Abstract
The class III phosphatidylinositol 3-kinase (PI3KC3) plays a central role in autophagy. Rubicon, a RUN domain-containing protein, is newly identified as a PI3KC3 subunit through its association with Beclin 1. Rubicon serves as a negative regulator of PI3KC3 and autophagosome maturation. The molecular mechanism underlying the PI3KC3 and autophagy inhibition by Rubicon is largely unknown. Here, we demonstrate that Rubicon interacts with the PI3KC3 catalytic subunit hVps34 via its RUN domain. The RUN domain contributes to the efficient inhibition of PI3KC3 lipid kinase activity by Rubicon. Furthermore, a Rubicon RUN domain deletion mutant fails to complement the autophagy deficiency in Rubicon-depleted cells. Hence, these results reveal a critical role of the Rubicon RUN domain in PI3KC3 and autophagy regulation.
Keywords: Autophagy, Multifunctional Protein, Phosphatidylinositol 3-Kinase (PI3K), Protein Domains, Beclin 1, RUN Domain, Rubicon, UVRAG, Vps34
Introduction
The class III PI3K (PI3KC3)2 is essential for autophagy, protein sorting, phagosome maturation, and cytokinesis (1–4). PI3KC3 is composed of the human Vps34 (hVps34) catalytic subunit and two regulatory subunits (p150 and Beclin 1) that are highly conserved from yeast to human. Functional equivalents of Beclin 1/hVps34/p150 in yeast (Atg6/Vps15/Vps34) are essential for autophagy and vacuolar protein sorting (3, 5). The specificity of PI3KC3 in yeast is determined by different complex compositions. Two regulatory proteins, Atg14 and Vps38, direct the core PI3K complex to either the pre-autophagosome structure for autophagy or the endosome for vacuolar protein sorting (3, 6), respectively.
Along with several other groups, we have recently purified a mammalian PI3KC3 holocomplex that includes hVps34, p150, Beclin 1, UVRAG, Barkor/Atg14(L), and Rubicon (also called p120 and Baron) (7–12). PI3KC3 forms two mutually exclusive protein subcomplexes that localize to different subcellular organelles and execute distinct functions. One complex is composed of the PI3KC3 core complex (hVps34, p150, and Beclin 1) and Barkor/Atg14(L). Barkor/Atg14(L) is the targeting factor for this complex to nascent autophagosomes (7). The other complex consists of the PI3KC3 core complex and UVRAG. UVRAG positively regulates PI3KC3 activity and autophagy maturation (13). UVRAG is also required for endocytosis through its interaction with C-VPS/HOPS (13, 14). Rubicon (RUN domain protein as Beclin 1-interacting and cysteine-rich containing) is a newly identified PI3KC3 subunit (7–12). It serves as a negative regulator of autophagosome and endosome maturation (9, 10, 12).
Understanding the functional mode and structure basis of Rubicon in autophagy is crucial. Here, we demonstrate that Rubicon resides in the UVRAG subcomplex of PI3KC3. Interestingly, we detected no direct interaction between Rubicon and Beclin 1, although it was originally purified in the Beclin 1 complex. Instead, Rubicon physically interacted with UVRAG and hVps34. We have mapped the interaction domain of Rubicon with hVps34 to the RUN domain. The RUN domain is critical for the function of Rubicon in suppressing PI3KC3 lipid kinase activity and autophagy maturation.
EXPERIMENTAL PROCEDURES
Cell Culture
293T and U2OS cells were cultured in DMEM supplemented with 10% FBS. For tetracycline-inducible cells, Tet system approved FBS (Clontech) was used. For immunoprecipitation, cell extracts were incubated with 2 μg of antibody for 4 h and collected with protein A-Sepharose beads (Amersham Biosciences) for 4 h at 4 °C. The immunocomplexes were then washed three times and subjected to SDS-PAGE. Immunoblotting was performed following standard procedures.
