Summary
Here we investigate the mechanisms that underlie the induction of developmental potential and establishment of cell fate during early hematopoiesis. A cascade of lineage-affiliated gene expression signatures, primed in hematopoietic stem cells (HSC) and differentially propagated in lineage-restricted progenitors, is identified. First evidence is provided for a stochastic sampling of lymphoid, erythroid and myeloid transcripts in HSC and multipotent progenitors (MPP). Multi-lineage priming is subsequently resolved upon lineage restrictions. Nonetheless, an unexpected association of lymphoid and myeloid signatures is detected past a nominal myeloid restriction point and a previously unappreciated lymphoid potential is revealed for this stage in development. New insight is provided into Ikaros' role as a bivalent regulator of multi-lineage priming during early hematopoiesis. Whereas Ikaros is responsible for activation of a cascade of lymphoid signatures in the HSC, at subsequent restriction points it is also involved in the repression of lineage-inappropriate signatures including stem cell-specific genes.
Introduction
Hematopoiesis is viewed as a numerically expanding hierarchy of cell types with progressively restricted self-renewal and increasing potential for differentiation into a specific blood or immune cell type (Lemischka and Moore, 2003; Weissman, 2000). Lineage restrictions in hematopoiesis have been extensively investigated using both cellular and genetic approaches (Busslinger, 2004; Cantor and Orkin, 2002; Rosenbauer and Tenen, 2007; Rothenberg, 2007). These studies have defined major steps in the lymphoid, myeloid and erythroid pathways, identified key signaling molecules and transcription regulators, and generated models for lineage differentiation. Nonetheless, the mechanisms that induce and modulate multi-lineage potential at the earliest steps of this developmental pathway remain unknown. One issue confounding these efforts is that the early hematopoietic hierarchy is more complex than previously perceived.
The prospective isolation of HSC and lineage-restricted progenitors based on differential expression of cell surface markers, or with surrogate markers driven by hematopoietic-specific regulatory cassettes has identified rare cells with defined lineage activities (reviewed by (Iwasaki and Akashi, 2007). These have been used to infer past and current models of hematopoietic lineage restrictions. The HSC compartment was operationally defined within the Lin− Sca-1hic-Kithi (LSK) population in the bone marrow (Morrison and Weissman, 1994; Osawa et al., 1996; Spangrude et al., 1988). The use of additional markers, including CD34 and the tyrosine kinase receptor Flt3, has further subdivided the LSK compartment into long-term HSC, short-term HSC and MPP (Adolfsson et al., 2001; Christensen and Weissman, 2001). Recent studies have shown that a significant fraction (1/3–1/4) of the LSK consists of progenitors with strong lymphoid and myeloid potential, but with limited erythro-megakaryocyte potential. These progenitors, also referred to as lymphoid-primed multipotent progenitors (LMPP), were identified using independent approaches that subdivide the LSK population; i.e. by differential expression of Flt3 (Adolfsson et al., 2005), of an Ikaros-reporter (Yoshida et al., 2006) and of VCAM1 (Lai and Kondo, 2006). Importantly, these studies together with earlier reports on fetal hematopoiesis (Katsura, 2002; Kawamoto, 2006) have provided evidence for an obligate lympho-myeloid stage of differentiation as a key branch point that leads into the lymphoid and myeloid pathways. An early lymphoid progenitor (ELP), with strong lymphoid but reduced myeloid potential, a likely descendant of the LMPP, was also identified in low numbers within the LSK using a Rag1-GFP knock-in reporter (Igarashi et al., 2002; Medina et al., 2001).
Downstream of the LSK, within the Lin− Sca-1loc-KitloIL-7Rα+ population, a common lymphoid progenitor (CLP) with strong in vitro potential for B cell, T cell and NK cell differentiation was described (Kondo et al., 1997). Recent studies have shown that some CLPs are still active in myeloid differentiation (Mansson et al., 2008; Rumfelt et al., 2006). Lineage restricted megakaryo-erythrocyte progenitors (MEP; CD34−FcRlo) and granulo-monocyte progenitors (GMP; CD34+FcRhi) were identified within the Lin− Sca-1−c-Kithi (LK) population (Akashi et al., 2000). A rare progenitor was also reported here, the common myeloid progenitor (CMP; CD34+FcRlo), with combined erythroid and myeloid potential. However, the claim that this progenitor is a major contributor of myeloid differentiation, is currently disputed (Pronk et al., 2007) and current investigation).
Studies that address the activation and restriction of lineage-specific transcriptional programs are providing an alternative molecular view into the earliest stages of hematopoiesis. Multipotent progenitors were reported to express low levels of genes affiliated with disparate differentiation programs prior to lineage restriction, a process known as lineage priming (Enver and Greaves, 1998; Hu et al., 1997). The low level co-expression of genes from disparate lineages has been taken as evidence of multi-lineage priming through chromatin accessibility, a step that is considered to be key for the rapid induction of lineage-specific gene expression programs upon selection of the affiliated cell fate (Bernstein et al., 2006). Earlier reports on lineage priming indicated that myeloid- and erythroid-, but not lymphoid-specific transcripts were co-expressed in single HSC (Hu et al., 1997; Miyamoto et al., 2002). Lymphoid transcripts were only detected in lineage restricted progenitors such as the CLP (Miyamoto et al., 2002). More recent studies have shown that lymphoid transcriptional priming can occur in a fraction of the earlier progenitor population, the LMPP, in combination with myeloid lineage transcripts (Mansson et al., 2007).
Nuclear regulators expressed in early progenitors may control cell fate by modulating expression of lineage-specific genes either stochastically or in response to environmental cues (Chang et al., 2008). The Krüppel-type zinc finger DNA-binding factor Ikaros is expressed in the HSC and is essential for normal lymphocyte development, maturation and homeostasis (Georgopoulos et al., 1994; Nichogiannopoulou et al., 1999; Wang et al., 1996; Winandy et al., 1995). Mutations in Ikaros indicate that it is essential for development of the lymphoid lineage and that its effects are manifested prior to the emergence of lymphoid-restricted progenitors such as the CLP and the proB (Allman et al., 2003; Wang et al., 1996). More recent studies have shown that Ikaros is not required for the initial segregation of the lympho-myeloid restricted progenitor, the LMPP, from the HSC, but it is required for the LMPP’s subsequent progression into the lymphoid pathway (Yoshida et al., 2006). Ikaros and its family members are thought to regulate the expression of lineage-specific genes by guiding key epigenetic and transcriptional events and by thus contributing to a state of multi-lineage epigenetic competence in the HSC and its progeny (Georgopoulos, 2002; Kioussis and Georgopoulos, 2007). This hypothesis is borne out in part by biochemical studies that have shown a stable association of Ikaros and its family members with the Nucleosome Remodeling and Deacetylase complex (NuRD) (Kim et al., 1999; O'Neill et al., 2000; Sridharan and Smale, 2007) and in part by Ikaros’ association with the chromatin of lineage-specific genes (Harker et al., 2002; Naito et al., 2007; Williams et al., 2004). The cellular and molecular effects observed upon deficiency of the chromatin remodeler Mi-2β of the NuRD complex in the hematopoietic and lymphoid systems provide support for this hypothesis (Naito et al., 2007; Williams et al., 2004; Yoshida et al., 2008).
Seeking to establish the molecular mechanisms that underlie early hematopoiesis, we examined an HSC-enriched population and its early lineage-restricted progeny for expression of lineage-affiliated transcriptional programs, referred to as signatures. By comparing HSC, LMPP, GMP and MEP populations, an HSC signature that is strongly affiliated with self-renewal and three layers of lineage-affiliated signatures were deduced. Using transcripts deduced from this analysis we show by single cell analysis that in contrast to previous reports, extensive transcriptional priming for lymphoid genes is detected in the HSC together with stem cell- as well as myeloid- and erythroid-affiliated transcripts. Unexpectedly, lymphoid transcriptional priming is detected in the GMP, which also exhibits latent potential for lymphoid differentiation under both in vitro and in vivo conditions. Finally, we demonstrate that induction and maintenance of lymphoid lineage priming in the HSC compartment and in lympho-myeloid-restricted progenitors are dependent on Ikaros. Downstream of the HSC, Ikaros is also required for the active repression of genetic programs that are normally compatible with the multipotent HSC state.
Results
Identification of a cascade of lineage-affiliated signatures in early hematopoiesis
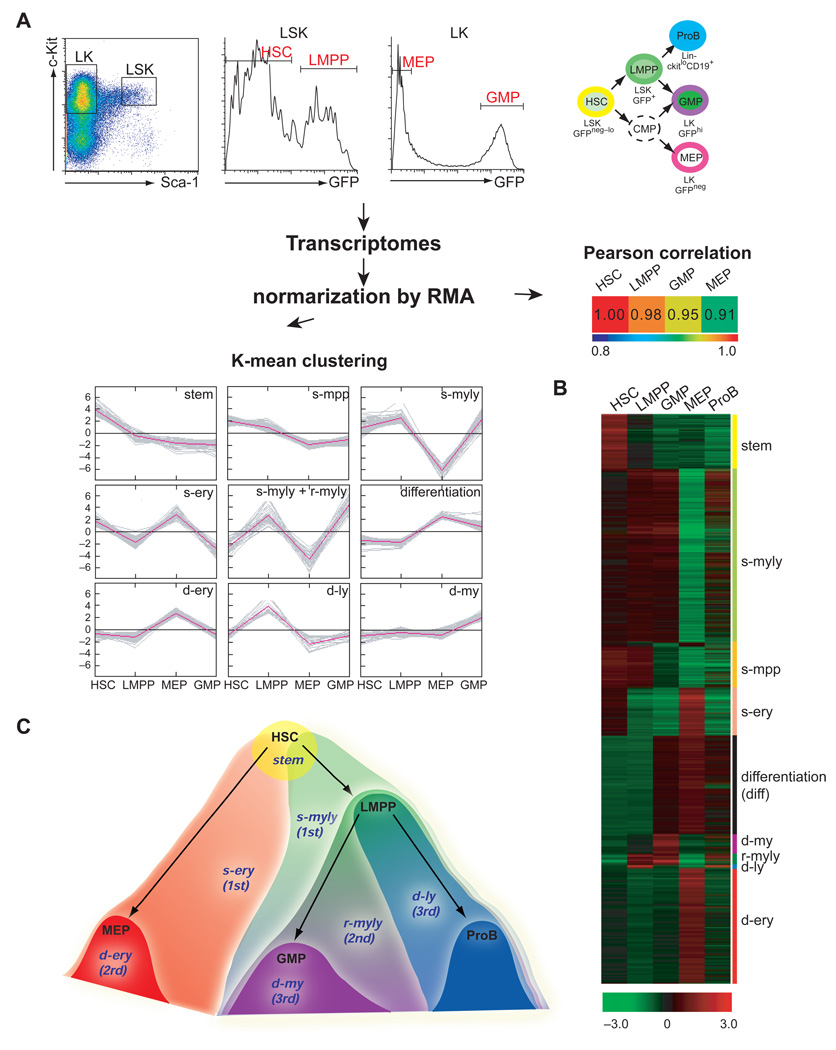
Given new insights into early hematopoietic progenitors and their unexpected lineage affiliations we examined these early steps of the hematopoietic hierarchy for expression of lineage-affiliated transcriptional programs. An Ikaros-based GFP reporter that provides a clean separation of the HSC-enriched population (LSK GFPneg–loFlt3neg-lo) from the LMPP (LSK GFP+Flt3lo-hi) and the GMP (LK GFPhi) from the MEP (LK GFPneg) was used for cell isolation and gene profiling (Figure 1A and Figure S1) (Yoshida et al., 2006). The population referred to as HSC (LSK GFPneg–lo) in our studies consists of ~80% LT-HSC+ ST-HSC defined by the LSK Flt3neg profile and of ~20% MPP defined by the LSK GFPneg–loFlt3lo profile (Figure S1B) (Christensen and Weissman, 2001; Yang et al., 2005). Transcriptomes deduced from three independent sets of these cell types were normalized and subjected to Pearson correlation analysis (Figure 1A). Pearson correlation has been widely used as a similarity measure between samples with similar expression patterns. A similarity order from HSC to LMPP to GMP with MEP being the most dissimilar was established. The normalized transcriptomes of HSC, LMPP, GMP and MEP were also subjected to K-means clustering that puts more weight on the pattern of gene expression changes across groups rather than on the magnitude of changes between individual populations. K-means clustering revealed 49 clusters of affiliated genes that fit into nine major signatures shown in Figure 1A–B and Table 1. The expression of these nine signatures was also examined in the more lymphoid-restricted proB (Lin− c-Kit+ CD19+) for further insight into their lineage affiliation (Figure 1B).
