Abstract
Here, we report the identification of the RNA binding motif protein RBM15B/OTT3 as a new CDK11p110 binding partner that alters the effects of CDK11 on splicing. RBM15B was initially identified as a binding partner of the Epstein-Barr virus mRNA export factor and, more recently, as a cofactor of the nuclear export receptor NXF1. In this study, we found that RBM15B co-elutes with CDK11p110, cyclin L2α, and serine-arginine (SR) proteins, including SF2/ASF, in a large nuclear complex of ∼1-MDa molecular mass following size exclusion chromatography. Using co-immunoprecipitation experiments and in vitro pulldown assays, we mapped two distinct domains of RBM15B that are essential for its direct interaction with the N-terminal extension of CDK11p110, cyclin L2α, and SR proteins such as 9G8 and SF2/ASF. Finally, we established that RBM15B is a functional competitor of the SR proteins SF2/ASF and 9G8, inhibits formation of the functional spliceosomal E complex, and antagonizes the positive effect of the CDK11p110-cyclin L2α complex on splicing both in vitro and in vivo.
Keywords: Cell Cycle, Cyclins, RNA Processing, RNA Splicing, RNA Transport
Introduction
The human cell division control 2-like genes Cdc2L1 and Cdc2L2 encode two related protein kinases, denoted CDK11B and -A, respectively, which are expressed as two predominant protein isoforms designated by their apparent molecular mass (p110 and p58 for the 110- and 58-kDa isoforms, respectively) (1, 2). Because current data indicate that the products of the two genes are functionally redundant, the term CDK11 will refer to products from both genes hereafter. The CDK11p110 and CDK11p58 isoforms are produced from the same mRNAs through the use of an internal ribosome entry site and two different AUG codons located in the coding sequence of the CDK11p110A and -B mRNAs (3).
The cyclin L proteins are the regulatory partners of CDK11p110 and CDK11p58 (4–8). These proteins are encoded by two genes, cyclin L1 and L2, which produce six distinct protein isoforms of various apparent molecular masses by alternative splicing (8). The ∼70-kDa cyclin L1α and L2α proteins contain an N-terminal cyclin box and a C-terminal arginine-serine (RS)-rich domain very similar to that of splicing-regulating SR proteins, whereas the short 20–35 kDa cyclin L1β, L2β A/B, and L1γ proteins contain the cyclin box but lack the RS domain.
Expression of the large CDK11p110 protein kinase isoforms is ubiquitous and constant throughout the cell cycle. CDK11p110 protein is a nuclear protein present in two macromolecular complexes of 1–2 MDa and ∼800 kDa that contain the cyclin Ls, the largest subunit of RNA polymerase II, the SSRP1 and SPT6 subunits of the transcription elongation factor FACT (facilitates chromatin transcription), CK2,5 and the Rap30 and Rap74 subunits of general transcription factor IIF (9). Using the yeast two-hybrid strategy, we identified the splicing factors RNPS1 (10) and 9G8 (11) as the first direct CDK11p110 binding partners. Both RNPS1 and 9G8 belong to the SR protein family, which stimulate excision of introns from pre-RNAs and regulate alternative splicing (12). RNPS1 and 9G8 co-immunoprecipitate with CDK11p110 and are phosphorylated by CK2 (13) and CDK11p110 (11), respectively. Taken together, these data suggested that CDK11p110 was involved in splicing and/or transcription. In contrast, recent reports have demonstrated that the mitosis-specific CDK11p58 protein is required for centrosome maturation, bipolar spindle formation, and maintenance of sister chromatid cohesion (14, 15).
The involvement of CDK11p110 in the regulation of transcription was first demonstrated by studies from our laboratory that established that anti-CDK11p110 catalytic domain antibodies reduced the synthesis of RNA transcripts produced from both TATA-like and GC-rich promoters in standard in vitro transcription assays (9). More recently, CDK11p110 was also identified as a positive regulator of hedgehog signaling in both fly and vertebrate cells (16, 17) and as a modulator of the Wnt/β-catenin signaling cascade (18).
Several lines of evidence also confirmed the role of the CDK11p110-cyclin L complexes in pre-mRNA splicing. Immunodepletion of the CDK11p110 kinase from nuclear extracts greatly reduced the in vitro splicing activity, whereas readdition of the CDK11p110 immunoprecipitates rescued the splicing activity (11). In addition, overexpression of CDK11p110 in cultured cells increased in vivo splicing, whereas overexpression of a catalytically inactive form of CDK11p110 inhibited splicing (8). Similarly, preincubation of nuclear extracts with purified cyclin Lα and Lβ proteins bound to Sepharose beads depletes the extract of in vitro splicing activity (8). We also demonstrated the direct role of CDK11p110-cyclin L complexes in the regulation of pre-mRNA splicing by showing that ectopic expression of cyclin Ls individually enhances in vivo splicing activity using a β-galactosidase/luciferase reporter construct (8). Moreover, enforced expression of cyclin L proteins alone or in combination with active or catalytically inactive CDK11p110 strongly affects in vivo alternative splicing of an E1A minigene reporter construct (8). In addition, others have shown that cyclin L1α is an immobile component of the splicing factor compartment (19) that is associated with hyperphosphorylated RNA polymerase II (5) and that cyclin L2α is a substrate of the nuclear protein kinase DYRK1A (7), which is a dual specificity protein kinase that phosphorylates several transcription factors and induces SR protein redistribution. Together, these data demonstrate that CDK11p110 is part of macromolecular complexes regulating RNA synthesis at the interface of transcription and splicing, two tightly linked processes occurring concomitantly and reciprocally influencing each other. Recently, it was also shown that CDK11 and 9G8 interact with the eukaryotic initiation factor 3 subunit f (eIF3f) and together alter the 3′ processing of the HIV-1 pre-mRNA (20). The involvement of CDK11p110 in RNA maturation most likely requires its indirect interaction with the nascent pre-RNAs via RNA-binding proteins such as the SR proteins RNPS1 and 9G8, hnRNPs, and other proteins containing RNA recognition motifs that remain to be identified.
Here, we report the identification of a new CDK11p110 and cyclin L2α binding partner, the RNA binding motif protein RBM15B/OTT3 (one-twenty-two 3). RBM15B was initially identified as a binding partner of the Epstein-Barr virus mRNA export factor (21) and more recently was found to be a cofactor of the nuclear export receptor NXF1 (22). In the present study, we showed that RBM15B is located in a nuclear macromolecular complex through its direct binding to CDK11p110-cyclin L complexes and the splicing factor 9G8. Using in vitro and in vivo splicing assays, we established that RBM15B is a functional competitor of the SR proteins, capable of inhibiting the formation of the spliceosomal E complex and antagonizing the positive effect of the CDK11p110-cyclin L2α complex on splicing.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen
The yeast two-hybrid screen was performed as described previously (10) using the full-length CDK11Bp110 (Cdc2L1) fused to the GAL4 DNA binding domain and cloned into the PAS1CYH2 plasmid and a human B-cell cDNA library subcloned into the pACT plasmid containing the GAL4 activation domain (kindly provided by Dr. S. Elledge).
Cell Culture and Transfection
HEK293T and human foreskin fibroblasts were maintained in DMEM supplemented with 10% fetal calf serum (FCS) and 2% l-glutamine. Transfections of HEK293T cells with expression vectors encoding CDK11p110, cyclins L1α and L2α, 9G8, SF2/ASF, and various RBM15B constructs were performed using the transfection reagents FuGENE 6 (Roche Applied Science), JetPEI (Qbiogene), or KLN47 (23), and cells were harvested 48 h after transfection.
Recombinant Protein Production
His6-tagged (pFast Bac expression vector, Invitrogen) full-length RBM15B and 9G8 (11) were expressed in Sf9 insect cells and purified using a nickel-nitrilotriacetic acid affinity column under native conditions according to the manufacturer's protocol (The QIAexpressionistTM, Qiagen). GST-RBM15B fusion proteins (supplemental Data S8) were expressed in BL21 bacteria (Invitrogen) and purified with GSH-SepharoseTM 4B beads (Amersham Biosciences) using the manufacturer's protocol. In vitro transcribed and translated CDK11, RBM15B, and cyclin L2α proteins were produced using the TnT kit (Promega) and [35S]methionine (PerkinElmer Life Sciences).
Antibodies
The CDK11 antibodies P2N100 (10) and P1C (24) and cyclin L (8) antibodies have been described previously. The RBM15B antibody was produced by immunization of rabbits with the purified GST-tagged N-terminal domain (amino acids 1–154; supplemental Data S2B). Specific anti-RBM15B immunoglobulins were affinity-purified using the GST N-terminal domain covalently bound to Sepharose 4B beads (supplemental Data S2, C and D). Rat monoclonal anti-HA (Roche Applied Science), mouse M2 monoclonal anti-FLAG (Sigma), anti-CK2α (C-18), 9G8 (H-120) and SF2/ASF (P-15) (Santa Cruz Biotechnology), anti-9G8-ZnK polyclonal antibody (a kind gift from Dr. Marie-Louise Hammarskjold), anti-SF2/ASF polyclonal antibody (38813, Abcam), proliferating cell nuclear antigen (PC10, Dako), and mouse monoclonal anti-SR proteins SRp75, SRp55, SRp40, SRp30a/b, and SRp20 (1H4, Zymed Laboratories Inc.) were used according to the manufacturers' instructions.