Plasmids and Antibodies
The full-length cDNA for human Rubicon (KIAA0226) was generated by PCR on the basis of a partial cDNA clone purchased from Kazusa. Anti-Rubicon antibodies were generated by Abmart, Abgent, and Cell Signaling. Other antibodies used in this study include anti-FLAG (M2, Sigma), anti-HA (Roche Applied Science), anti-Myc (9E10, Santa Cruz Biotechnology), anti-tubulin (Santa Cruz Biotechnology), anti-UVRAG (Abgent), and anti-LC3 (Sigma). pcDNA5/FRT/TO (Invitrogen) was modified by introducing the coding sequence of a 3×FLAG tag (between NotI and XhoI) and designated as pcDNA5-FLAG. The Rubicon coding sequence was inserted between BamHI and NotI to generate the reading frame of Rubicon-FLAG, which was used for setting up a stable cell line. pcDNA4/TO (Invitrogen) was modified by introducing the coding sequence of a Myc tag (between NotI and XhoI) and designated as pcDNA4-Myc, into which Rubicon coding sequence were cloned between BamHI and NotI.
PI3K Kinase Assay
The procedure was modified from a protocol described previously (13). FLAG-tagged hVps34 expressed in cells was immunoprecipitated with M2 beads (Sigma) and extensively washed and preincubated at room temperature for 10 min in 60 μl of reaction containing 2 μg of sonicated phosphatidylinositol. The reaction was initiated by the addition of 5 μl of ATP mixture (1 μl of 10 mm unlabeled ATP, 1 μl of [γ-32P]ATP, and 3 μl of H2O) and incubated for 15 min at room temperature. The reaction was terminated, dried using a SpeedVac system, and resuspended in 20 μl of CHCl3/MeOH (1:1). The resultant assay product was separated on dried silica alumina thin liquid chromatography plates with running buffer (9:7:2 CHCl3/MeOH and 4 m NH4OH). Radiolabeled phosphatidylinositol 3-phosphate (PI(3)P) was visualized with a Fuji phosphorimaging scanner.
Recombinant Protein Purification
The protocol for recombinant protein production from insect cells is similar to a tandem affinity purification procedure described previously (7). For the recombinant proteins purified from mammalian cells, whole cell lysates from transfected 293T cells were prepared in tandem affinity purification buffer, and recombinant proteins were collected with M2 beads. After extensive washing, the proteins were eluted with FLAG peptide (0.1 mg/ml) and analyzed.
Immunostaining
Cells were grown on 6-well plates with coverslips. One day later, cells were fixed with 4% paraformaldehyde solution in PBS at room temperature for 10 min. After permeabilization with PBS buffer containing 0.1% Triton X-100 at room temperature for 20 min, cells were incubated with primary antibodies at 37 °C for 2 h. Upon extensive washing, cells were incubated with different fluorescent dye-conjugated secondary antibodies at 37 °C for 2 h. Slides were examined using a laser scanning confocal microscope (Zeiss LSM 510 META VIS/UV).
Inducible shRNA and Complementation Cell Lines
The shRNA sequences were designed with siRNA Target Finder provided by Ambion and were cloned into BglII and HindIII sites of the pSUPERIOR.puro vector (Oligoengine). The inducible shRNA cell lines were established according to the protocol. The shRNA coding sequence for human Rubicon knockdown is GATCCCCGCAGTGGAACAGAACAACCTTCAAAGAGGTTGTTCTGTTCCACTGCTTTTTA (with the targeted sequence of Rubicon underlined). The inducible knockdown stable cell lines were generated from U2OSTetR cells (7). To measure the knockdown efficiency, cells were treated with 500 ng/ml doxycycline for 3 days, followed by Western blotting against Rubicon. In complementation assays, RNAi-resistant wild-type Rubicon and mutants (in pcDNA4-GFP-FLAG) were generated by site-specific mutagenesis with primer p1 (catgctgcagtgcctggaagcCgtCgaGcaAaaTaaTccccgcctcctggctcagatc) and p2 (gatctgagccaggaggcggggAttAttTtgCtcGacGgcttccaggcactgcagcatg). These constructs were expressed in Rubicon-depleted cells, and stable cell lines were established by selection against 200 μg/ml Zeocin, 0.5 μg/ml blasticidin, and 1 μg/ml puromycin.
Transmission Electron Microscopy Analysis
For electron microscopy, cells were fixed with 2% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.2) for 2 h, followed by 1% osmium tetroxide in 0.1 m sodium cacodylate buffer (pH 7.2) for 2 h. Samples were blocked with 0.5% aqueous uranyl acetate overnight and subjected to low temperature dehydration and infiltration with a graded series of Epon/Araldite, followed by embedding in 100% Epon/Araldite. Thin sections (60 nm) were cut and stained with Reynolds lead citrate using an FEI Tecnai 12 transmission electron microscope.