Figure 1. A cascade of lineage-specific transcriptional signatures primed in the HSC and propagated into appropriate lineage-restricted progeny.
(A) An Ikaros-GFP reporter that displays a bimodal distribution in the LSK and LK compartments (Yoshida et al., 2006), was used to isolate HSC-enriched (~80% LT-+ST-HSC and ~20% MPP; LSK GFPneg-lo as in Figure S1B), LMPP (LSK GFP+), MEP (LK GFPneg) and GMP (LK GFPhi) populations for global gene profiling. The developmental relationship between progenitors used for this study is indicated. Progenitor expression profiles were subjected to Pearson correlation coefficient analysis and K means clustering that deduced nine differentially expressed signatures (Table 1). (B) Heat map of signature expression in HSC, LMPP, MEP, GMP and ProB. Signature designation and lineage affiliation is provided on the right. (C) A graphic representation of signature distribution at the early steps of the hematopoietic hierarchy.
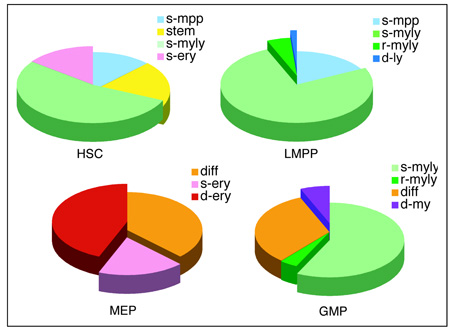
Table 1. Lineage-specific signatures deduced by K-mean clustering.
Lineage-affiliated signatures deduced by K-means clustering of progenitor. samples. Signature designation, definition, expression pattern and gene probe set sizes are provided. The relative distribution of each signature in HSC-enriched, LMPP, GMP and MEP populations is shown by a pie chart for each developmental stage. Lineage affiliation is color-coded.
| Signature | Definition | Expression | Gene probe # |
|---|---|---|---|
| stem | contains self-renewing genes | HSC (LT- + ST-) | 483 |
| s-mpp | no significant expression of lineage specific genes | HSC/MPP, LMPP | 373 |
| s-ery | erythroid lineage specific genes primed in HSC. 1st wave of erythroid lineage specific expression program | HSC/MPP, MEP | 315 |
| s-myly | lymphoid and myeloid lineage specific genes primed in HSC. 1st wave of lymphoid and myeloid lineage specific expression program | HSC/MPP, LMPP, GMP, ProB | 1340 |
| r-myly | lymphoid and myeloid progenitors-primed lineage specific genes. 2nd wave of lymphoid and myeloid lineage specific expression program | LMPP, GMP, ProB | 92 |
| diff | no expression with lineage specific genes. demarcating a progenitor-restricted state | GMP, MEP, ProB | 761 |
| d-ery | erythroid progenitor-specific. 2rd wave of erythroid lineage specific expression program | MEP | 888 |
| d-my | myeloid progenitor-specific. 3rd wave of myeloid lineage program | GMP | 151 |
| d-ly | lymphoid progenitor-specific. 3rd wave of lymphoid lineage program | LMPP, proB | 23 |
 | |||
The first set of signatures is restricted within the HSC and LMPP populations (Table 1, stem; 483 probes and s-mpp; 315 probes and Figure 1). The first signature (stem) is expressed in the HSC-enriched population but is down-regulated in the LMPP. The stem signature contains previously defined regulators of self-renewal and shows overlap with previously described LT-HSC-affiliated signatures (Ivanova et al., 2002; Ramalho-Santos et al., 2002). The second signature (s-mpp) is expressed in both the HSC-enriched population and in the LMPP. It lacks any lineage affiliation and is associated with the high proliferative potential of MPP. The s-mpp signature provides molecular evidence for the close relationship between LMPP and the HSC population and the relative primitiveness of the LMPP within the early progenitor hierarchy.
The second set of signatures is expressed in the HSC population and in some but not all of its lineage-restricted progeny, revealing the priming of lineage-specific genes potentially as early as the HSC. A major signature shared by the HSC, LMPP and GMP and down-regulated in the MEP is designated as stem-myelo-lymphoid (Table 1, s-myly; 1340 probes). This consists of both factors of myeloid differentiation such as Mpo, Csf3r, Lmo1, Gfi1, Cebpb and lymphoid differentiation such as Dntt, sterile Igh transcripts, Satb1, Sox4, Foxp1, Flt3 and Notch1. Notably, expression of both of the lineage-affiliated legs of the s-myly signature is maintained in the GMP and to a certain extent within the pro-B cell population in spite of nominal lineage restrictions. A stem-erythroid signature shared by the HSC and MEP but not by the LMPP, GMP or pro-B populations is also deduced here (Table 1, s-ery; 373 probes). The s-ery signature contains known erythroid lineage differentiation factors such as Gata1, Klf9, Eraf, Tgfbr3 and Gja1. Notably, there is no significant s-my signature (expressed by HSC and GMP and not by LMPP) or s-myery (expressed by HSC, GMP and MEP and not by LMPP) suggesting that within the HSC compartment myeloid gene expression is activated concomitantly with lymphoid gene expression. Both the lymphoid and myeloid gene expression programs are maintained but also augmented in the bi-potent lympho-myeloid progenitor (LMPP), a likely key step for subsequent differentiation decisions.
The next group of signatures contains the second and third layers of lineage-specific transcriptional priming that occurs downstream of the HSC compartment and underscore further lineage restrictions. A restricted myelo-lymphoid (Table 1, r-myly) signature represents a second layer of myelo-lymphoid lineage transcriptional priming that is specifically activated in the LMPP and GMP and consists of prominent lymphoid (Il7r, Irf8, Igh sterile transcripts) and myeloid (Csf1r, Ly6c, Ccr2) differentiation markers (Table 1, r-myly; 92 probes). The lymphoid but very few of the myeloid components of this signature are still expressed in proB cells. The third layer of lineage priming represents further restriction into either the erythroid or the myeloid or the lymphoid cell fate. The d-ery is numerically the largest progenitor-restricted signature, d-my the second, and d-ly the smallest (Table1, d-ery; 888 probes, d-my; 151 probes and d-ly; 21 probes). The relatively small size of the d-ly signature deduced from the LMPP is consistent with its limited lymphoid lineage-restricted nature. The LMPP although strongly primed for lymphoid differentiation in its majority retains bi-potentiality for both lymphoid and myeloid differentiation (Adolfsson et al., 2005; Lai and Kondo, 2006; Yoshida et al., 2006). Finally, a group of genes shared by the GMP, MEP and proB but not by the HSC and LMPP underscores the lineage-restricted state of hemo-lymphoid progenitors and is thus designated as a differentiation signature (Table 1, diff; 761).
By comparative bioinformatics analysis of progenitor-derived transcriptomes we have deduced a cascade of lineage-affiliated signatures that is activated within the HSC compartment and is propagated in a differential manner in lineage-restricted progenitors (Figure 1C). Importantly, early lineage transcriptional priming includes not only erythroid and myeloid but also lymphoid-affiliated transcripts. Lymphoid and myeloid gene expression programs appear to be activated concomitantly and to remain associated through several steps of lymphoid and myeloid differentiation. In contrast, restriction into the erythroid lineage appears to involve the rapid elimination of opposing genetic programs including both lymphoid and myeloid.
Priming of lymphoid, erythroid and myeloid gene expression in the HSC
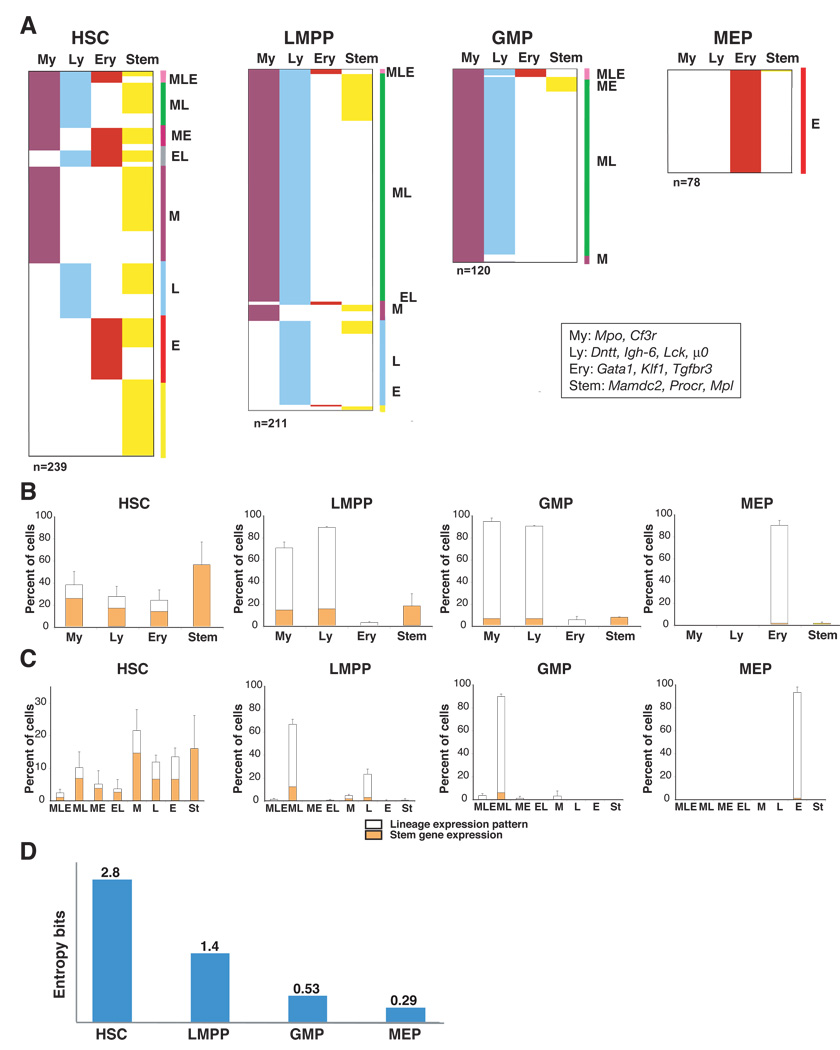
Whereas co-activation of myeloid- and erythroid-affiliated genes has been previously shown in HSC and MPP, activation of a lymphoid gene expression program is thought to occur much later in lymphoid-restricted or in lymphoid-primed progenitors (Mansson et al., 2007; Miyamoto et al., 2002). Nonetheless, the global cascade of lineage-affiliated signatures deduced from our studies (Figure 1) indicates that lymphoid transcriptional priming is active even earlier in multipotent progenitors and possibly in HSC. To further explore these findings obtained at the population level, we subjected single cells from the HSC-enriched population (LSK GFPneg–lo) to multiplex RT-PCR analysis for both HSC- and lineage-affiliated transcripts (Figure 2). Transcripts that belong to the first layer of lineage-affiliated signatures were chosen for this study. Gata1, Klf1 and Tgfbr3 were chosen from s-ery as representative of early erythroid transcriptional priming. The myeloid; Mpo, Csf3r and the lymphoid; Dntt, Igh6, Lck, µ0, components of the s-myly signature were chosen as representative of early myeloid and lymphoid priming respectively. Mamdc2, Procr, and Mpl deduced from the stem-signature were used to identify which cells expressed HSC-affiliated transcripts. These three transcripts were previously shown to be either involved or correlated with HSC’s long-term reconstituting potential (Balazs et al., 2006; Ivanova et al., 2002; Tong et al., 2007; Yoshihara et al., 2007). The expression of selected genes was first confirmed by real-time RT-PCR analysis in bulk progenitor populations (Figure S2). cDNAs generated from single cells (n=239) were used in multiplex RT-PCR reactions. An example of primary data obtained from multiplex RT-PCR of single progenitors as well as individual frequencies of transcript expression within each population are provided in Figure S3.