Chromatography/Size Fractionation
Size fractionation analysis was carried out using a Superose 6 column (Amersham Biosciences). Protein standards of 737, 460, 170, and 65 kDa were used to calibrate the column in 50 mm phosphate (pH 7), 150 mm NaCl, 0.2 mm EDTA, and 0.1% Nonidet P-40. One milligram (∼0.1 ml) of HeLaScribe® nuclear extract (Promega) was loaded onto the Superose 6 column, and 1-ml fractions were collected. Immunoblot analyses using 40 μl from each fraction were performed.
Immunoblots and Immunoprecipitations
For immunoblots, human cell lines were washed in PBS and lysed by brief sonication in 50 mm Tris-HCl (pH 8), 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 15 mm MgCl2, 60 mm β-glycerophosphate, 1 mm DTT, 0.1 mm sodium vanadate, 0.1 mm sodium fluoride, 15 mm p-nitrophenyl phosphate, and 0.5% Nonidet P-40. Immunoprecipitation of endogenous CDK11 and cyclin L proteins was performed using HEK293T cell lysates. In all immunoprecipitation experiments, cell lines were lysed in 50 mm Hepes (pH 7.9), 150 mm NaCl, 0.1 mm EDTA, 10% glycerol, 0.5% Tween 20, 10 mm β-glycerophosphate, 0.1 mm sodium vanadate, and 0.2 mm sodium fluoride. Lysis buffers were supplemented with 1× Complete protease inhibitors (Complete EDTA-free, Roche Applied Science). Lysates were centrifuged at 14,000 rpm in a microcentrifuge for 20 min prior to incubation overnight with antibodies (500 μg of lysates and 1 μg of antibody) followed by a 2-h incubation with GammaBind Plus Sepharose beads (Amersham Biosciences) at 4 °C. Washes were performed four times with 1 ml of lysis buffer each time. Immunoblotting was performed as described previously (10).
GST Pulldown Assay
Pulldown assays were performed by incubating GST-RBM15B fusion proteins (supplemental Data S5) with [35S]methionine-labeled in vitro transcribed and translated (IVTT) products or total lysates of either non-transfected HEK293T to detect endogenous CDK11p110 and SR proteins or transiently transfected HEK293T to detect cyclins L1α and L2α, SF2/ASF, and 9G8. The lysis was performed using immunoprecipitation buffer, and incubations were carried out for 2 h at 4 °C under gentle rotation. Beads were then washed four times with the same buffer prior to immunoblotting analysis or autoradiography of [35S]methionine-labeled IVTT products.
In Vitro Splicing Assay
The 32P-labeled β-globin pre-mRNA substrate was prepared by in vitro transcription using SP6 RNA polymerase and BamHI-linearized pSP64-HβΔE6 plasmid as the template (25). The HeLa nuclear extracts (NEs) used for the in vitro splicing reactions were obtained from Promega or prepared according to Dr. Mayeda's protocol (supplemental Data S7). In vitro splicing reactions were carried out in a final volume of 25 μl with 25 μg of NE and 20 fmol of 32P-labeled β-globin pre-mRNA substrate followed by incubation at 30 °C for 4 h. The spliced RNA products were analyzed by electrophoresis on a denaturing 5.5% polyacrylamide, 7 m urea gel (25). Statistical analysis was performed using the Statview software: the non-parametric Mann-Whitney test was used, and p < 0.05 was considered significant. For detection of functional spliceosomal complexes E, A, B, and C, β-globin RNA-protein complexes were resolved on native horizontal agarose minigels as reported previously (26, 49).
In Vivo β-Galactosidase/Luciferase Splicing Assay
HEK293T cells were transfected using FuGENE 6 (Roche Applied Science) as described by the manufacturer. The optimal amounts of DNA expression vectors for RBM15B, CDK11, cyclin L2α, SF2/ASF, and 9G8 used in the transfections were established in pilot experiments that were performed to optimize protein expression (data not shown). The cyclin L2α cDNA (1 μg) containing an N-terminal FLAG tag was cloned into the pFlex vector. CDK11Bp110 (Cdc2L1, 6 μg) and CDK11Bp110 DN (catalytically inactive due to mutation of amino acid Asp-567 to Asn) (3 μg) containing a C-terminal FLAG tag were in the pUHD 10-3 vector. N-terminal HA-tagged RBM15B wild type (WT) (0.5 μg) was in the pCMV vector, whereas RBM15B Core (amino acids 1–609; 0.5 μg) and RBM15B Core/NLS constructs (amino acids 1–609 + the putative nuclear localization signal (NLS); 0.5 μg) were in the pcDNA 3.1 vector. All RBM15B truncated mutants are depicted in supplemental Data S8 and contain an N-terminal HA tag. The C-terminal His6-tagged 9G8 and the N-terminal His6-tagged SF2/ASF constructs (0.5 μg) were in the pCEP4-E1A vector (gifts from Dr. W.-Y. Tarn). The splicing reporter vector pTN24 (a gift from Dr. I. C. Eperon) (27) was used (1 μg) in all experiments. Cells were harvested 24 h after transfection for the enzymatic assays. β-Galactosidase and luciferase activities were measured using the Dual-Light System (Applied Biosystems), Bright-Glo Luciferase Assay System (Promega), and Beta-Glo Assay System (Promega). All measurements were within the linear range that was established using luciferase and β-galactosidase standards. All samples were measured in quadruplicate, and 3–10 independent transfected cultures were assayed per data point. Immunoblot analyses of equal volume cell lysates were performed using anti-CDK11, -cyclin L2α, -RBM15B, -9G8, and -SF2/ASF polyclonal antibodies. 9G8 construct enforced expression was also detected by quantitative RT-PCR using 9G8-specific primers (5′-GTTGGTAACCTGGGAACTGG-3′ and (5′-CGAATTCCACAAAGGCAAAT-3′) and SYBR Green (Applied Biosystems). PCR samples were incubated for 10 min at 95 °C followed by the following cycling parameters: 95 °C for 15 s, 60 °C for 1 min for 40 cycles. For the statistical significance analyses of this in vivo splicing assay, Drs. W. Zhao and M. Kocak (Department of Biostatistics, St. Jude Children's Research Hospital, Memphis, TN) have developed a specific non-parametric rank-based test as described previously (8) that is available upon request.
RESULTS
RNA Binding Motif Protein RBM15B Interacts with CDK11p110 and Is Present in Large Nuclear Complex
To identify new CDK11p110 binding partners, a yeast two-hybrid screen was performed using the full-length CDK11p110 as bait and a human B-cell library. In addition to RNPS1, a known interactor of CDK11p110 (10), one of the proteins we identified in this screen was a partial cDNA (clone 33) that encoded the majority of the RNA binding motif 15B protein (RBM15B/OTT3) but lacked the 5′-end of the full-length cDNA (supplemental Data S1). RBM15B is an intronless gene that produces a 110-kDa protein (supplemental Data S1C and S2C) that was initially identified through its physical interaction with the EB2 protein of the Epstein-Barr virus (21). RBM15B is highly similar to RBM15/OTT1 (supplemental Data S3), which was originally discovered as a fusion partner with the gene megakaryoblastic leukemia 1 (MKL1) in the translocation t(1;22)(p13;q13) associated with childhood acute megakaryocytic leukemia (28, 29). RBM15/OTT1 and RBM15B proteins both contain three RNA recognition motifs (RRMs) localized within the N-terminal half, a consensus nuclear localization signal, and a (Spen (split end) paralog and ortholog C-terminal) (SPOC) domain (supplemental Data S1 and S3), which mediates interaction with the co-repressors of transcription silencing mediator for retinoic and thyroid receptors (SMRT) and nuclear receptor co-repressor (NCoR). Data from Hiriart et al. (21) suggested that the RBM15B gene encodes two transcripts containing the same open reading frame but using alternative poly(A) signals: a major 3.5-kb short mRNA and a minor 6.6-kb variant including a 3.1-kb 3′-UTR. In our experimental conditions, the 3.5-kb mRNA was the only transcript detected by Northern blot in human foreskin fibroblast and HepaRG cells (data not shown). However, RT-PCR experiments confirmed the production of two RBM15B mRNA variants from mouse, rat, and human in normal liver as well as HepaRG and human foreskin fibroblast cell lines (supplemental Data S1B).
