J 2008. Bacterial actin: Architecture of the ParMRC plasmid DNA partitioning complex. EMBO J 27: 2230–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR 1984. Spindle microtubule dynamics in sea urchin embryos: Analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J Cell Biol 99: 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedock J, Mccormick JR, Angert ER, Nodwell JR, Losick R 1997. Assembly of the cell division protein Ftsz into ladder like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol 25: 847–858 [DOI] [PubMed] [Google Scholar]
- Small E, Marrington R, Rodger A, Scott DJ, Sloan K, Roper D, Dafforn TR, Addinall SG 2007. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J Mol Biol 369: 210–221 [DOI] [PubMed] [Google Scholar]
- Stryer L 1978. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem 47: 819–846 [DOI] [PubMed] [Google Scholar]
- Traag BA, van Wezel GP 2008. The SsgA-like proteins in actinomycetes: Small proteins up to a big task. Antonie van Leeuwenhoek 94: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag BA, Kelemen GH, Van Wezel GP 2004. Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2). Mol Microbiol 53: 985–1000 [DOI] [PubMed] [Google Scholar]
- van Wezel GP, van der Meulen J, Kawamoto S, Luiten RG, Koerten HK, Kraal B 2000a. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J Bacteriol 182: 5653–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel GP, van der Meulen J, Taal E, Koerten H, Kraal B 2000b. Effects of increased and deregulated expression of cell division genes on the morphology and on antibiotic production of streptomycetes. Antonie van Leeuwenhoek 78: 269–276 [DOI] [PubMed] [Google Scholar]
- van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol 72: 5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth H, Hopwood DA 1970. Septation during sporulation in Streptomyces coelicolor. J Gen Microbiol 60: 51–59 [DOI] [PubMed] [Google Scholar]
- Willemse J, van Wezel GP 2009. Imaging of Streptomyces coelicolor A3(2) with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS One 4: e4242 doi: 10.1371/journal.pone.0004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
- Xu Q, Traag BA, Willemse J, McMullan D, Miller MD, Elsliger MA, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, et al. 2009. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J Biol Chem 284: 25268–25279 [DOI] [PMC free article] [PubMed] [Google Scholar]