Figure 2. Multiplex single cell expression analysis of lineage-affiliated transcripts in HSC and progeny.
A. Single cells from HSC, LMPP, GMP and MEP populations were sorted into 96 well plates and subjected to reverse transcription followed by a two-step nested PCR for Actb and lineage-affiliated transcripts (My; Mpo, Csf3r, Ly; Dntt, Igh-6, Lck and µ0, Ery; Gata1, Klf1, Tgfbr3 and Stem; Mpl, Mamdc2 and Procr). Progenitors expressing at least one lineage-specific transcript are color-coded appropriately in each panel. Co-expressed patterns of lineage transcripts are identified on the right side of each panel. The total number of cells used in 2–4 experiments is indicated below each panel. Cells were provided from two or more independent sorts. The percentage of overall lineage-affiliated transcript distribution (B) and the percentage of co-expression of lineage- affiliated transcripts (C), are provided for cells in each progenitor population. The % of progenitors that express HSC-affiliated genes is shown for both the whole population and for subsets primed with lineage-specific genes. Mean +/− SD on percent distribution for progenitor experiments is shown. (D) The single progenitor expression data was analyzed by information theory (Shannon entropy) to provide an independent measure of the differentiation uncertainty (entropy value in bits) of each subset.
60% of the cells in the HSC-enriched population expressed genes affiliated with self-renewal and were thus classified as self-renewing HSC (Figure 2B and Figure S3, HSC). This number is actually lower than the number of cells (~80%) in this population with a ST-+LT-HSC surface phenotype (LSK GFP–ve/loFlt3−ve, Figure S1B) indicating that a smaller fraction of these cells is actually genetically wired for self-renewal. Lymphoid transcripts were detected in 29% of these HSC, erythroid transcripts in 24% and myeloid transcripts in 45% of this population (Figure 2B). Lineage transcripts were also detected in cells within the HSC-enriched population that did not express self-renewal-affiliated transcripts and were thus classified as MPP. Co-priming of lymphoid with myeloid (10%), lymphoid with erythroid (4%), lymphoid with erythroid and myeloid (3%) as well as myeloid with erythroid transcripts (5%) were detected in the HSC population. Within this population low levels of lineage co-priming were detected in both the self-renewing HSC and MPP subsets with no apparent bias (Figure 2C).
Thus, single cell analysis reveals that lymphoid transcriptional priming occurs in both HSC and MPP at a level that is comparable to that of other hematopoietic lineages. Importantly, co-priming of lymphoid, erythroid, and myeloid transcripts is detected at similar low frequencies in HSC and MPP, indicating that this process is stochastic in nature. The extensive co-expression of HSC- and lineage-affiliated genes in early hematopoietic progenitors, suggests that priming for lineage differentiation can occur concomitantly with a genetic program that supports self-renewal.
Differential resolution of multi-lineage priming in erythroid vs. myeloid progenitors
We next examined how multi-lineage priming detected in the HSC and MPP is resolved in its lineage-restricted progeny the MEP, LMPP and GMP. The MEP (n=78), unlike the HSC, MPP, LMPP and GMP populations expressed only erythroid transcripts. No lymphoid or myeloid transcripts were present in this progenitor population (Figure 2A–C). Additionally, HSC-affiliated transcripts were almost absent.
Downstream of the HSC and MPP, the LMPP with very little erythroid potential is considered to be the first major restriction point leading into the lymphoid and myeloid pathways. Consistent with this notion, single cell analysis of LMPP (n=211) revealed that, in their vast majority expressed early lymphoid (93%) and myeloid transcripts (73%) primed for expression in the HSC population (Figure 2A–C, HSC vs. LMPP). Co-expression of lymphoid and myeloid transcripts was detected in the majority of LMPP (67%) whereas a substantial number of lymphoid-only transcript-expressing cells (23%) were also detected (Figure 2A–C, LMPP). These numbers are comparable to the previously reported frequency of myelo-lymphoid or lymphoid-only progenitor activities within the LMPP (Adolfsson et al., 2005; Yoshida et al., 2006). In contrast to the increase in lymphoid and myeloid gene expression observed in the LMPP, expression of HSC- and erythroid–affiliated transcripts was diminished. Expression of erythroid transcripts was reduced from 24% in the HSC to 2.7% in LMPP (Figure 2A–C), consistent with the reduction in erythroid potential of the latter population and a previous report (Mansson et al., 2007). HSC-affiliated transcripts were also reduced from 60% in the HSC to 18% in the LMPP consistent with a further loss in self-renewal in the latter population.
We also analyzed transcript expression in the myeloid-restricted progenitor the GMP at the single cell level (n=120). GMP analysis demonstrated that 97% of these cells expressed myeloid transcripts compared to 73% of the LMPP (Figure 2A–C, GMP). Surprisingly, lymphoid lineage priming was also widespread in this population with 93% of the cells expressing some of the lymphoid transcripts detected in the LMPP (Figure 2A–C). Nonetheless, expression of transcripts, such as Dntt (LMPP: 64%, GMP: 20%) and Lck (LMPP: 32%, GMP: 3%) was greatly reduced whereas Igh6 (LMPP: 87%, GMP: 92%) and µ0 (LMPP: 18%, GMP: 28%) was increased (Figure S3, GMP). Thus, although specific components of an early lymphoid lineage program were down-regulated in the GMP others remained expressed at significant levels. As expected, the frequency of expression of myeloid transcripts; Mpo (LMPP: 62%, GMP: 98%) and Csf3r (LMPP: 42%, GMP 52%) was increased (Figure S3). As with the LMPP, HSC- (7%) and erythroid-affiliated (5%) transcripts were diminished in the GMP population (Figure 2A–C).
To obtain an independent measure of progenitor multi-potency, the single cell type-specific transcript data was also analyzed by Shannon information theory (see methods). Based on transcript expression in single cells, this method calculates the differentiation uncertainty for each progenitor population in entropy bits. The HSC population displayed the highest uncertainty at 2.9 entropy bits, LMPP was next with 1.4 bits followed by GMP at 0.58 bits. Finally, MEP exhibited the least differentiation uncertainty at 0.29 entropy bits. Thus the derivation of lineage-affiliated signatures in HSC and early progeny combined with lineage transcript analysis at the single progenitor level has provided us with new unexpected insights into lineage priming and a measure of developmental plasticity.
Latent lymphoid potential in the GMP
The unexpected expression of lymphoid-affiliated genes in the GMP prompted us to further investigate its nature and potential for differentiation. First, to understand how differences in progenitor isolation protocols may contribute to differences in progenitor composition, we compared our GMP isolation protocol to that previously reported (Akashi et al., 2000) (Figure S4A). Our protocol, which excludes Mac-1+ cells, consistently yielded a lower number of GMP (~2–4 fold) and, likely, a more primitive myeloid progenitor population. The presence of IL-7Rα+ cells within the GMP was found to be insufficiently low to explain its high frequency of lymphoid expression (Figure S4B).