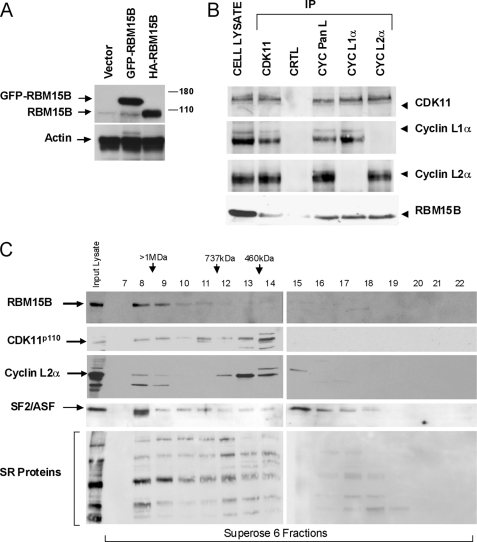
To confirm the interaction between CDK11p110 and RBM15B, transient transfections were performed in HEK293T using expression vectors encoding HA- and GFP-tagged RBM15B followed by immunoprecipitation (IP) of endogenous CDK11p110 and immunoblotting of RBM15B using anti-HA and -GFP antibodies (supplemental Data S2A). RBM15B was found in CDK11p110 IP, demonstrating that these two proteins co-immunoprecipitated. The reciprocal IPs using anti-HA and -GFP antibodies further confirmed the interaction between RMB15B and CDK11p110 (supplemental Data S2A). In addition, an anti-RBM15B-specific antibody was raised (Fig. 1A and supplemental Data S2, B–D) to verify the interaction of endogenous CDK11p110 and RBM15B. Endogenous CDK11p110 and cyclin L proteins were immunoprecipitated from HEK293T cells, and the specific anti-RBM15B antibody was used for immunoblotting. RBM15B associated with both CDK11p110 and cyclin L protein IPs but not in the control IP using total immunoglobulins (Fig. 1B).
FIGURE 1.
CDK11p110-cyclin L and RBM15B co-immunoprecipitate and are present in large nuclear macromolecular complexes. A, characterization of RBM15B antibody. An anti-RBM15B-specific rabbit polyclonal antibody was raised against the GST-N-term domain of RBM15B (amino acids 1–154; supplemental Data S8) and affinity-purified (supplemental Data S2, B–D). Immunoblots were performed with this affinity-purified antibody using cell lysates from HEK293T cells transfected with empty vector or GFP- or HA-tagged RBM15B expression vectors. Actin was used as loading control. B, co-immunoprecipitation of CDK11p110 and cyclin L with RBM15B. Endogenous CDK11p110 and cyclin L proteins were immunoprecipitated from HEK293T cell lysates with anti-CDK11 (P1C), -cyclin (CYC) pan-L, -cyclin L1α, and -cyclin L2α antibodies. Purified rabbit IgG was used as a negative control (CTRL). Immunoprecipitates were analyzed by immunoblotting using CDK11 (P1C), cyclin L1α, cyclin L2α, and RBM15B antibodies. C, co-elution of CDK11p110, cyclin L2α, SR proteins, and RBM15B following size exclusion chromatography. Extracts from HeLa nuclei (NE; 1 mg) were fractionated using a Superose 6 column and analyzed by immunoblotting using the CDK11 (P1C), cyclin L2α, SF2/ASF, anti-SR protein (1H4 mAb), and RBM15B antibodies. The anti-SR antibody recognizes SRp75, SRp55, SRp40, SRp30a/b, and SRp20 proteins. Input lanes contain 20 μg of HeLa NE, whereas other lanes contain 40 μl from each 1-ml fraction.
In situ detection of HA-RBM15B fusion protein and indirect immunofluorescence of endogenous RBM15B revealed a pattern of diffuse nucleoplasmic staining and an intense staining in large speckles (supplemental Data S4 and S5B). Interestingly, we did not find any enrichment of RMB15B at the nuclear envelope as had been reported for the highly related protein RMB15/OTT1 (30). However, we observed a partial nuclear co-localization of RBM15B with both CDK11p110 and the SR proteins SRp75, SRp55, and SRp20 (supplemental Data S4). By Western blotting, we found that expression of both CDK11p110 and RBM15B was constant throughout the cell cycle of synchronized human foreskin fibroblasts, whereas there was a slight increase in CDK11p110 levels during S and G2/M phases (supplemental Data S5A). The nuclear co-localization of these two proteins was observed throughout interphase, whereas they were evenly distributed in the cytoplasm during mitosis (supplemental Data S5B).
Previously, we reported that CDK11p110 and cyclin Ls are present in two different macromolecular complexes containing transcription and splicing factors (8, 9, 11). Our co-IP and nuclear co-localization of RBM15B with CDK11p110 suggested that this RNA-binding protein should also be present in these nuclear complexes. To confirm this hypothesis, HeLa nuclear extract was fractionated using Superose 6 size exclusion chromatography (Fig. 1C). As expected, CDK11p110 and cyclin L2α were detected in two large complexes of ∼1 MDa and ∼500 kDa and co-eluted with SR proteins including SF2/ASF. Interestingly, RBM15B was found predominately in a large complex of >1 MDa, indicating that only a fraction of CDK11p110 was associated with RBM15B in a macromolecular nuclear complex.
Two Distinct Domains of RBM15B Are Involved in Binding to CDK11p110, Cyclin L2α, and 9G8
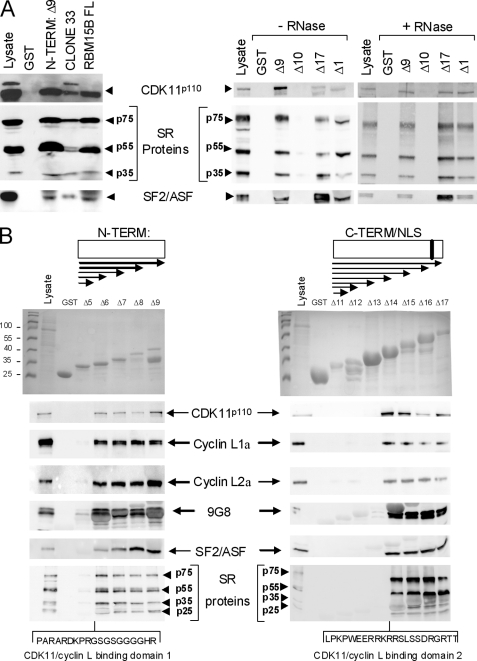
We next performed pulldown assays using bacterially expressed GST-RBM15B fusion proteins to map the domain of interaction with CDK11p110. In the initial two-hybrid screening, we pulled out a partial RBM15B cDNA that lacked the N-terminal segment containing the first 154 amino acids (clone 33; see supplemental Data S1 and S8) using CDK11p110 as bait. This strongly suggested that the binding between RBM15B and CDK11p110 involved an RBM15B motif downstream of the N-terminal domain. To confirm this hypothesis, we produced a series of recombinant proteins containing the N-terminal domain peptide, a protein lacking the N-terminal domain (clone 33), and full-length RBM15B proteins fused to the GST to perform pulldown assays (supplemental Data S8 and Fig. 2A). These recombinant proteins, bound to Sepharose beads, were incubated with HEK293T total cell extracts. The presence of interacting proteins including CDK11p110, SR proteins SRp75/55/35, and SF2/ASF was then detected by immunoblot analysis (Fig. 2A, left panel). As expected, CDK11p110; SR proteins SRp75, SRp55, and SRp20; and SF2/ASF were found associated with the clone 33 and the full-length RBM15B proteins, but surprisingly, these proteins were also detected in the N-term RBM15B pulldown, suggesting that the N-terminal domain of RBM15B contained another binding motif for CDK11p110. No proteins were detected in the negative control pulldown using the GST protein.
FIGURE 2.
Domain mapping of RBM15B for its interaction with CDK11p110. GST pulldown assays were performed using a series of GST-RBM15B fusion peptides (supplemental Data S8) incubated with lysates from HEK293T cells transfected with expression vectors for cyclins L1α and L2α, 9G8, and SF2/ASF or lysates of non-transfected cells for CDK11p110 and SR protein SRp75, SRp55, and SRp35 binding assays. Detection of proteins bound to GST and GST-RBM15B beads was performed using specific antibodies. A, GST pulldown assays with the RBM15B FL, N-term (Δ9; amino acid 1–154), clone 33 (amino acids 249–890), RRM (Δ10; amino acids 141–489), C-term/NLS (Δ17; amino acids 490–720), and ΔSPOC (Δ1; amino acids 1–720). GST pulldown assays using recombinant GST-RBM15B mutants Δ9, Δ10, Δ17, and Δ1 were performed in the absence (−RNase) or presence (+RNase) of RNase (20 units/ml Benzonase®, Merck). B, GST pulldown assays with deletion constructs for the N-term (Δ5–Δ9) and C-term/NLS regions (Δ11–Δ17) of RBM15B and mapping of the two CDK11-cyclin Lα binding domains.
To further localize the interaction sites, we divided RBM15B into four distinct domains (supplemental Data S8): the N-terminal end (N-term; amino acids 1–154), the three RRMs (amino acids 141–489), the C-terminal domain containing a putative NLS (C-term/NLS; amino acids 490–720), and the SPOC domain at the very C-terminal end of the protein (amino acids 721–890). In addition, we generated a RBM15 fusion protein lacking only the SPOC domain (ΔSPOC mutant; amino acids 1–720). To demonstrate that two CDK11p110 binding motifs were present in RBM15B, we used these GST fusion proteins to perform the pulldown assay (Fig. 2A, right panel). CDK11p110, SR proteins SRp75/55/35, and SF2/ASF were found associated with the N-term and C-term/NLS domains and the ΔSPOC mutant but not with the RRM domain. These data suggested that CDK11p110 binding motifs were present in both the N- and C-terminal domains of RBM15B and that the SPOC domain is dispensable for these interactions. Similar experiments were performed using RNase-treated HEK293T cell extracts, which gave the same interactions, indicating that binding of CDK11p110, SR proteins SRp75/55/35, and SF2/ASF to RBM15B was not mediated by RNA.
To further map the motifs of interaction in RBM15B, we generated additional GST-N-term and -C-term fusion proteins of various lengths (supplemental Data S8) that were used for pulldown assays to detect CDK11p110, cyclins L1 and L2α, 9G8, SF2/ASF, and SR proteins p75/55/35 by immunoblot analysis (Fig. 2B). No proteins were detected in the negative control pulldown using the GST protein alone, and only very minimal binding of cyclin L and SR proteins was seen in the shortest GST-N-term RBM15B protein (Δ5), whereas all of the proteins were bound to the other N-term constructs (Δ6–Δ9; Fig. 2B, left panel). Similarly, CDK11p110, cyclins L1α and L2α, 9G8, SF2/ASF, and SR proteins p75/55/35 were not detected in the shortest three GST-C-term RBM15B proteins (Δ11–Δ13) but were all present in the longest truncation proteins (Δ14–Δ17; Fig. 2B, right panel). These data confirmed that two domains of RBM15B are involved in the interaction with CDK11p110 and its associated proteins and suggested that the segments containing the amino acids 48–67 (C terminus of Δ6) and 585–607 (C terminus of Δ14), respectively, localized within the N-terminal and C-terminal domains of RBM15B (Fig. 2) are essential either for the direct binding to CDK11p110 or required for proper folding of the associated amino acids into the interaction domains. These results also indicated that the peptide (amino acids 1–607) including the N-term, RRM domains, and amino acids 490–607 from the C-terminal domain (supplemental Data S8, RBM15B “Core”) is sufficient for the interaction with the splicing regulators such as SR proteins and CDK11p110-cyclin L complexes.
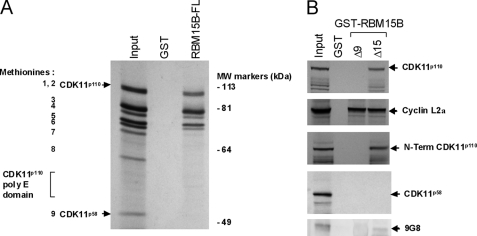
To delineate the domain of CDK11p110 required for its interaction with RBM15B, we set up pulldown assays using the GST-RBM15B full length (FL) and IVTT products of CDK11p110 labeled with [35S]methionine (Fig. 3). In vitro transcription and translation of CDK11Ap110 cDNA generated nine products (Fig. 3A) as observed previously (10) because of the initiation of translation from the eight internal AUG codons upstream of the CDK11p58 AUG. Pulldown assays using these IVTT reaction products showed that the full-length CDK11p110 proteins and at least five truncated IVTT products bound to the GST-RBM15B full length but not with the GST control protein (Fig. 3A). In this experiment, we did not detect the IVTT product of CDK11p58, suggesting that RBM15B specifically binds to the N-terminal domain of the CDK11p110 form.
FIGURE 3.
RBM15B interacts with N-terminal extension of CDK11p110, cyclin L2α, and 9G8. A, [35S]methionine-labeled CDK11p110 IVTT products were generated from translation initiating at several internal AUG codons indicated as their corresponding methionines (1–9). Methionine (Met) 1 corresponds to the translation start for full-length CDK11p110, whereas Met-9 is the translation start for the CDK11p58 isoform. The polyglutamine (poly E) domain of CDK11p110 is located between methionines 8 and 9. The GST pulldown assay was performed using CDK11p110 IVTT products incubated with GST-RBM15B FL and GST as control. B, pulldown assay using the GST-RBM15B N-term (Δ9) and C-term RBM15B (Δ15) fusion proteins incubated with [35S]methionine-labeled IVTT products of CDK11p110, the N-term domain of CDK11p110 upstream of the sequence of CDK11p58, CDK11p58, cyclin L2α, and bacterially produced recombinant 9G8 protein. IVTT products were visualized by autoradiography, whereas 9G8 was detected by immunoblotting.
To confirm this hypothesis and to determine whether RBM15B interacted directly with other proteins in the complex such as cyclin L2α and 9G8, we set up pulldown assays using the GST-RBM15B N-term (Δ9) and C-term RBM15B (Δ15) fusion proteins and IVTT products of CDK11p110, the N-terminal domain of CDK11p110 upstream of the CDK11p58 sequence, CDK11p58, cyclin L2α, and bacterially produced recombinant 9G8 (Fig. 3B). Full-length CDK11p110 IVTT product interacted with the C-terminal domain of RBM15B, but unexpectedly, it did not bind the N-terminal motif in contrast to the interaction data obtained from the pulldown assays performed with total cell lysates (Fig. 2B). Similarly, the N-terminal CDK11p110 IVTT product was found associated with the C-terminal domain of RBM15B and not the N-terminal motif, whereas the CDK11p58 IVTT product did not bind either of the two RBM15B recombinant peptides. We also found that cyclin L2α IVTT protein interacted with both N- and C-terminal domains of RBM15B, whereas recombinant 9G8 bound only to the RBM15B C-terminal motif. These data indicated that only the N-terminal extension of CDK11p110 interacts with the C-terminal domain of RBM15B, whereas the cyclin L2α binds to both domains of RBM15B and potentially bridged CDK11p110 to the N-terminal domain of RBM15B in the total cell lysate assays (Fig. 2B).
RBM15B Inhibits Pre-mRNA Splicing in Vitro
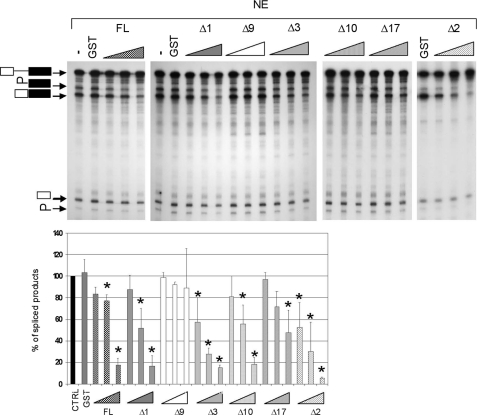
Because Hiriart et al. (21) reported that RBM15B affected the alternative splicing of a β-thalassemia reporter pre-RNA (containing a G-to-A transition at position 1 of intron 1 that results in the usage of three cryptic 5′-splice sites and expression of three mRNA variants by alternative splicing) in transfected cells, indicating that the RBM15B protein could regulate pre-mRNA splicing, we investigated whether RBM15B could affect splicing in vitro using the radiolabeled human β-globin pre-RNA, HeLa NE, and recombinant GST-RBM15B fusion proteins (Fig. 4). Addition of increasing amounts of GST-RBM15B full length, GST-RBM15B lacking the C-terminal SPOC domain (Δ1 or ΔSPOC; amino acids 1–720), or the truncated GST-RBM15B Core (Δ3 mutant) proteins resulted in a dose-dependent decrease in the ability of the NE to catalyze the excision of the pre-RNA intron (Fig. 4), whereas addition of GST alone did not affect the splicing rate. Supplementation of splicing assays with the N-term domain (Δ9; amino acids 1–159) had no significant effect, whereas GST-RRM (Δ10) and GST-C-term/NLS (Δ17) domains separately showed a ∼40–60% decrease in the amount of spliced β-globin RNA. Furthermore, addition of the GST-RRM + C-term/NLS domains (Δ2) resulted in a strong inhibition of β-globin splicing (Fig. 4), demonstrating that the most potent inhibitory effects were observed with the RRM associated with the N- and/or C-terminal/NLS domains. Together, these results demonstrate that RBM15B is a strong inhibitor of pre-mRNA splicing.
FIGURE 4.
RBM15B inhibits in vitro splicing of β-globin pre-RNA. HeLa NE was used for an in vitro splicing assay using the pre-RNA of the human β-globin gene as a substrate. Five well characterized β-globin RNA species with different electrophoretic mobilities (25) are represented: white and black rectangles indicate exons 1 and 2, respectively, whereas the line represents the intron separating the two exons. The splicing reaction was supplemented with GST (5 pmol/25-μl splicing reaction) and increasing concentrations (0.5, 2.5, and 5 pmol/splicing reaction) of GST-RBM15B fusion proteins (supplemental Data S5): FL, RBM15B Core (Δ3), ΔSPOC (Δ1), N-term (Δ9), RRM (Δ10), C-term/NLS (Δ17), and RRM + C-term/NLS (Δ2). The data in the chart represent the mean of three independent experiments. Standard deviation from the mean is shown by the error bars. Statistical analysis was done using the non-parametric Mann-Whitney test: *, p (probability value) < 0.05 versus GST control condition (CTRL).