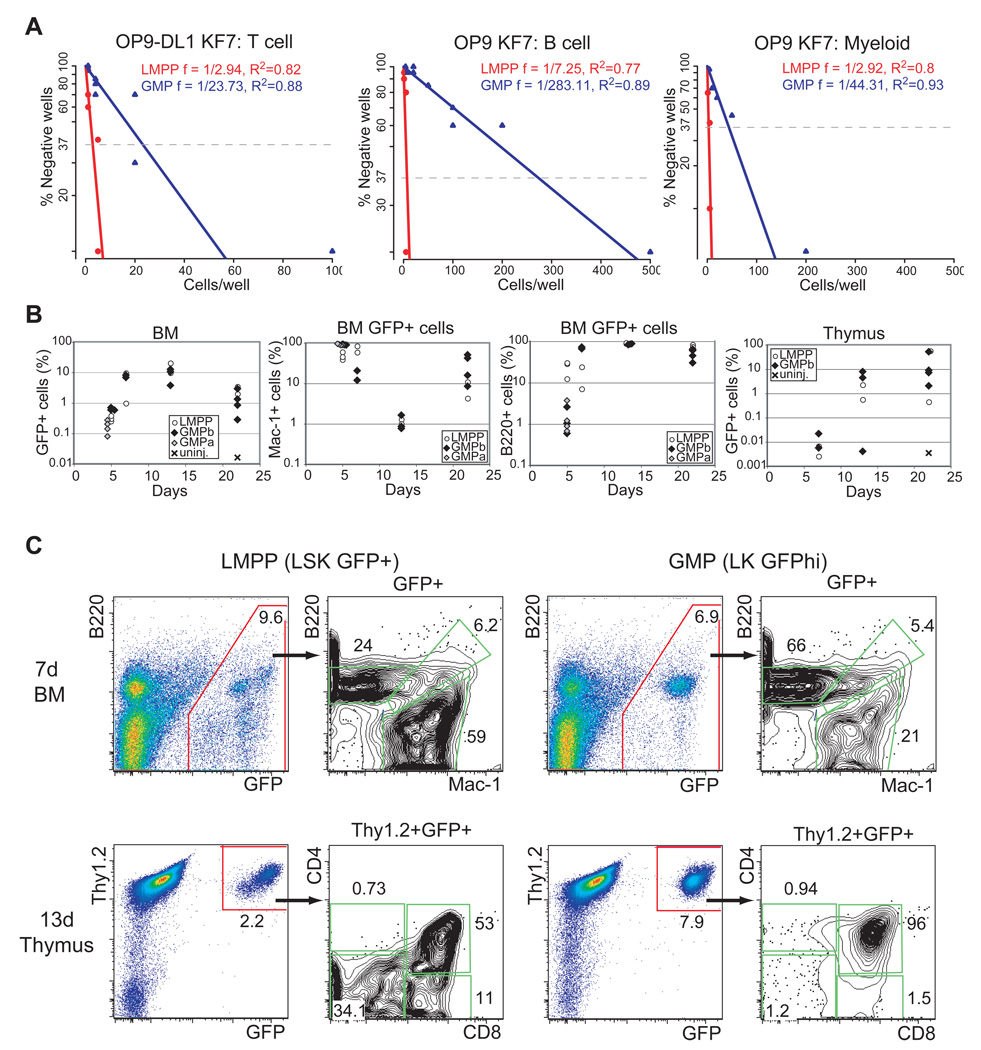
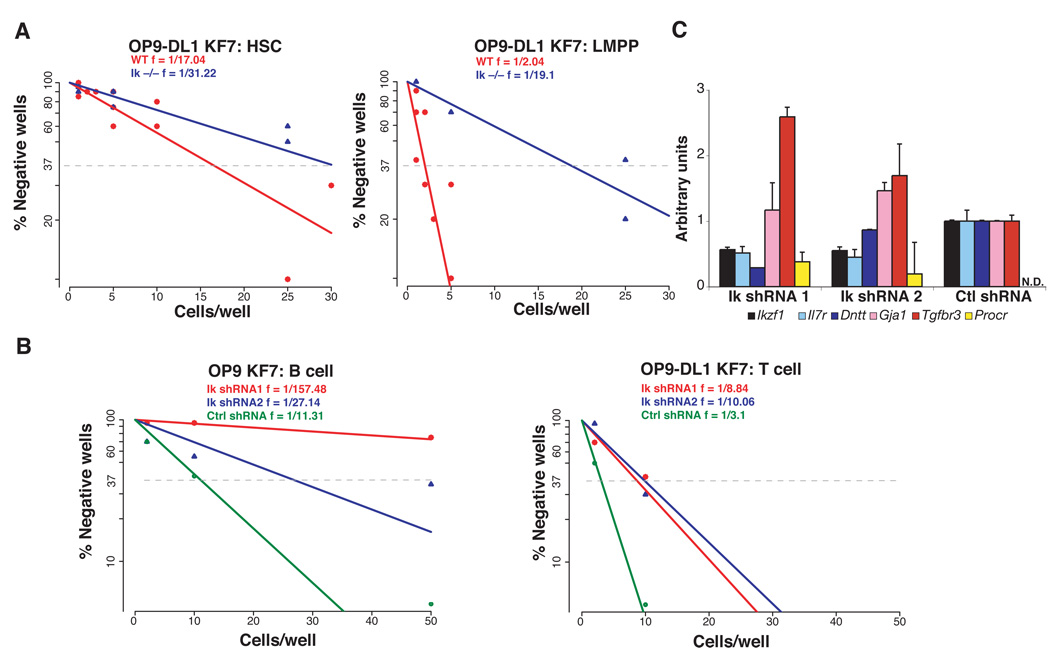
Next we evaluated the GMP’s potential for lymphoid differentiation first under in vitro conditions. Limiting dilution analysis of GMP and LMPP was performed on OP9 stroma under conditions that allow the generation of both B cells (B220+CD19+) and myeloid cells (Mac1+Gr1+) and on OP9-DL1 stroma under conditions that promote T cell differentiation (Figure 3A and Table S1). Whereas LMPP exhibited similar potential for B, myeloid and T cell differentiation (~frequencies of 1:7, 1:3 and 1:3), GMP was distinguished by unexpected differences in lineage potential (~frequencies of B 1:283, M 1:44, T 1:24). The apparent reduction in GMP’s frequency for myeloid differentiation compared to LMPP (1: 44 vs. 1:3) is likely due to a reduction in clonability and plating efficiency. The greater reduction in the GMP’s frequency for B cell differentiation (1:283 vs. 1:7) indicates an additional loss in B cell potential (Figure 3A). Notably, the reduction in GMP’s T cell frequency (1:24 vs 1:3) was by far smaller and similar in range to the reduction in myeloid frequency. The differentiation potential of single GMP were also investigated under T cell differentiation conditions (Figure S5A). Whereas all GMP capable of clonal expansion on OP9-DL1 gave rise to T cells a fraction of these gave rise to both T cells (DN3; Thy1+, CD44−CD25+) and myeloid (Mac1+ Gr1+) cells.
Figure 3. Latent lymphoid potential in the GMP.
(A) Limiting dilution analysis of GMP and LMPP for T cell, B cell and myeloid differentiation potential. Cells were sorted at the indicated doses and co-cultured with OP9-DL1 for 14–18 days and with OP9 for 8–11 days under conditions that promote T cell, B cell and myeloid differentiation before FACS analysis for lineage markers. Frequencies of T cell, B cell differentiation were calculated using Pöisson statistics. Analysis of combined data from four independent experiments is shown. R2 values are provided for each progenitor analysis. (B) 2000 LMPP (white circle), 7,500 GMP (a-grey diamond) or 30,000 GMP (b-black diamond,) were intravenously injected into sublethally irradiated recipient mice. Total and lineage- specific (Mac-1+, B220+, Thy1.2+) donor contribution (GFP+) was measured at days 5, 7, 13 and 22 post-injection. For every time point 2–4 mice per group were analyzed. (C) Representative LMPP and GMP donor contributions in the myeloid and B cell lineage (Mac-1+ vs. B220+) in the bone marrow at day 7 post-transplantation. LMPP and GMP contributions (GFP+Thy1.2+) to CD4 vs. CD8 profiles in the thymus is also shown at day 13 post-transplantation.
The GMP’s potential for differentiation was also evaluated under in vivo differentiation conditions. The GMP differentiation output was compared to that of the LMPP after direct placement into a thymic microenvironment (Figure S5B). Six days after intra-thymic injection of GMP (1000 LK GFPhi) or LMPP (25 LSK GFP+) into sub-lethally irradiated recipients (GFP−) donor-derived myeloid cells (Mac1+GFP+) were detected in thymuses populated by either progenitor population. At 21 days, donor-derived (GFP+) double positive (CD4+CD8+) thymocytes developed from GMP or LMPP.
The ability of GMP to migrate and differentiate into the bone marrow and thymus was also tested relative to the LMPP (Figure 3B). LMPP (2000 LSK GFP+) and GMP (a-7,500 or b-30,000 LK GFPhi) were injected intravenously into sub-lethally irradiated recipients and total donor contribution as well as contributions into the myeloid, B cell and T cell lineages were measured from 5 to 22 days after transplantation (Figure 3B). Total donor contribution from either progenitor peaked at 2 weeks in the bone marrow and at 3 weeks in the thymus. Donor-derived myeloid differentiation peaked during the 1st week whereas B cell differentiation during the 2nd week (Figure 3B, Mac1+vs. B220+). The kinetics of myeloid, B cell and T cell development were faster in GMP-derived cells compared to LMPP-derived cells consistent with the GMP’s more advanced stage in development. Whereas both the high and lower dose of GMP gave donor-derived myeloid and B cell contributions in the bone marrow only the high GMP dose contributed consistently to T cells in the thymus (although some contribution was detected by the low dose of GMP-data not shown).
Taken together these studies demonstrate that the GMP not only displays significant expression for lymphoid genes but it also possesses significant potential for T cell differentiation. Differences in the GMP’s potential for T cell differentiation revealed by in vivo vs. in vitro assays highlight this progenitor’s normal homing to the bone marrow vs. an intrinsic capacity for T cell differentiation when provided with appropriate signals.
Lymphoid lineage transcriptional priming is dependent on Ikaros
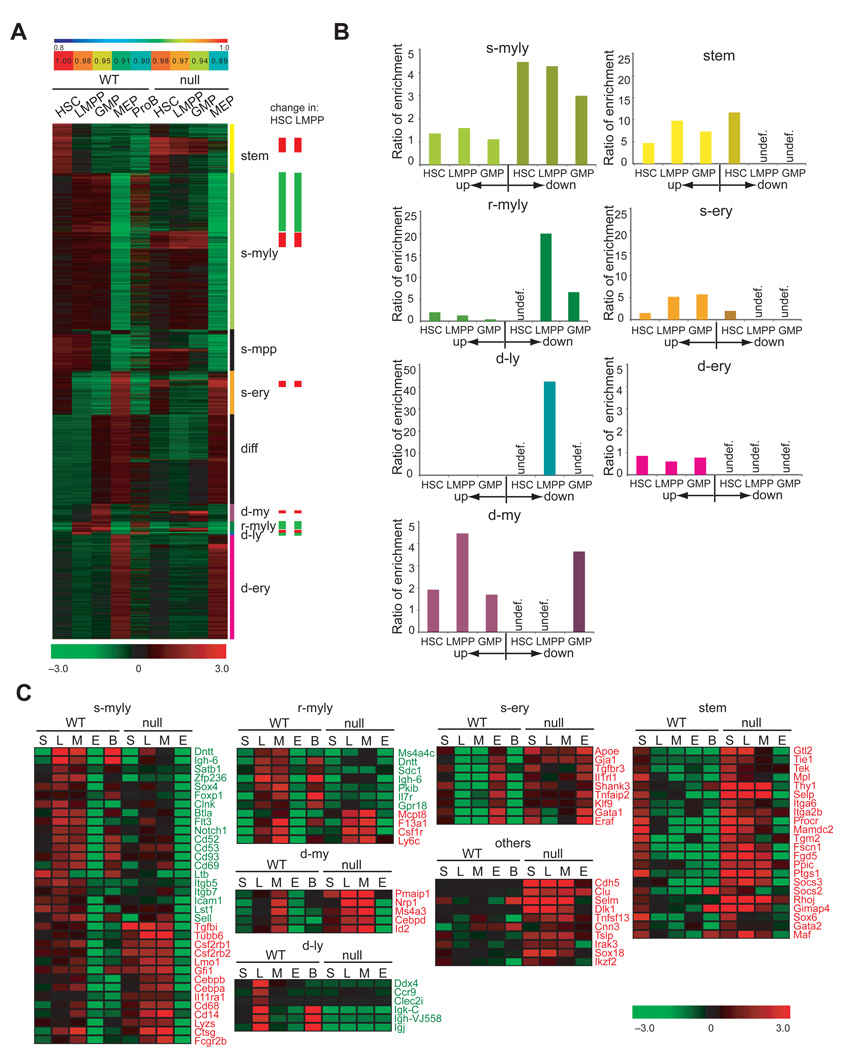
We have previously shown that LMPP’s differentiation into the lymphoid branch of the lympho-myeloid pathway is uniquely dependent on the zinc finger DNA binding factor Ikaros (Yoshida et al., 2006). Given new insight into lymphoid lineage transcriptional priming starting in the HSC and the global network of lineage-affiliated genes involved (Figure 1D), we investigated the role of Ikaros in this process. HSC-enriched and LMPP populations were isolated from Ikaros-null mice using the Ikaros GFP reporter (Yoshida et al., 2006), and subjected to a comparative global gene expression analysis with their wild type counterparts. Pearson analysis of mutant and wild-type progenitor subsets revealed a strong correlation supporting similar cellular composition and lineage relations (Figure 4A).
Figure 4. Ikaros effects on lineage-specific signatures during early hematopoiesis.
(A) Graphical representation of Pearson correlation coefficient analysis and heat map of signature expression in Ikaros-null vs. wild type progenitors. Signature designation and Ikaros effects (down-green, up-red) are indicated on the right. (B) Ratio of enrichment of gene signature sets with respect to Ikaros-differentially regulated gene lists in mutant HSC, LMPP and GMP as calculated by ratio-of-ratios method. Down-regulation of signature sets is not shown for progenitors where they are normally not expressed (undef). (C) Effects of Ikaros deletion on select members of the stem, s-myly, s-ery, r-myly, d-my and d-ly signatures. Genes exhibiting down-regulation (green) and up-regulation (red) are shown. A heat map of the average expression level (base2 log transformed mean centered; −3.0 to + 3.0) deduced from three independent samples is shown.
In the Ikaros-null HSC population, a similar number of up- (276) and down-regulated (280) gene probes were detected (Figure S6A). As mutant HSC became restricted to the LMPP, a 2-fold increase in the number of de-regulated gene probes (632 up and 463 down) was seen (Figure S6B), correlating with the previously reported increase in endogenous Ikzf1 expression during this developmental transition (Yoshida et al., 2006). We next examined how these changes in gene expression were distributed within the lineage-affiliated signatures deduced from HSC and progeny (Figure 4B–C). The majority of down-regulated genes in the Ikaros-null HSC and LMPP were distributed within the three layers of the myelo-lymphoid signatures primed progressively from the HSC to the LMPP and GMP (s-myly, r-myly, d-ly, Figure 4A–B and Figure S6). The earliest primed s-myly signature was enriched by 4.5 fold among the down-regulated genes of the mutant HSC and LMPP and by 3 fold in the mutant GMP. The r-myly signature representing the second layer of myelo-lymphoid gene priming in the LMPP and GMP, exhibited a 20 fold enrichment among the down-regulated genes in the mutant LMPP and a 6.7 fold enrichment in the mutant GMP. Notably, most of the components of the late d-ly signature expressed only in LMPP and proB were deregulated in the mutant LMPP exhibiting a 42.4 fold enrichment. Changes in gene expression in the mutant HSC and LMPP were also subjected to an unbiased hierarchical clustering across all WT and mutant progenitors providing us with an independent evaluation of their lineage affiliation (Figure S6A–B).
The lymphoid leg of the myelo-lymphoid signatures lineage factors was prominently down-regulated within the mutant progenitors (Figure 4A). Among the first layer of lymphoid-affiliated genes down-regulated in the absence of Ikaros were Flt3, Notch1, Satb1, Btla, Dntt, Igh-6 and Ltb (Figure 4C, s-myly). These include growth factor receptors required for lymphocyte differentiation and a growth factor important for the development and maintenance of secondary lymphoid organs which are absent in Ikaros-null mice (Alvarez et al., 2000; Gilfillan et al., 1993; Komori et al., 1993; Kuprash et al., 1999; Nakayama et al., 2005; Radtke et al., 2004; Sitnicka et al., 2003; Sitnicka et al., 2002; Tumanov et al., 2002; Watanabe et al., 2003). The growth factor receptors Il7r (r-myly), and Ccr9 (d-ly), components of the second and third layers of lymphoid lineage priming, were also dependent on Ikaros for expression (Figure 4C). The decrease in lymphoid-affiliated gene expression was also manifested in the GMP that normally maintains priming for some of these factors (Figure 4C).
Thus, Ikaros is required for the induction and propagation of a cascade of lymphoid-lineage gene expression events from the HSC to its downstream lympho-myeloid restricted progeny, the LMPP and GMP. Several of the down-regulated factors are known effectors of lymphoid development. Others are novel and may provide us with new insight into the regulation of this process.
Ikaros-dependent repression of multi-lineage transcriptional priming during lympho-myeloid restrictions
Among the genes up-regulated in early progenitors upon Ikaros inactivation, a significant number was affiliated with inappropriate cell fates (Figure 4 and Figure S6). Most strikingly, loss of Ikaros resulted in the up-regulation of numerous HSC-affiliated genes in the LMPP and the GMP. Among the genes that were up-regulated in the LMPP and GMP a respective ~10 fold and 7.5 fold enrichment was observed in the HSC-affiliated signature (Figure 4A–B, stem). HSC-affiliated transcripts with increased expression included those of Procr, Mamdc2, Fgd5, Fcn1, Socs2 and Socs3 and the receptor tyrosine kinases Tie1, Tek and Mpl implicated in self-renewal (Figure 4C, stem). A ~5 to 6 fold enrichment of early-primed erythroid factors (s-ery), including Gja1. Tgfbr3, Il1rl1, Apoe, Gata1, Klf9, was detected, which are not normally expressed in the LMPP or GMP. Finally, an enrichment (2–4.5 fold) of late myeloid genes, such as Csf1r, Cebpd and Id2, normally enriched in the GMP was detected in the mutant HSC and LMPP indicating their premature induction (Figure 4A–C, r-myly and d-my).
Thus, Ikaros in addition to promoting the priming and establishment of lymphoid gene expression in the HSC and its early lympho-myeloid progeny, it is also involved in extinguishing the expression of stem cell and erythroid genes and in preventing the premature induction of late myeloid genes.
Single cell progenitor analysis of Ikaros-mediated changes in lineage priming
Loss of Ikaros in the early hematopoietic hierarchy deregulates the activation as well as the restriction of lineage-specific transcriptional programs. These apparent defects in lineage-specific gene expression may in part reflect changes in the cellular makeup of mutant progenitors. Nevertheless, Pearson correlation analysis indicated that the mutant HSC and mutant LMPP populations were close to their wild counterparts (Figure 4A). Ikaros-null HSC and LMPP were further examined by single cell multiplex RT-PCR for expression of lineage affiliated transcripts (Figure 5). This line of study provided independent support that the cellular composition of these populations was not significantly altered although priming of lymphoid transcripts was reduced.
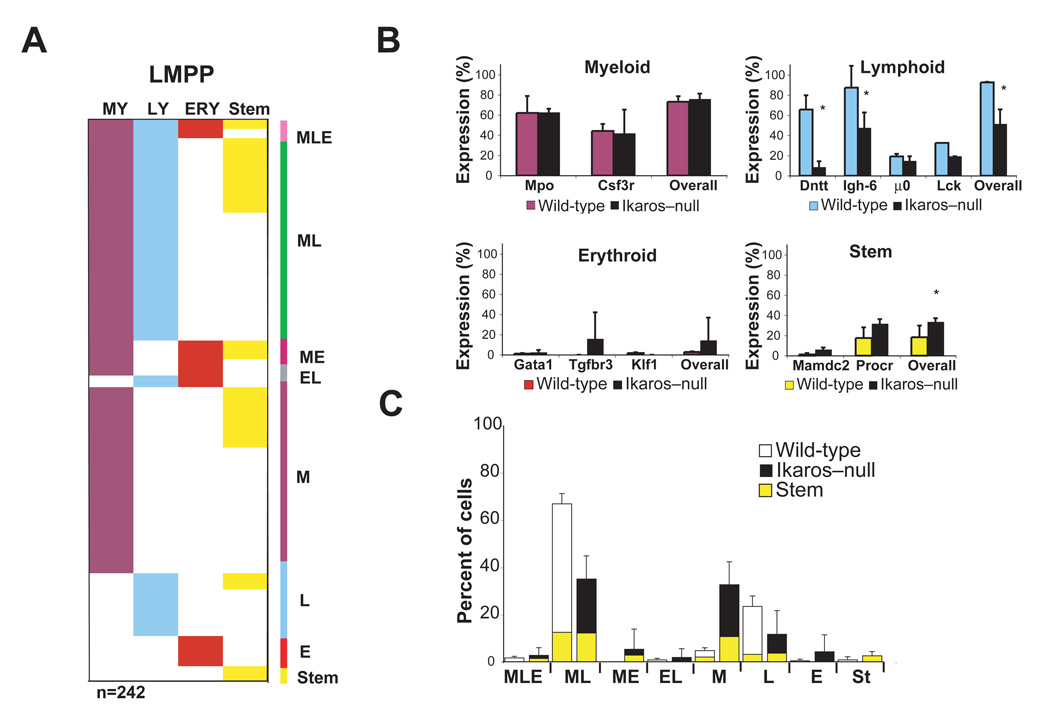
Figure 5. Multiplex single-cell gene expression analysis of Ikaros–null LMPP.
(A) Single progenitor analysis for lineage-affiliated transcripts was performed as described in Figure 2. Mutant LMPP (n=242) generated from two independent sorts are shown. (B) The percentage of overall lineage transcript distribution as well as of individual lineage-affiliated transcripts is provided for WT and mutant populations. *p<0.05. (C) A comparison of lineage co-expression patterns between WT and KO LMPP is provided. HSC-affiliated gene expression manifested within each co-expression pattern is shown. Mean +/− SD on percent distribution for the experiments performed per progenitor population is indicated.
Of all lineage-affiliated transcripts, the frequency of lymphoid transcripts detected was the most reduced in the Ikaros-null HSC population (overall: WT: 28%, Ik–null: 15%, p<0.1). Of the lymphoid transcripts analyzed, Dntt was the most severely affected (Figure S7). Smaller reductions in the frequency of myeloid (overall: WT: 39%, Ik–null: 24%, p=0.22) and erythroid (overall WT: 25%, Ik–null: 22%, p=0.71) transcripts were observed (Figure S7). Although some differences were noted in lineage transcript distribution and co-expression, their overall pattern was not dissimilar to wild type.
The transition from an HSC to a lympho-myeloid restricted LMPP is accompanied by an augmentation in the frequency of myeloid and lymphoid transcripts. An increase in the frequency of lymphoid transcripts was observed in the mutant LMPP compared to the mutant HSC (Overall; Ik–null LMPP 56% vs. Ik–null HSC 15%, Figure 5 and Figure S7). However, this frequency was significantly lower compared to that observed in the wild-type LMPP (Figure 5A–B, Overall; Ik–null LMPP: 52% vs. WT LMPP: 93% p=0.01). A prominent decrease in the frequency of certain lymphoid transcripts was again observed in mutant LMPP as in mutant HSC. A decrease in the frequency of detection of Dntt (Wild-type: 66%, Ik–null 8%) followed by Igh-6 (Wild-type: 87%, Ik–null 48%), Lck (Wild-type: 32%, Ik–null 19%), and µ0 (Wild-type: 19%, Ik–null 15%) was evident (Figure 5B). The frequency of myeloid transcript expression in the mutant LMPP (Overall; Ik–null: 76%, WT: 73% p=0.55) was unchanged compared to wild-type (Figure 5A–B), however, a significant decrease in cells co-expressing myeloid and lymphoid transcripts (Ik–null: 35%, WT: 67% p<0.05) was detected (Figure 5C). An increase in the frequency of HSC- (Overall; Ik–null: 33%, WT: 18%) and erythroid-affiliated (Overall; Ik–null: 14%, WT: 2.7%) transcripts was observed in the mutant LMPP compared to its wild type counterpart (Figure 5B). Notably, Shannon entropy analysis of the Ikaros-null HSC and LMPP single cell RT-PCR data provided similar entropy values for the two populations (2.6 and 2.5 bits respectively), suggesting that the mutant LMPP retains a differentiation-uncertain HSC-like phenotype.