RBM15B Inhibits Pre-mRNA Splicing in Vivo
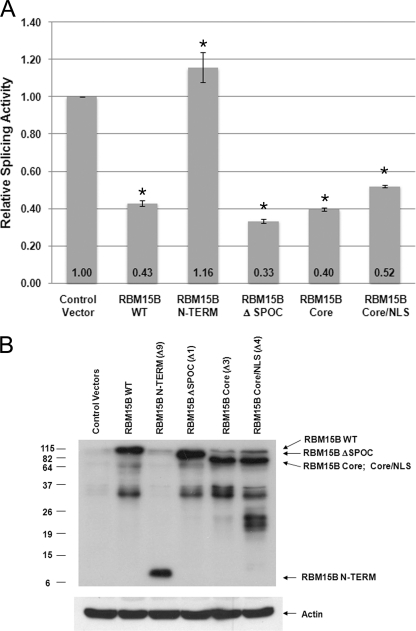
The double β-galactosidase/luciferase reporter system, designed to measure splicing activity in vivo (27), was next used to ascertain whether RBM15B full length or mutants affect splicing in vivo and to see which regions of RBM15B are important for its ability to regulate splicing. Unspliced transcripts produced by transiently transfecting the pTN24 construct into HEK293T cells yields a protein with only the β-galactosidase activity, whereas properly spliced intronless mRNAs are translated into a fusion protein with both β-galactosidase and luciferase activities. Thus, the ratio of luciferase to β-galactosidase activity in the cell lysate allows us to measure the proportion of spliced mRNA. Co-transfection of RBM15B expression constructs was conducted to evaluate the effects of RBM15B on splicing in comparison with co-transfection of pTN24 with control empty expression vector (data were normalized to base-line control activity seen upon co-transfection with the empty vector, which was arbitrarily set as 1). Titration experiments showed that transfections performed using 0.5 μg of RBM15B full length and mutant expression vector plasmid DNAs did not significantly affect the β-galactosidase expression, whereas higher amounts (>1 μg) significantly decreased expression (data not shown). Ectopic expression of full-length RBM15B (WT), the GST-RBM15B protein lacking the SPOC domain (ΔSPOC: Δ1), the RBM15B Core (Δ3), or the core peptide plus an exogenous nuclear localization domain (RBM15B Core/NLS: Δ4) all significantly repressed the relative splicing activity compared with the control cells transfected with empty expression vectors (Fig. 5A). Conversely, enforced expression of the RBM15B-N-term (Δ9; amino acids 1–154) showed no effect on splicing (Fig. 5A). For these experiments, protein expression was verified by immunoblotting (Fig. 5B). As seen in the in vitro splicing assays, the RRM domains were necessary for inhibition of splicing. Thus, expression of RBM15B WT or the truncated RBM15B constructs containing the RRM domains inhibit excision of the intron from the β-galactosidase/luciferase reporter pre-mRNA in vivo. To determine whether inhibition of splicing by RBM15B was specific to β-globin or whether RBM15B could affect splicing of other pre-mRNAs, we used an in vivo E1A minigene reporter system (supplemental Data S6) that encodes an unspliced RNA and five splice variants (13, 12, 11, 10, and 9 S). In these experiments, HepaRG cells were transiently transfected with 2 μg of E1A encoding vector, increasing amounts of RBM15B expression vectors, and appropriate amounts of empty vector to reach equal concentrations of plasmid in all conditions (supplemental Data S6). RBM15B expression was verified by immunoblotting (supplemental Data S2C). RT-PCR and gel electrophoresis were performed to detect E1A RNAs (supplemental Data S6B). We found that enforced expression of RBM15B led to the increase in unspliced E1A pre-RNA content and a decrease in the levels of the 12 and 9 S splice variants. Interestingly, the amounts of the 13 and 10 S forms were not significantly affected by expression of RBM15B, whereas the 11 S variant was not detected in these cells. Together, these in vivo data indicate that RBM15B most likely affects the overall exon splicing and confirm that RBM15B affects alternative splicing (21).
FIGURE 5.
RBM15B inhibits in vivo splicing of β-galactosidase/luciferase reporter system. Expression constructs for RBM15B WT, N-term, ΔSPOC, Core, and Core/NLS and empty vector were transiently co-transfected into HEK293T cells with the pTN24 splicing reporter plasmid for 24 h. RBM15B expression vector was titered to determine the optimal amount for in vivo splicing assays (0.5 μg). A, relative splicing activities were calculated using the ratios of luciferase to β-galactosidase activities with the activity of the control cells transfected with empty expression vectors (Control Vector) set at 1. The results shown represent three to six independent transfection experiments in which one plate was transfected, and each lysate was measured in quadruplicate. The error bars represent the S.E. and (p) denotes probability values. * denotes p < 0.001 compared with empty vector calculated using a non-parametric rank model. B, cell lysates were analyzed by immunoblotting for expression of RBM15B proteins (anti-RBM15B antibody). Relative protein loading was confirmed by immunoblotting with anti-actin antibody.
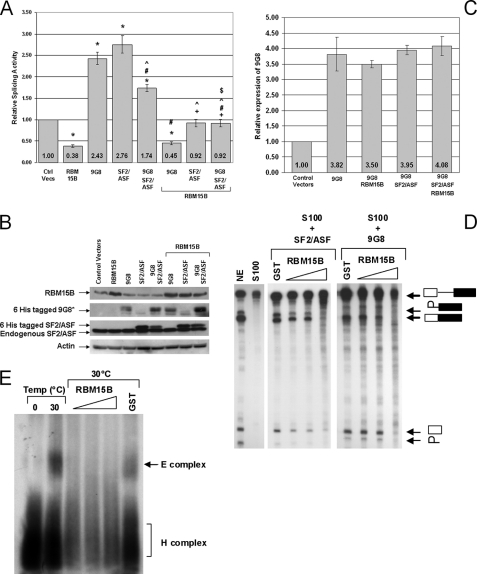
RBM15B Inhibits Splicing by Competition with SR Proteins
The double β-galactosidase/luciferase reporter system was also used to determine whether RBM15B could antagonize the stimulation of splicing induced by the SR proteins 9G8 and SF2/ASF. Expression vectors encoding RBM15B, 9G8, and SF2/ASF and control expression vectors were co-transfected with pTN24. As expected, a strong increase in the ratio of luciferase to β-galactosidase activity was observed in cells expressing 9G8 and/or SF2/ASF (Fig. 6A). Consistent with the results shown above, ectopic expression of RBM15B alone decreased splicing activity. Importantly, the relative splicing activity was significantly reduced in cells co-expressing RBM15B with 9G8 and/or SF2/ASF compared with cells expressing the SR proteins alone (Fig. 6A), demonstrating that RBM15B was indeed capable of antagonizing the positive effects of SR proteins on splicing. For these experiments, protein expression was verified by immunoblot (Fig. 6B), and the overall increase in 9G8 expression was confirmed by quantitative RT-PCR (Fig. 6C). To further demonstrate that RBM15B was a direct competitor of SR proteins, we performed in vitro splicing assays using the β-globin construct and S100 cell extract supplemented with recombinant SF2/ASF and 9G8 (Fig. 6D). S100 cytoplasmic extracts contain components of the spliceosome but lack the SR proteins needed for recruiting the factors required to catalyze the intron excision of the β-globin pre-RNA and to generate the spliced product (Fig. 6D, lane 2). Addition of 50 ng (∼2 pmol) of recombinant purified SF2/ASF or 9G8 to the S100 extract allowed the splicing reaction to occur (Fig. 6D, lanes 3 and 7). Importantly, when increasing amounts of GST-RBM15B were added to the S100 supplemented with SF2/ASF or 9G8, a dose-dependent inhibition of the splicing process was observed (Fig. 6D, lanes 4–6 and 8–10), whereas addition of control GST did not affect the amount of spliced product (Fig. 6D, lanes 3 and 7). Together, these data demonstrate that RBM15B is able to inhibit SF2/ASF- and 9G8-mediated splicing, indicating a direct competition between the positive splicing factors SF2/ASF and 9G8 and the splicing inhibitor RBM15B. Importantly, these in vitro data also allow us to conclude that RMB15B directly affects splicing regardless of its effect on RNA export (30) because detection of the spliced product in this assay does not require transport of the RNA.
FIGURE 6.