These studies establish a bivalent role for Ikaros in the transcription of lineage-affiliated genetic programs downstream of the HSC. Ikaros is on one hand suppressing multi-potency-affiliated genetic programs while on the other it is activating lymphoid-promoting genetic programs.
Priming of the HSC’s lymphoid lineage potential is dependent on Ikaros
Whereas B cell differentiation is not detected in Ikaros null mice, T cell differentiation takes place albeit at a reduced frequency (~10 fold) compared to WT (Wang et al., 1996; Winandy et al., 1999; Yoshida et al., 2006). The T cell differentiation potential of HSC and LMPP was compared in vitro under limiting dilution conditions in the presence and absence of Ikaros (Figure 6A and Table S2). Under these conditions, an increase in T cell differentiation is normally detected from the HSC to the LMPP (Figure 6A, WT HSC 1:17, vs. WT LMPP 1:2) possibly reflecting an increase in Notch1 expression (Figure 4C). A comparison of WT to Ikaros-null HSC revealed an ~2 fold reduction in T cell activity in the mutant population (Figure 6A, 1:17 vs. 1:31). A greater reduction (~10 fold) in T cell potential was observed when comparing Ikaros-null to WT LMPP (Figure 6A, 1:19 vs. 1:2). This is in line with their failure to induce Notch1 expression (Figure 4C) and the previously reported 10 fold reduction in thymic progenitors observed in Ikaros-null mice (Wang et al., 1996; Winandy et al., 1999; Yoshida et al., 2006)
Figure 6. Ikaros is required for priming, augmentation and maintenance of lymphoid potential.
(A) T cell differentiation potential of HSC and LMPP from WT and Ikaros-null mice as revealed by limiting dilution analysis. Cells were sorted at the indicated doses and co-cultured with OP9-DL1 for 14–21 days under lymphoid conditions before analyzed for expression of lineage-specific differentiation markers. Frequencies of T cell differentiation were calculated using linear regression analysis of data from five combined experiments (Table S2). R2 values are provided for each progenitor analysis. (B) The differentiation potential of LMPP transduced with lentivirus-expressing shRNAs against Ikzf1 (1 and 2) or control shRNA was determined. Transduced LMPP were cocultured with OP9 to test B and myeloid frequency and with OP9-DL1 to test T cell frequency. Frequencies of T and B cell differentiation were calculated using Pöisson statistics. Data from one of two representative experiments is shown (Table S3). (C) Real-time RT-PCR of lymphoid (Ikzf1, Dntt, IL7R), erythroid (Gja1, Tgfbr3) and stem (Procr) transcripts amplified from at least 5000 lentivirally-transduced LMPPs harvested 48 hours after lentiviral transduction. The data is representative of two independent experiments.
Thus, the priming and establishment of lymphoid lineage potential in the early hematopoietic hierarchy directly correlates with the activation of a cascade of lymphoid gene expression events. The combination of these cellular and molecular events is dependent on Ikaros.
Maintenance of lymphoid potential is dependent on Ikaros
The role of Ikaros in actively maintaining lymphoid potential in the LMPP was further investigated by knock-down studies. WT LMPP were transduced with lentiviruses that produce Ikaros-specific short hairpin RNAs (shRNAs) and a GFP reporter for 48 hours. GFP-expressing cells were sorted onto B cell and T cell differentiation cultures and assayed under limiting dilution conditions (Figure 6B and Table S3). Sorted cells were also analyzed for gene expression (Figure 6C). Real-time RT-PCR analysis of sorted cells indicated that Ikaros expression was decreased by 44–45% in LMPP transduced with IkshRNA (1 and 2) compared to cells transduced with control shRNA. In addition to the reduction in Ikaros expression, a change in expression of previously described Ikaros gene targets in the LMPP, such as Dntt, Procr and Tgfbr3 was observed.
After 8–10 days under B cell promoting culture conditions, the progeny of transduced LMPP were analyzed for differentiation into lymphoid (B220+CD19+) and myeloid (Mac1+Gr1+) cells. B cell production was reduced in Ikaros shRNA transduced LMPP (~ frequencies; Ik shRNA1 1:157, Ik shRNA2 1:27) compared to LMPP transduced with control shRNA (1:11). Under these culture conditions, Ikaros shRNA-transduced LMPP produced myeloid cells at higher frequencies (Ik shRNA1; 1:2.5, Ik shRNA2; 1:4) compared to LMPP transduced with control shRNA (1:68). A reduction in T cell potential (Ik shRNA1; 1:9 and Ik shRNA2; 1:10 vs. ctl shRNA 1:3) was also observed in Ikaros shRNA-transduced LMPP grown under T cell differentiation conditions (Figure 6B).
Thus, Ikaros is not only required for establishment but also for maintenance of lymphoid lineage potential downstream of the HSC.
Discussion
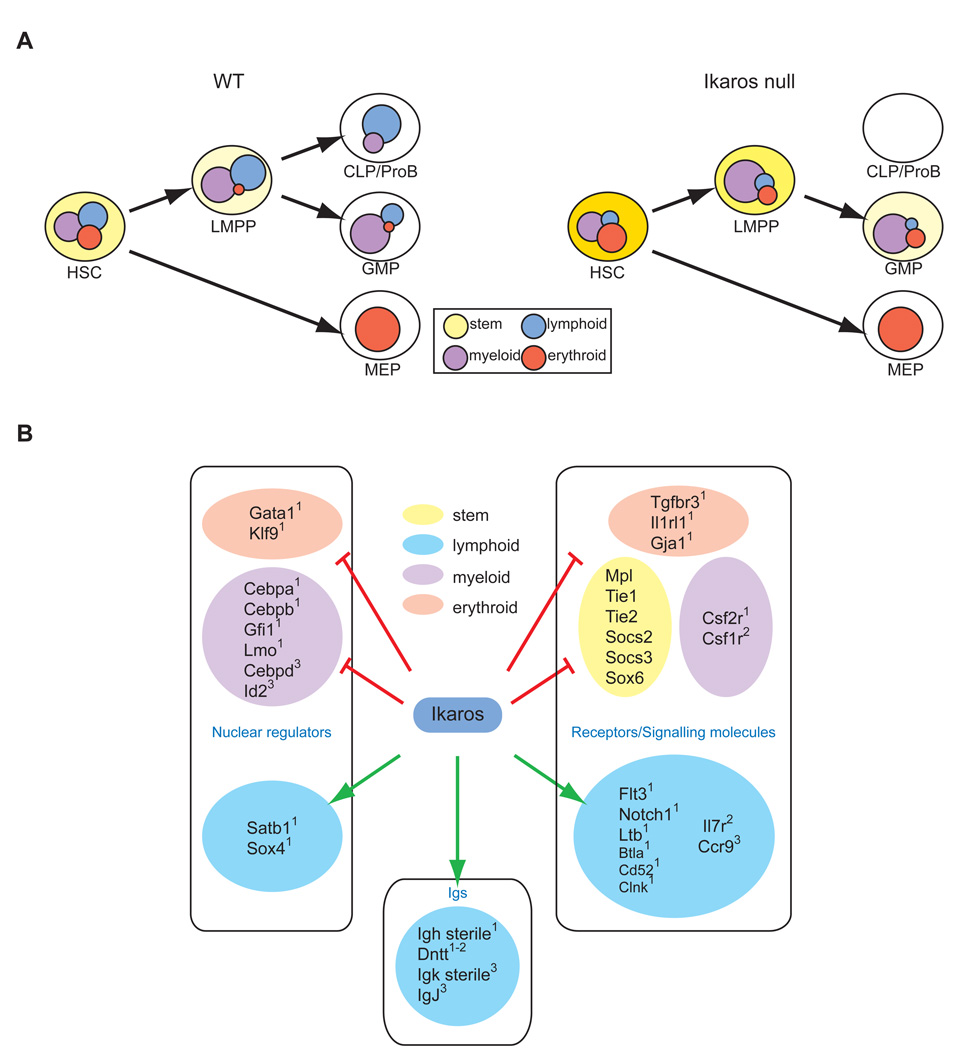
Here we provide new insights into the molecular events that modulate lineage potential in the HSC and its early progeny. An early genetic network that underscores cell fate decisions at the earliest steps of hematopoiesis is defined providing us with new important revisions in lineage transcriptional priming and its regulation by Ikaros. These studies provide us with a major step towards delineating the epigenetic regulation of stem cell biology and lineage plasticity.
A comparative analysis of global transcription profiles, deduced from HSC and progeny, established a cascade of lineage-specific gene expression programs that underlie respective progression into the erythroid or myeloid and lymphoid pathways. Lineage-affiliated transcripts deduced from this cascade and examined for expression in single HSC revealed priming of myeloid, erythroid and lymphoid transcripts at a similar robust frequency (~1/3). Co-expression of lymphoid, myeloid and erythroid transcripts, in different combinations, was detected at lower frequencies supporting a stochastic co-priming of opposing genetic programs in the HSC and MPP compartment. Subsequent lineage restrictions were demarcated by augmentation of HSC-primed, lineage-appropriate genetic programs and by the rapid extinction of opposing genetic programs. For example upon erythroid lineage restriction, a concomitant augmentation in the expression of erythroid transcripts primed in the HSC, and extinction of transcripts affiliated with the lymphoid, myeloid, and stem cell fates was observed. Conversely, upon HSC restriction into an LMPP, a concomitant establishment of lymphoid and myeloid transcriptional programs and extinction of erythroid and stem cell programs was detected. Unexpectedly, a significant expression of lymphoid genes was maintained in the LMPP’s myeloid-restricted progeny, the GMP.
Recent models have suggested that lymphoid lineage development is initiated downstream of the HSC and after establishment of a myeloid genetic program (Laiosa et al., 2006; Rothenberg and Pant, 2004). This assertion was partly based on the late evolutionary ontogeny of lymphocytes and on recent evidence that lymphoid lineage priming is first detected in a fraction (30%) of the LMPP that displays robust myeloid gene expression (Akashi et al., 2003; Mansson et al., 2007). If myeloid gene expression positively reinforces myeloid differentiation, then this developmental outcome should prevail most of the time. However, the balanced lympho-myeloid differentiation potential reported for the LMPP does not support this hypothesis (Mansson et al., 2007; Yoshida et al., 2006). Studies that interrogated lymphoid priming in the HSC and the LMPP did so with genes such as Il7r and Rag1 (Mansson et al., 2007; Miyamoto et al., 2002). Although these genes are readily expressed in committed lymphoid progenitors such as the CLP, they are not part of the earliest layer of lymphoid transcription primed in the HSC (i.e. s-myly). Instead they are representative of later layers of lymphoid transcription (i.e. r-myly and d-ly) described here. Thus, in contrast to previous reports, our studies identify an early and extensive lymphoid genetic program that is activated in the HSC, and reveal equal access to the erythroid, lymphoid, and myeloid pathways at the earliest point of hematopoiesis.
Multi-lineage priming detected in the HSC is resolved at subsequent lineage restriction points. However, a continued association of lymphoid and myeloid genetic programs and differentiation potential was apparent not only in the LMPP but also unexpectedly, in its nominal myeloid-restricted progeny, the GMP. The lack of erythroid potential and prominent myeloid differentiation properties of this progenitor population were previously described (Yoshida et al., 2006). Unexpectedly, our current transcriptional analysis has demonstrated a widespread expression of lymphoid genes throughout this population. The implication that the myeloid-committed GMP retains a latent lymphoid lineage potential under both in vitro and in vivo differentiation conditions was confirmed empirically here. In vivo transplantation studies, although not quantitative, have demonstrated that the GMP has latent potential for lymphoid differentiation. The GMP or its progeny can migrate into the thymus and undergo T cell differentiation at a low frequency. In sharp contrast, in vitro, the GMP displays a robust potential for T cell but not for B cell differentiation. Differences in the GMP’s potential for T cell differentiation revealed under in vitro vs. in vivo settings highlight the progenitor’s normal bone marrow homing properties and an intrinsic capacity for T cell differentiation when presented with appropriate signals. In this regard, it is noteworthy that the Notch1 receptor, normally primed in the HSC and up-regulated in the LMPP, is still expressed in the GMP and may promote the observed T cell differentiation on OP9-DL1 stroma. Taken together our GMP studies and recent reports on the ETP predict a similarity in the lineage restriction processes along the myeloid and T cell pathways (Bell and Bhandoola, 2008; Benz and Bleul, 2005; Rumfelt et al., 2006; Wada et al., 2008). Both appear to involve a rapid loss in B cell potential and a gradual loss in T cell or myeloid potential respectively.
The lymphoid potential of an HSC is augmented during restriction to an LMPP and this gain is dependent on Ikaros. In line with this biological effect, Ikaros is responsible for the activation and propagation of a cascade of lymphoid lineage-promoting genetic programs from the HSC to the LMPP (Figure 7A). Loss of Ikaros uniquely reports both known regulators of early lymphopoiesis and genes that are potentially novel regulators of this process (Figure 7B). The nuclear factors Sox4, Satb1, FoxP1 previously implicated in B cell and T cell development (Alvarez et al., 2000; Hu et al., 2006; Schilham et al., 1996), are in the first line of regulators downstream of Ikaros. These may work to augment expression of lymphoid genes as well as to repress competing genetic programs. Signaling receptors such as Flt3, IL-7Rα and Notch1, expressed in the HSC and LMPP and required for lymphocyte development (Radtke et al., 1999; Sitnicka et al., 2003; Sitnicka et al., 2002) are also dependent on Ikaros for normal expression. Increased expression of the signaling adaptors Socs2 and Socs3, involved in the negative regulation of STAT signaling (Hennighausen and Robinson, 2008; O'Sullivan et al., 2007), may provide additional interference to residual Flt3 or IL-7R signaling manifested in mutant progenitors. Signaling molecules such as Btla, Clnk, Pkib, CD52, shown to be important for functional responses of mature lymphocytes (Greenwald et al., 2005; Kumar and Walsh, 2002; Watanabe et al., 2003; Wu and Koretzky, 2004), are also expressed in the LMPP and their dependence on Ikaros suggests that these may also contribute to early lymphoid development. CCR9 expression in the LMPP supports progenitor migration into the thymus (Benz and Bleul, 2005; Uehara et al., 2002) and its loss in the mutant progenitors may explain the reduced number of thymic progenitors reported in Ikaros-null mice (Winandy et al., 1999). Finally, lack of lymph node structures in Ikaros-null mice (Wang et al., 1996) correlates with the loss of Ltb expression from mutant progenitors. Ltb expression in hematopoietic progenitors is required for lymph node structure development (reviewed by (Cupedo and Mebius, 2005).
Figure 7. Regulation of multi-lineage transcriptional priming during early hematopoiesis.
(A) Priming of myeloid (purple), erythroid (red) and lymphoid (blue) transcriptional programs occur at a similar frequency (~1/3) in the HSC. A similar low level of overlap between disparate genetic programs supports their stochastic co-priming at the multipotent state. Subsequent lineage restrictions are demarcated by an increase in lineage-appropriate transcription and decrease in lineage-inappropriate counterparts. Ikaros is a key regulator of this process. In the HSC compartment, Ikaros promotes priming and establishment of lymphoid genetic programs while in lympho-myeloid restricted progenitors it represses expression of lineage-inappropriate transcripts. Many of the genes repressed by Ikaros in the LMPP and GMP underscore a multipotent differentiation state. (B) Ikaros, as a bivalent regulator of a genetic network that controls lineage output in the HSC compartment. Select Ikaros-dependent components of this network that influence the myeloid (purple), lymphoid (blue), erythroid (pink) and stem cell (yellow) fate are shown. Superscripted numbers indicate placement of these factors in the cascade of lineage-specific and stem cell signatures described in Figure 1 and Table 1.
Loss of nuclear factors and signaling pathways that promote lymphocyte differentiation from the LMPP is expected to unbalance the lympho-myeloid genetic network operating in this progenitor that controls its lymphoid vs. myeloid output. A premature augmentation in the expression of myeloid factors, such as Csf1r, Csf2r, C/EBPα, β, δ, Id2, normally elevated upon LMPP’s restriction into a GMP may result from such a network imbalance. Thus Ikaros is a key coordinator in a lympho-myeloid genetic network that balances development of the innate and adaptive immune systems at the earliest steps of hematopoiesis. Loss of Ikaros does not deregulate expression of nuclear factors that have been previously reported to control lymphocyte development at its earliest stages, such as PU.1 and E2A (reviewed by (Nutt and Kee, 2007). E2A has been recently shown to also regulate lymphoid lineage priming in the LMPP in a manner that is likely parallel to Ikaros (Dias et al., 2008; Kondo, 2008).
Ikaros also regulates a series of genetic events that contribute to antigen receptor rearrangement and progression through the later stages of the lymphoid pathway. As shown here, sterile transcripts from the Igh locus and the end-nucleotide addition enzyme, Dntt, are in the first wave of lymphoid lineage transcriptional priming activated in the HSC, propagated in the LMPP, and dependent on Ikaros for expression. Priming of sterile transcripts from the Igk locus and IgJ occurs downstream of the HSC in a fraction of the LMPP and is also dependent on Ikaros. Although expression of these genes does not influence lymphoid lineage potential their deregulation suggest a role for Ikaros at subsequent stages of lymphoid development that are dependent on antigen receptor signaling.
Notably, of the genes that are negatively regulated by Ikaros, a significant fraction consists of HSC-affiliated genes (Figure 7B). Several of these have been implicated in self-renewal. The failure to extinguish stem cell transcripts such as Tie1, Tie2 and Mpl (Arai et al., 2004; Moore and Lemischka, 2004; Puri and Bernstein, 2003; Qian et al., 2007; Yoshihara et al., 2007), in Ikaros deficient LMPP and GMP may result in the abnormal acquisition of stem cell features, most intriguingly self-renewal that may contribute to a pre-leukemic status and drug resistance that may eventually contribute to the development of a highly malignant state as observed in human B cell precursor acute lymphoblastic leukemias (Georgopoulos, 2009; Mullighan et al., 2009). An increase in early erythroid lineage genes was also observed (Gata1, Klf9, Gja1), however, this did not appear to have an overt effect on the mutant LMPP’s differentiation towards the erythroid pathway (Yoshida et al., 2006). The pre-established expression of myeloid factors in the mutant progenitor may readily overcome this gene expression effect. Future studies on the genetic and epigenetic networks in operation at the earliest stages of hematopoiesis will provide us with new means of manipulating self-renewal and the choice of cell fate during normal and aberrant manifestations of hematopoiesis with important implications to both basic and clinical research.
Experimental procedures
Mice
Transgenic mice (B-p-GFP-C line, C57BL/6 × C3H)(Kaufmann et al., 2003) and Ikaros-null mice (I74 line, C57BL/6 × 129SV)(Wang et al., 1996) were bred and maintained under specific pathogen free condition in the animal facility at Massachusetts General Hospital, Bldg. 149. Mice were 4 to 12 weeks of age at the time of analysis. All animal experiments were done according to protocols approved by the Subcommittee on Research and Animal Care at Massachusetts General Hospital (Charlestown, MA) and in accordance with the guidelines set forth by the National Institutes of Health.
Intrathymic injections and analysis
Congenic recipient mice (CD45.1) were sub-lethally irradiated 6 hours prior to injection. 1×103 of LK GFPhi (GMP) or 25 of LSK GFP+ (LMPP) suspended in 10 µl of PBS were injected into the thymi of each host. Six or 21 days post injection, thymi were harvested and analyzed by flow cytometry with Mac-1 PE-Cy7, CD25 APC-Cy7, CD44-APC, CD4 APC-Cy5.5, CD8a PE or CD25 APC-Cy7, CD44-APC, Thy1.2-PE and Mac-1 PE-Cy7, B220-APC-Cy7, Gr-1 APC, CD19 PerCP-Cy5.5, and Thy1.2-PE along with the donor GFP marker.
Intravenous injections and analysis
Recipient mice (C57BL/6) were sub-lethally irradiated 6 hours prior to injection. 2000 of LSK GFP+ (LMPP), 7,500 of LK GFPhi (GMPa) or 30,000 of LK GFPhi (GMPb) cells sorted from the Ikaros-GFP reporter transgenic line (B-p-GFP-C) were injected into recipient mice retro-orbitally along with 2×105 BM competitor cells of host origin. Mice were maintained on acidified water for the duration of the study. Donor contribution to various hematopoietic lineages was measured in the bone marrow and thymus from 5 to 22 days post injection using GFP as a donor marker and by cell surface staining for myeloid (Mac-1, Gr-1), B cell (B220, CD19) and T cell lineage markers (Thy1.2, CD44, CD25, CD4, CD8a).
Antibodies
Antibodies were purchased from BD PharMingen, Invitrogen-Caltag or eBioscience. In some cases, hybridoma supernatant containing antibodies against B220, CD19, Mac-1, Gr-1, TER119, and CD3ε were used. Antibodies and the specific clones used were: CD3 (17A2), CD4 (L3T4), CD5 (53-7.3), CD8α (53-6.7), CD8β (H35-17.2), TCRβ (Η57−597), TCRγδ (GL3), Flt3 (A2F10.1), c-Kit (2B8, ACK2), IL-7Rα (A7R34), Sca-1 (D7 or E13-161.7), Mac-1 (M1/70), CD25 (PC61), CD19 (1D3), CD44 (IM7), Thy1.2 (53-2.1), B220 (RA3-6B2), DX5, Gr-1 (RB6-8C5), CD34 (RAM34), FcγRII/III (2.4G2), Ter119, NK1.1 (PK136) and 7/4.
Flow cytometry and cell sorting
Bone marrow (BM) cells were isolated and immuno-labeled as previously described (Yoshida et al., 2006). Briefly, BM cells were harvested from femurs and tibias and subjected to red blood cell (RBC) lysis using ACK buffer (0.15 M Ammonium Choloride, 10 mM Potassium Bicarbonate, 0.1 mM EDTA). Lineage positive cells were subsequently labeled with antibodies against the lineage markers TER119, B220, Mac-1, Gr-1, 7/4, CD3, CD5, CD8α , CD8β, CD19, TCRβ, TCRγδ, and DX5 and were removed with magnetic beads conjugated to goat anti-rat IgG (Qiagen). The remaining cells were labeled with R-phycoerythrin-Cy5.5 conjugated (PE-Cy5.5)- anti-rat-IgG to label any remaining lineage positive cells or biotin conjugated-anti-rat-IgG. Cells were then labeled with allophycocyanin conjugated (APC)-c-Kit and R-phycoerythrin conjugated (PE)-Sca-1 or PE-Cy7 conjugated Sca-1 and streptavidin conjugated APC-Cy7 prior to FACS analysis and cell sorting. For proB cell isolation, wild-type bone marrow cells were depleted with antibodies against Ter119, Mac-1, Gr-1, IgM, CD3, CD8a, TCRβ, TCRγδ, DX5 and the remaining cells with a c-Kitlo CD19+ phenotype were sorted.