RBM15B suppresses in vivo and in vitro splicing mediated by SR protein SF2/ASF. Expression constructs for RBM15B WT and the SR proteins SF2/ASF and 9G8 were transiently co-transfected with the splicing reporter plasmid pTN24 for 24 h. A, relative splicing activities were calculated using the ratios of luciferase to β-galactosidase activities with the activity of the control cells transfected with empty expression vectors (Ctrl Vecs) set at 1. The results shown represent six independent transfection experiments in which one plate was transfected, and each lysate was measured in quadruplicate. The error bars represent the S.E. Statistically significant comparison data are indicated by the following symbols: *, p < 0.0007 for comparisons with control vectors; +, p < 0.01 for RBM15B with RBM15B/SF2 and RBM15B/9G8/SF2; #, p < 0.01 for 9G8 with 9G8/SF2, RBM15B/9G8, and RBM15B/9G8/SF2; ^, p < 0.01 for SF2 with 9G8/SF2, RBM15B/SF2, and RBM15B/9G8/SF2; and $, p < 0.01 9G8/SF2 with RBM15B/9G8/SF2. B, cell lysates were analyzed by immunoblotting for expression of the exogenous and endogenous proteins. *, His6-tagged 9G8 was detected by immunoprecipitation from cell lysates using nickel-nitrilotriacetic acid-agarose (Qiagen) and immunoblotting with anti-9G8-ZnK polyclonal antibody. Actin detection demonstrated that equal lysate volume loading resulted in comparable total protein levels. C, overall enforced expression of 9G8 was confirmed by quantitative RT-PCR. D, cytoplasmic S100 cell extract was used for an in vitro splicing assay using the pre-RNA of the human β-globin gene as substrate. S100 extract does not catalyze excision of the β-globin intron without the addition of SR proteins. The accumulation of the mature β-globin spliced product in the presence of HeLa NE (lane 1) is not observed using the S100 fraction alone (lane 2). Addition of 2 pmol/splicing reaction recombinant SF2/ASF (lane 3) or 9G8 (lane 7) proteins to the S100 fraction is sufficient to activate splicing of the β-globin pre-RNA. Increasing amounts (0.5, 2.5, and 5 pmol/splicing reaction) of GST-RBM15B FL protein inhibited the SF2/ASF- (lanes 4–6) and the 9G8-mediated splicing (lanes 8–10), whereas addition of GST alone (5 pmol/splicing reaction; lanes 3 and 7) had no effect. In vitro splicing assays using SF2/ASF were performed with HeLa nuclear extract produced according Mayeda and Krainer (48), whereas splicing reactions with 9G8 were performed using HeLaScribe nuclear extract (Promega). These two extracts differed in splicing activities (supplemental Data S7A). Higher activity was observed in the HeLaScribe as demonstrated by the low abundance of the intermediate splicing product (intron-exon 2). However, RBM15B inhibited splicing with both extracts. E, detection of the spliceosomal complex E was performed using the β-globin substrate and native horizontal agarose minigels. The ATP-independent spliceosomal complex E was detected after a 40-min incubation of the β-globin pre-RNA with HeLa nuclear extract at 30 °C (lane 2). At a temperature of 0 °C (lane 1), this complex did not form. Addition of increasing amounts (0.5, 2.5, and 5 pmol/splicing reaction) of GST-RBM15B FL protein inhibited formation of the E complex at 30 °C, whereas 5 pmol of GST did not affect this splicing step. The H complex is a nonspecific complex resulting from binding of hnRNP proteins to pre-RNAs.
To further elucidate whether RBM15B inhibited spliceosomal assembly, the formation of spliceosomal complexes (E, A, B, and C complexes) corresponding the sequential recruitment of the spliceosome subunits (26) was investigated using the β-globin pre-RNA and nuclear extract supplemented with recombinant GST and GST-RBM15B proteins (Fig. 6E and supplemental Data S7B). Without any recombinant proteins added, we detected the sequential formation of E (Fig. 6E), A, B, and C (supplemental Data S7B) complexes. Addition of 5 pmol of GST did not affect formation of spliceosomal complexes, whereas increasing amounts (0.5–5 pmol) of RBM15B strongly prevented formation of all complexes, demonstrating that RBM15B inhibited the first step of spliceosome assembly, the formation of the E complex corresponding to the recruitment of the U1 small nuclear ribonucleoprotein onto pre-RNAs.
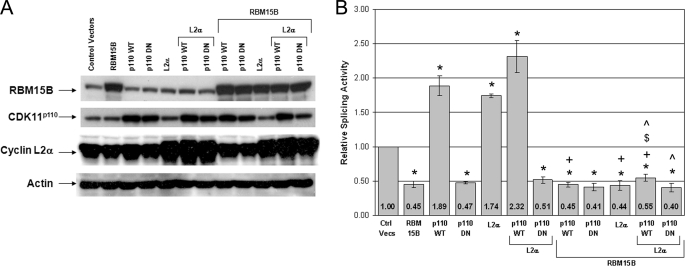
RBM15B Antagonizes Positive Effect of CDK11p110-Cyclin L2α Complex on Pre-mRNA Splicing
We have recently established that CDK11p110 interacts physically and functionally with cyclin L proteins to regulate splicing (8). We also showed that ectopic expression of CDK11p110 and cyclin Ls individually enhances splicing activity and, importantly, that co-expression of CDK11p110 and cyclin L2α resulted in a greater splicing activity than either of these two proteins alone (8). Because RBM15B interacts with and is found in a nuclear complex with CDK11 and cyclin L2α, we next addressed the question of whether RBM15B could modulate splicing promoted by the CDK11p110 and cyclin L2α complex. Using the same in vivo assay shown in Figs. 5 and 6, splicing activity was measured following co-expression of wild-type or kinase-dead CDK11p110 with cyclin L2α and RBM15B (Fig. 7B). Each protein was expressed individually and in combination to be able to compare the effects of individual expression and combined co-expression on the relative splicing activities. In these experimental conditions, the total amount of plasmid DNA in the transfection was equalized by addition of the appropriate empty vector. Protein expression levels for the “paired” transfections are shown in Fig. 7A. As expected, individual expression of CDK11p110 WT and cyclin L2α and their co-expression enhanced splicing, whereas expression of RBM15B and kinase-dead CDK11p110 (DN mutant) alone or in combination with cyclin L2α repressed splicing. Finally, co-expression of RBM15B with CDK11p110 WT and/or cyclin L2α inhibited splicing activities by 4–5-fold compared with the conditions without enforced expression of RBM15B. Thus, the protein RBM15B antagonizes the positive effect of the CDK11p110-cyclin L2α complex on splicing in vivo.
FIGURE 7.
Expression of RBM15B suppresses in vivo splicing enhancement by CDK11p110WT-cyclin L2α. WT or kinase-dead (DN) CDK11p110, cyclin L2α, RBM15B, and empty vector (Control Vectors) were transiently expressed alone or in combination with the splicing reporter construct pTN24 and harvested after 24 h as indicated in the figure. In all conditions, the total amount of plasmid DNA in the transfection was equivalent. A, cell lysates were analyzed by immunoblotting for expression of CDK11, cyclin L2α, and RBM15B. Actin detection demonstrated that equal lysate volume loading resulted in comparable total protein levels. B, relative splicing activities were calculated using the ratios of luciferase to β-galactosidase activities with the activity of the control cells transfected with empty expression vectors (Ctrl Vecs) set at 1. The results shown represent 5–10 independent transfection experiments in which one plate was transfected, and each lysate was measured in quadruplicate. The error bars represent the S.E. Statistically significant comparison data are indicated by the following symbols. The probability value is denoted as p. *, p < 0.003 for comparisons with the control vectors; +, p < 0.05 for RBM15B/CDK11p110WT with CDK11p110WT, RBM15B/CDK11p110WT-cyclin L2α with CDK11p110WT-cyclin L2α, RBM15B-cyclin L2α with cyclin L2α; $, p < 0.03 for RBM15B/CDK11p110WT-cyclin L2α with CDK11p110WT; ^, p < 0.02 for RBM15B/CDK11p110WT-cyclin L2α with cyclin L2α and RBM15B/CDK11p110DN-cyclin L2α with cyclin L2α.
DISCUSSION
Previous data from our laboratory demonstrated that CDK11p110 and its regulatory cyclin L subunits interact with several splicing factors and regulators of transcription to form at least two large macromolecular complexes involved in the regulation of transcription and RNA splicing (8, 9, 11). In addition, we established that CDK11p110-cyclin L complexes promote splicing (8, 11) most likely through phosphorylation of splicing factors such as 9G8 (11). Here we report the identification of RBM15B/OTT3 as a new binding partner for CDK11p110, cyclins L1α and L2α, and the splicing factor 9G8. We also demonstrated that RBM15B/OTT3 and CDK11p110-cyclin L complexes co-immunoprecipitate and that RBM15B/OTT3 co-elutes with CDK11p110, the cyclins L1α and L2α, and SR proteins including SF2/ASF in a large nuclear complex of ∼1-MDa molecular mass following size exclusion chromatography. Using in vitro pulldown assays, we mapped two distinct domains of RBM15B responsible for its direct interaction with the N-terminal extension of CDK11p110 as well as the cyclin Lα isoforms and the splicing factor 9G8.
RBM15B/OTT3 is a member of the Spen family of proteins (31). This family includes huSHARP (SMRT/HDAC1-associated repressor protein), muMINT (Msx2-interacting nuclear target), RBM15/OTT1, and RBM15B/OTT3 (supplemental Data S3). RBM15B (21) is the most highly related to RBM15/OTT1 (supplemental Data S3), which was originally discovered as a fusion partner with the gene MKL1 in the translocation t(1;22)(p13;q13) associated with childhood acute megakaryocytic leukemia (28, 29). All of these proteins contain three highly conserved N-terminal RNA binding motifs, suggesting that they play a role in mRNA synthesis, processing, or export, and a C-terminal SPOC domain, which mediates interaction with the co-repressors of transcription SMRT and NCoR. SHARP binds these co-repressors with a high affinity and strongly represses transcription (32, 33). Repression of transcription mediated by SHARP and its associated co-repressors SMRT and NCoR (32, 33) is thought to involve modification of chromatin structure by recruitment of histone deacetylases (HDACs), especially HDAC3, that directly bind to both co-repressors SMRT and NCoR (34). RBM15/OTT1 and RBM15B also interact with each other (22), strongly suggesting their involvement in the same biochemical processes. Importantly, Spen proteins also function in several signaling cascades including mitogen-activated protein kinase (MAPK) (35), Wnt (36), Notch (37), Hox10 (38), and epidermal growth factor receptor (39).
In the first report investigating the function of RBM15/OTT1 and RBM15B, their involvement in the regulation of transcription was not clear because neither of these proteins strongly represses transcription of reporter constructs in transfected cells (21). In agreement, the SPOC domain of these proteins interacts weakly with SMRT, and they lack the domains of SHARP (nuclear receptor-interacting domain and recombination signal binding protein-Jκ interaction domain) that target SHARP to specific promoters to repress transcription (33). However, these two proteins do interact with HDAC3 like the other family members (40). In addition, recent data indicated that RBM15 and the RMB15-MKL1 fusion protein can modulate the levels of multiple mRNAs including the proto-oncogene c-myc (41), although this study did not conclusively establish that these changes in gene expression resulted from the direct effects of RBM15 or RBM15-MKL-1 on transcription (41). Taken together, these recent data suggest that the two Spen family members RBM15/OTT1 and RBM15B may play some role in the regulation of transcription.
In contrast to the inconclusive data on the role of RBM15/OTT1 and RBM15B in transcription, existing data and the studies presented herein demonstrate that RBM15/OTT1 and RBM15B are involved in RNA splicing and/or mRNA export. The conclusion that RBM15/OTT1 plays a role in mRNA export is based on studies showing that RBM15/OTT1 binds to the RNA transport element and the mRNA export factor NFX1 (42). Furthermore, recent data by Zolotukhin et al. (30) support the hypothesis that RBM15/OTT1 promotes the recognition of spliced RNA-NFX1 complexes by the RNA helicase Dbp5, thereby contributing to efficient messenger ribonucleoprotein complex export through the nuclear pore complex. Similarly, RBM15B associates with NFX1 and contributes to mRNA export (30). In fact, RBM15B was initially isolated as an interactor of the Epstein-Barr virus early protein EB2, a viral protein that exhibits properties of an mRNA export factor (43). Of particular relevance to our study, Hiriart et al. (21) demonstrated that RBM15B/OTT3 regulates alternative splicing.
The fact that RBM15B is part of a large nuclear complex containing CDK11p110-cyclin L complexes as well as several SR proteins strongly reinforced the hypothesis that this RNA-binding protein could also be involved in RNA maturation upstream of its involvement in export. Here, we used in vitro and in vivo splicing assays to further investigate the involvement of RBM15B in constitutive exon splicing. We demonstrated for the first time that recombinant RBM15B/OTT3 added to nuclear extract strongly inhibits in vitro splicing of a human β-globin construct. Similarly, enforced expression of RBM15B in transiently transfected mammalian cells led to a significant reduction of pre-RNA reporter systems that monitor constitutive splicing, adding to the conclusion that RBM15B is a potent inhibitor of splicing. RBM15B possesses three RNA recognition motifs very similar to hnRNPs (31). Most hnRNPs inhibit splicing by competition with SR proteins (12, 44). Although SR proteins bound to exonic or intronic splicing enhancer sequences of the pre-RNA generally promote recruitment of the spliceosome subunits, many hnRNPs bind to exonic or intronic splicing silencer sequences and inhibit spliceosome activation or cancel the positive effects of SR proteins on splicing (12, 44). Thus, we tested whether RBM15B was able to antagonize the positive effect of SR proteins on splicing. For these studies, we performed in vitro experiments using S100 cytoplasmic extract, which is unable to catalyze in vitro splicing of the β-globin reporter pre-RNA unless it is supplemented with SR proteins. In our assay, S100 extract was supplemented with the recombinant SF2/ASF and 9G8 SR proteins, and addition of recombinant RBM15B decreased splicing mediated by both SF2/ASF and 9G8. Moreover, using the β-galactosidase/luciferase reporter system, we confirmed that RBM15B inhibited splicing promoted by SF2/ASF and 9G8 in transfected cells. These data led us to determine whether RBM15B could affect the assembly of spliceosome subunits. Using the β-globin in vitro splicing assay, we showed that RBM15B prevented formation of the E spliceosomal complex, the first step of the assembly. Altogether, these data indicate that RBM15B negatively regulates splicing by acting as a functional competitor of SR proteins.
Because we previously established that CDK11p110-cyclin L complexes enhance splicing (8), we also investigated whether RBM15B could negatively regulate the splicing mediated by CDK11p110 and/or cyclin L proteins. Using β-galactosidase/luciferase reporter in vivo splicing assays, we established that RBM15B antagonizes the positive effect on splicing of CDK11p110 or cyclin L2α expressed separately or co-expressed in HEK293T cells. Thus, the large molecular complex containing CDK11p110 either contains both positive and negative regulators of splicing and/or CDK11p110 forms multiple complexes alternatively containing either SR proteins or RBM15B. Based on our pulldown and co-IP experiments using CDK11, RBM15B, and SR proteins, we favor the hypothesis that both positive and negative regulators of splicing co-exist in CDK11p110 protein complexes, but we cannot rule out the possibility of multiple complexes. The synthesis of mRNA is indeed a highly dynamic process that recruits multiple positive and negative factors at different steps of transcription, elongation, and splicing. In addition, cyclin Ls also bind to CDK12 (45) and CDK13 (46, 47) to regulate splicing. Thus, it appears important to further investigate whether these other CDKs can also bind RBM15B via their cyclin L partners. The multiple potential interactions between CDKs, cyclin Ls, SR proteins, and hnRNPs such as RBM15B also raise the questions of relevance of these complexes in the regulation of the multiple processes occurring during mRNA synthesis. Beyond the description of these interactions, a major challenge remains to decipher how the CDKs affect pre-mRNA processing by the identification of specific phosphorylation substrates and by characterization of responses to various signaling pathways.
Supplementary Material
Acknowledgments
We especially thank Dr. Eperon for the pTN24 plasmid, Dr. Elledge for the yeast two-hybrid library, Dr. Hammarskjold for the 9G8 antibody, Dr. Bourgeois for the SF2/ASF plasmid, Prof. Hardy and Dr. Mereau for advice regarding the detection of spliceosomal complexes, Dr. A. Mayeda for helping to set up the β-globin in vitro splicing assay, and Dr. Mohmet Kocak and members of the Lahti laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM044088 and Cancer Center Support Grant P30CA021765. This work was also supported by the American Lebanese Syrian Associated Charities and INSERM.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Data S1–S8.
- CK2
- casein kinase II
- CDK
- cyclin-dependent kinase
- RNPS1
- RNA-binding protein with serine-rich domain
- IP
- immunoprecipitation
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- IVTT
- in vitro transcribed and translated
- NE
- nuclear extract
- NLS
- nuclear localization signal
- RRM
- RNA recognition motif
- SPOC
- Spen paralog and ortholog C-terminal
- SMRT
- silencing mediator for retinoic and thyroid receptors
- NCoR
- nuclear receptor co-repressor
- OTT
- one-twenty-two
- FL
- full length
- MKL1
- megakaryoblastic leukemia 1
- SHARP
- SMRT/HDAC1-associated repressor protein
- HDAC
- histone deacetylase
- Spen
- split end.