Flow cytometric analysis was performed using a two-laser FACSCanto™ (BD), a two-laser FACSCalibur™ (BD) or a three-laser MoFlo® (Dako Cytomation). Cell sorting was performed using a three-laser MoFlo®. The resulting files were uploaded to FlowJo (Tree Star) for further analysis.
Microarray analysis
LSK GFPneg-lo (HSC), LSK GFP+ (LMPP), LK GFPneg (MEP) and LK GFPhi (GMP) populations were isolated from wt or Ikaros-null B-p-GFP-C mice (Yoshida et al., 2006). ProB were isolated as described in the previous section. Total RNA from at least 2×104 sorted cells were prepared using TriZol reagent (Invitrogen) followed by purification using MEGAclear (Ambion). Biotinylated aRNA probes were synthesized by two round of amplification using the MessageAmp™II aRNA Amplification kit (Ambion). The probes were hybridized with Affymetrix Mouse Genome 430_2.0 array chips. Hybridization was done in triplicates using three independently sorted samples from each population. Affymetrix .DAT files were converted to .CEL files and the quality check of hybridization, normalization of the data and comparison tests were performed as previously described (Yoshida et al., 2008).
Standard Pearson correlation coefficients were determined for each sample versus all other samples based on their logarithm (base 2) (Rodgers, 1988). K-means clustering was performed using R software suite, resulting in 49 clusters. Clusters were categorized into twelve groups based on patterns of expression to derive nine molecular signatures associated with each stem and progenitor population (Kanungo et al., 2002).
Data from microarray analysis will be made available through appropriate public databases in compliance with Immunity guidelines.
Ratio of enrichment of gene signature sets in Ikaros-null progenitors
Ratio of enrichment of gene signature sets with respect to Ikaros differentially regulated gene lists was calculated by ratio-of-ratios method. For example, a 424 “stem” signature probe set was identified from 26587 quality passed probe sets on 430_2 array chip. A 21 “stem” probe set was identified among the 276 differentially up-regulated probe set in the Ikaros-null HSC-enriched population. The ratio enrichment of “stem” signature in mutant HSC can be calculated as ratio of ratios, 21/276 (i.e. 7.6%) versus the background information 424/26587 (i.e. 1.6%), roughly 7.6% / 1.6% = 4.8 fold enrichment. The Fisher exact probability for over-representation is calculated using the hypergeometric probability distribution that describes sampling without replacement from a finite population consisting of two types of elements.
Shannon entropy analysis
Uncertainty or “purity” for each cell population was measured by Shannon entropy based on single cell RT-PCR results, which detect the cell type distribution, such as frequency of “MLE”, “ML”, “ME”, “EL”, “M-only”, “L-only”, “E-only”, “Stem-only”, and “Unpr”, in each cell population. The information entropy of cell type X, that can take on possible values {x1, …, xn} (i.e. “MLE”, “ML”, “ME”, “EL”, “M-only”, “L-only”, “E-only”, “Stem-only”, and “Unpr”) is
where
I(X) is the information content of X; and
p(xi) is the frequency of xi; and 0log0 is taken to be 0; and
the logarithm is taken to base 2 (giving an entropy value in bits).
Multiplex single-cell RT-PCR analysis
Single cells were sorted into 96-well plates, with each well containing: 2 µl Invitrogen 5X First-strand buffer™, 2% Triton X-100, 10 mM dNTPs, 8 U RNAseOUT™, 30 U SuperScript™ II (Invitrogen), 0.1 µg BSA, and 0.005 µg Oligo-dT12–18 primer in a total volume of 10 µl. The reaction mix was incubated at 42° for 60 minutes followed by 70° for 15 minutes. cDNAs generated from single cells were split into two multiplex PCR reactions, utilizing a combination of 4–6 sets of PCR primers (1 mM) per reaction. Each reaction was subsequently re-amplified with a nested pair of primers used individually and products were resolved by electrophoresis.
Single cell and limiting dilution analysis of progenitors
OP9 and OP9-DL1 cells (Schmitt and Zuniga-Pflucker, 2002) were maintained as previously described. Prior to initiation of co-culture, cells were irradiated and plated into 96-well plates at a concentration of 2×103 cells/well. Cells were directly sorted onto plates for limiting dilution assays and maintained in the presence of stem cell factor, Flt3 ligand, and IL-7 as described in (Yoshida et al., 2006). Media was refreshed every 4 days during culture. Cultures were checked for growth on day 8–11 (OP9) or day 18–21 (OP9-DL1). Wells with >30 hematopoietic cells were scored as positive and their lineage identities were confirmed by FACS. FACS stains used were Mac-1 PE-Cy7, B220-APC-Cy7, Gr-1 APC, CD19 PerCP-Cy5.5, and Thy1.2-PE for OP9-GFP. FACS stains used were Mac-1 PE-Cy7, CD25 APC-Cy7, CD44-APC, CD4 APC-Cy5.5, CD8a PE or CD25 APC-Cy7, CD44-APC, Thy1.2-PE for OP9-DL1.
Statistical analysis
Linear regression analysis was performed with the R statistical software package. Frequencies, 95% confidence intervals, and R2 values for combined and individual experiments are shown in Figures 3 and 5 and Supplemental Tables 1–3. Two-tailed Student’s t-test was applied to obtain P values for single progenitor transcript analysis in Microsoft Excel software.
Supplementary Material
Acknowledgments
This work was supported by NIH R37 AI33062-16 and NIH-R01-AI42254-11 to KG. We thank; Bob Czyzewski for mouse husbandry and the KG lab and Bruce Morgan for a critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T, Mebius RE. Cellular interactions in lymph node development. J Immunol. 2005;174:21–25. doi: 10.4049/jimmunol.174.1.21. [DOI] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Greaves M. Loops, lineage, and leukemia. Cell. 1998;94:9–12. doi: 10.1016/s0092-8674(00)81215-5. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Acute Lymphoblastic Leukemia -- On the Wings of IKAROS. N Engl J Med. 2009 doi: 10.1056/NEJMe0809819. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Kanungo T, Mount DM, Netanyahu N, Piatko C, Silverman R, Wu AY. An efficient k-means clustering algorithm: Analysis and implementation. IEEE Trans Pattern Analysis and Machine Intelligence. 2002;24:881–892. [Google Scholar]
- Katsura Y. Redefinition of lymphoid progenitors. Nat Rev Immunol. 2002;2:127–132. doi: 10.1038/nri721. [DOI] [PubMed] [Google Scholar]
- Kaufmann C, Yoshida T, Perotti EA, Landhuis E, Wu P, Georgopoulos K. A complex network of regulatory elements in Ikaros and their activity during hemo-lymphopoiesis. Embo J. 2003;22:2211–2223. doi: 10.1093/emboj/cdg186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 2006;27:169–175. doi: 10.1016/j.it.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kioussis D, Georgopoulos K. Epigenetic flexibility underlying lineage choices in the adaptive immune system. Science. 2007;317:620–622. doi: 10.1126/science.1143777. [DOI] [PubMed] [Google Scholar]
- Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- Kondo M. Multitalented E2A: A New Role in Lymphoid-Lineage Priming. Immunity. 2008;29:169–170. doi: 10.1016/j.immuni.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kumar P, Walsh DA. A dual-specificity isoform of the protein kinase inhibitor PKI produced by alternate gene splicing. Biochem J. 2002;362:533–537. doi: 10.1042/0264-6021:3620533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuprash DV, Alimzhanov MB, Tumanov AV, Anderson AO, Pfeffer K, Nedospasov SA. TNF and lymphotoxin beta cooperate in the maintenance of secondary lymphoid tissue microarchitecture but not in the development of lymph nodes. J Immunol. 1999;163:6575–6580. [PubMed] [Google Scholar]
- Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- Lemischka IR, Moore KA. Stem cells: interactive niches. Nature. 2003;425:778–779. doi: 10.1038/425778a. [DOI] [PubMed] [Google Scholar]
- Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- Medina KL, Garrett KP, Thompson LF, Rossi MI, Payne KJ, Kincade PW. Identification of very early lymphoid precursors in bone marrow and their regulation by estrogen. Nat Immunol. 2001;2:718–724. doi: 10.1038/90659. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- Moore KA, Lemischka IR. "Tie-ing" down the hematopoietic niche. Cell. 2004;118:139–140. doi: 10.1016/j.cell.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. N Engl J Med. 2009 doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Mian IS, Kohwi-Shigematsu T, Ogawa T. A nuclear targeting determinant for SATB1, a genome organizer in the T cell lineage. Cell Cycle. 2005;4:1099–1106. [PubMed] [Google Scholar]
- Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan LA, Liongue C, Lewis RS, Stephenson SE, Ward AC. Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol Immunol. 2007;44:2497–2506. doi: 10.1016/j.molimm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Puri MC, Bernstein A. Requirement for the TIE family of receptor tyrosine kinases in adult but not fetal hematopoiesis. Proc Natl Acad Sci U S A. 2003;100:12753–12758. doi: 10.1073/pnas.2133552100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Buza-Vidas N, Hyland CD, Jensen CT, Antonchuk J, Mansson R, Thoren LA, Ekblom M, Alexander WS, Jacobsen SE. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rodgers JLaN, W A. Thirteen ways to look at the correlation coefficient. The American Statistician. 1988;42:59–66. [Google Scholar]
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Cell lineage regulators in B and T cell development. Nat Immunol. 2007;8:441–444. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Pant R. Origins of lymphocyte developmental programs: transcription factor evidence. Semin Immunol. 2004;16:227–238. doi: 10.1016/j.smim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Sitnicka E, Brakebusch C, Martensson IL, Svensson M, Agace WW, Sigvardsson M, Buza-Vidas N, Bryder D, Cilio CM, Ahlenius H, et al. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198:1495–1506. doi: 10.1084/jem.20031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- Tong W, Ibarra YM, Lodish HF. Signals emanating from the membrane proximal region of the thrombopoietin receptor (mpl) support hematopoietic stem cell self-renewal. Exp Hematol. 2007;35:1447–1455. doi: 10.1016/j.exphem.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumanov A, Kuprash D, Lagarkova M, Grivennikov S, Abe K, Shakhov A, Drutskaya L, Stewart C, Chervonsky A, Nedospasov S. Distinct role of surface lymphotoxin expressed by B cells in the organization of secondary lymphoid tissues. Immunity. 2002;17:239–250. doi: 10.1016/s1074-7613(02)00397-7. [DOI] [PubMed] [Google Scholar]
- Uehara S, Grinberg A, Farber JM, Love PE. A role for CCR9 in T lymphocyte development and migration. J Immunol. 2002;168:2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Wu JN, Koretzky GA. The SLP-76 family of adapter proteins. Semin Immunol. 2004;16:379–393. doi: 10.1016/j.smim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3-short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Zhang J, Hazan I, Naito T, Ng SY, Snippert HJ, Heller EJ, Lawton L, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multi-lineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara H, Arai F, Hosokawa K, Hagiwara T, Takubo K, Nakamura Y, Gomei Y, Iwasaki H, Matsuoka S, Miyamoto K, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.