REFERENCES
- 1. Trembley J. H., Loyer P., Hu D., Li T., Grenet J., Lahti J. M., Kidd V. J. (2004) Prog. Nucleic Acid Res. Mol. Biol. 77, 263–288 [DOI] [PubMed] [Google Scholar]
- 2. Malumbres M., Harlow E., Hunt T., Hunter T., Lahti J. M., Manning G., Morgan D. O., Tsai L. H., Wolgemuth D. J. (2009) Nat. Cell Biol. 11, 1275–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cornelis S., Bruynooghe Y., Denecker G., Van Huffel S., Tinton S., Beyaert R. (2000) Mol. Cell 5, 597–605 [DOI] [PubMed] [Google Scholar]
- 4. Berke J. D., Sgambato V., Zhu P. P., Lavoie B., Vincent M., Krause M., Hyman S. E. (2001) Neuron 32, 277–287 [DOI] [PubMed] [Google Scholar]
- 5. Dickinson L. A., Edgar A. J., Ehley J., Gottesfeld J. M. (2002) J. Biol. Chem. 277, 25465–25473 [DOI] [PubMed] [Google Scholar]
- 6. Yang L., Li N., Wang C., Yu Y., Yuan L., Zhang M., Cao X. (2004) J. Biol. Chem. 279, 11639–11648 [DOI] [PubMed] [Google Scholar]
- 7. de Graaf K., Hekerman P., Spelten O., Herrmann A., Packman L. C., Büssow K., Müller-Newen G., Becker W. (2004) J. Biol. Chem. 279, 4612–4624 [DOI] [PubMed] [Google Scholar]
- 8. Loyer P., Trembley J. H., Grenet J. A., Busson A., Corlu A., Zhao W., Kocak M., Kidd V. J., Lahti J. M. (2008) J. Biol. Chem. 283, 7721–7732 [DOI] [PubMed] [Google Scholar]
- 9. Trembley J. H., Hu D., Hsu L. C., Yeung C. Y., Slaughter C., Lahti J. M., Kidd V. J. (2002) J. Biol. Chem. 277, 2589–2596 [DOI] [PubMed] [Google Scholar]
- 10. Loyer P., Trembley J. H., Lahti J. M., Kidd V. J. (1998) J. Cell Sci. 111, 1495–1506 [DOI] [PubMed] [Google Scholar]
- 11. Hu D., Mayeda A., Trembley J. H., Lahti J. M., Kidd V. J. (2003) J. Biol. Chem. 278, 8623–8629 [DOI] [PubMed] [Google Scholar]
- 12. Cáceres J. F., Kornblihtt A. R. (2002) Trends Genet. 18, 186–193 [DOI] [PubMed] [Google Scholar]
- 13. Trembley J. H., Tatsumi S., Sakashita E., Loyer P., Slaughter C. A., Suzuki H., Endo H., Kidd V. J., Mayeda A. (2005) Mol. Cell. Biol. 25, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petretti C., Savoian M., Montembault E., Glover D. M., Prigent C., Giet R. (2006) EMBO Rep. 7, 418–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu D., Valentine M., Kidd V. J., Lahti J. M. (2007) J. Cell Sci. 120, 2424–2434 [DOI] [PubMed] [Google Scholar]
- 16. Nybakken K., Vokes S. A., Lin T. Y., McMahon A. P., Perrimon N. (2005) Nat. Genet. 37, 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evangelista M., Lim T. Y., Lee J., Parker L., Ashique A., Peterson A. S., Ye W., Davis D. P., de Sauvage F. J. (2008) Sci. Signal. 1, ra7 [DOI] [PubMed] [Google Scholar]
- 18. Naik S., Dothager R. S., Marasa J., Lewis C. L., Piwnica-Worms D. (2009) Clin. Cancer Res. 15, 7529–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrmann A., Fleischer K., Czajkowska H., Müller-Newen G., Becker W. (2007) FASEB J. 21, 3142–3152 [DOI] [PubMed] [Google Scholar]
- 20. Valente S. T., Gilmartin G. M., Venkatarama K., Arriagada G., Goff S. P. (2009) Mol. Cell 36, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hiriart E., Gruffat H., Buisson M., Mikaelian I., Keppler S., Meresse P., Mercher T., Bernard O. A., Sergeant A., Manet E. (2005) J. Biol. Chem. 280, 36935–36945 [DOI] [PubMed] [Google Scholar]
- 22. Uranishi H., Zolotukhin A. S., Lindtner S., Warming S., Zhang G. M., Bear J., Copeland N. G., Jenkins N. A., Pavlakis G. N., Felber B. K. (2009) J. Biol. Chem. 284, 26106–26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laurent V., Fraix A., Montier T., Cammas-Marion S., Ribault C., Benvegnu T., Jaffres P. A., Loyer P. (2010) Biotechnol. J. 5, 314–320 [DOI] [PubMed] [Google Scholar]
- 24. Xiang J., Lahti J. M., Grenet J., Easton J., Kidd V. J. (1994) J. Biol. Chem. 269, 15786–15794 [PubMed] [Google Scholar]
- 25. Mayeda A., Screaton G. R., Chandler S. D., Fu X. D., Krainer A. R. (1999) Mol. Cell. Biol. 19, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee C. S., Das R., Reed R. (2003) Current Protocol in Molecular Biology (Ausubel F. M., Brent R., Kingston R. G., Moore D. D., Seidman J. G., Smith J. A., Strahl K. eds) 63, 27.1.1–27.1.5, John Wiley and Sons, New York: [Google Scholar]
- 27. Nasim M. T., Chowdhury H. M., Eperon I. C. (2002) Nucleic Acids Res. 30, e109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mercher T., Coniat M. B., Monni R., Mauchauffe M., Nguyen Khac F., Gressin L., Mugneret F., Leblanc T., Dastugue N., Berger R., Bernard O. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5776–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Z., Morris S. W., Valentine V., Li M., Herbrick J. A., Cui X., Bouman D., Li Y., Mehta P. K., Nizetic D., Kaneko Y., Chan G. C., Chan L. C., Squire J., Scherer S. W., Hitzler J. K. (2001) Nat. Genet. 28, 220–221 [DOI] [PubMed] [Google Scholar]
- 30. Zolotukhin A. S., Uranishi H., Lindtner S., Bear J., Pavlakis G. N., Felber B. K. (2009) Nucleic Acids Res. 37, 7151–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Colley S. M., Iyer K. R., Leedman P. J. (2008) IUBMB Life 60, 159–164 [DOI] [PubMed] [Google Scholar]
- 32. Shi Y., Downes M., Xie W., Kao H. Y., Ordentlich P., Tsai C. C., Hon M., Evans R. M. (2001) Genes Dev. 15, 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ariyoshi M., Schwabe J. W. (2003) Genes Dev. 17, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rebay I., Chen F., Hsiao F., Kolodziej P. A., Kuang B. H., Laverty T., Suh C., Voas M., Williams A., Rubin G. M. (2000) Genetics 154, 695–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng Y., Bommer G. T., Zhai Y., Akyol A., Hinoi T., Winer I., Lin H. V., Cadigan K. M., Cho K. R., Fearon E. R. (2007) Cancer Res. 67, 482–491 [DOI] [PubMed] [Google Scholar]
- 37. Kuroda K., Han H., Tani S., Tanigaki K., Tun T., Furukawa T., Taniguchi Y., Kurooka H., Hamada Y., Toyokuni S., Honjo T. (2003) Immunity 18, 301–312 [DOI] [PubMed] [Google Scholar]
- 38. Wiellette E. L., Harding K. W., Mace K. A., Ronshaugen M. R., Wang F. Y., McGinnis W. (1999) Development 126, 5373–5385 [DOI] [PubMed] [Google Scholar]
- 39. Chen F., Rebay I. (2000) Curr. Biol. 10, 943–946 [DOI] [PubMed] [Google Scholar]
- 40. Sawada T., Nishiyama C., Kishi T., Sasazuki T., Komazawa-Sakon S., Xue X., Piao J. H., Ogata H., Nakayama J., Taki T., Hayashi Y., Watanabe M., Yagita H., Okumura K., Nakano H. (2008) J. Biol. Chem. 283, 26820–26828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Niu C., Zhang J., Breslin P., Onciu M., Ma Z., Morris S. W. (2009) Blood 114, 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindtner S., Zolotukhin A. S., Uranishi H., Bear J., Kulkarni V., Smulevitch S., Samiotaki M., Panayotou G., Felber B. K., Pavlakis G. N. (2006) J. Biol. Chem. 281, 36915–36928 [DOI] [PubMed] [Google Scholar]
- 43. Juillard F., Hiriart E., Sergeant N., Vingtdeux-Didier V., Drobecq H., Sergeant A., Manet E., Gruffat H. (2009) J. Virol. 83, 12759–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Licatalosi D. D., Darnell R. B. (2010) Nat. Rev. Genet. 11, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen H. H., Wang Y. C., Fann M. J. (2006) Mol. Cell. Biol. 26, 2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Even Y., Durieux S., Escande M. L., Lozano J. C., Peaucellier G., Weil D., Genevière A. M. (2006) J. Cell. Biochem. 99, 890–904 [DOI] [PubMed] [Google Scholar]
- 47. Chen H. H., Wong Y. H., Geneviere A. M., Fann M. J. (2007) Biochem. Biophys. Res. Commun. 354, 735–740 [DOI] [PubMed] [Google Scholar]
- 48. Mayeda A., Krainer A. R. (1999) Methods Mol. Biol. 118, 309–314 [DOI] [PubMed] [Google Scholar]
- 49. Das R., Reed R. (1999) RNA 5, 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.