RESULTS
Composition of the Rubicon-containing PI3KC3 Complex
The PI3KC3 complex has recently been purified in mammalian cells by several groups (7–11). In addition to the catalytic subunit hVps34, the regulatory subunit p150, Beclin 1, UVRAG, and Barkor/Atg14(L), a novel protein called Rubicon was also identified in this complex (7, 9, 10). Rubicon is involved in autophagosome maturation and is likely involved in endosome maturation also (9, 10). Rubicon contains 972 amino acids, with a coiled-coil domain, an FYVE-like domain, and a recognizable RUN domain that is shared by a group of proteins interacting with small GTPases (15–17).
The stoichiometry of the PI3KC3 complex is complicated by the formation of two mutually exclusive subcomplexes characterized by Barkor/Atg14(L) and UVRAG, respectively (7, 8). These two subcomplexes have organelle-specific distributions. The Barkor/Atg14(L) complex is localized to autophagosomes (7, 8), whereas the UVRAG complex resides predominantly on endosomes (14). Although Rubicon has been identified as a component of PI3KC3, it is still not clear which PI3KC3 subcomplex it associates with or whether it forms a unique subcomplex of PI3KC3. To clarify this question, we performed sequential affinity purifications of cellular complexes of Rubicon to elucidate its complex compositions.
Rubicon fused to a ZZ tag (two tandem repeats of the PrA IgG binding domain) at the amino terminus and a FLAG tag at the COOH terminus was used as bait (Fig. 1A) to purify its endogenous complex following a procedure described previously in U2OS cells (7, 11, 18). In the Rubicon complex, UVRAG, hVps34, p150, and Beclin 1 (but not Barkor/Atg14(L)) were detected by mass spectrometry (Fig. 1A). Rubicon consistently coprecipitated with hVps34 (Fig. 1B). These data confirmed the presence of Rubicon in the PI3KC3 complex and assigned Rubicon to the subcomplex with UVRAG.
FIGURE 1.
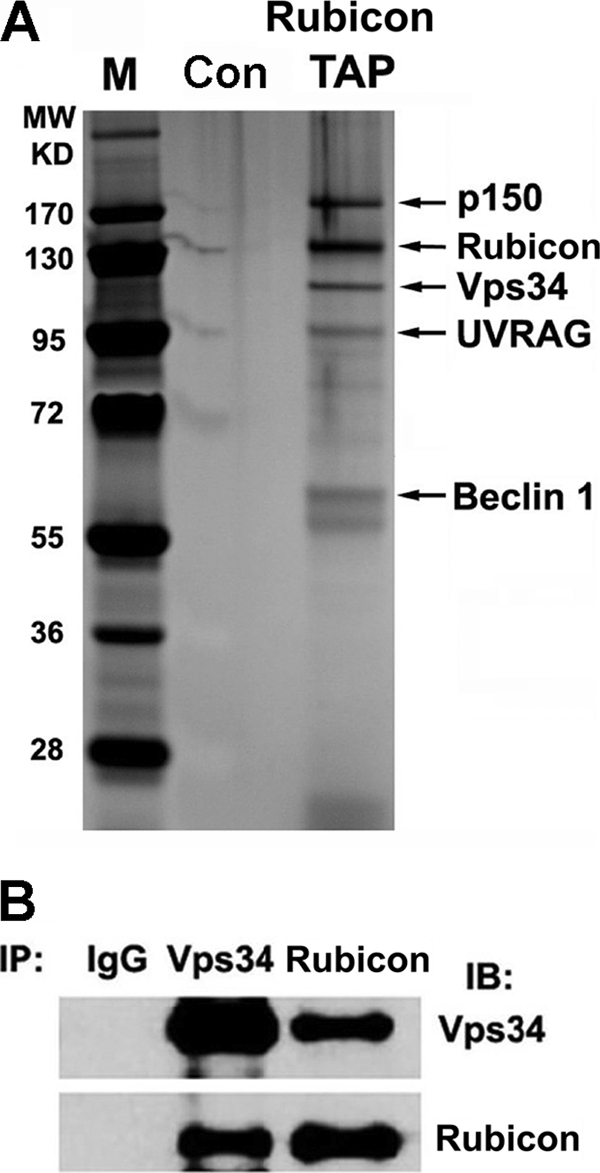
Rubicon is part of the PI3KC3 complex with UVRAG. A, cell extracts from U2OS cells expressing ZZ-Rubicon-FLAG at physiological levels or vector alone (control (Con)) were collected and purified sequentially through IgG beads and FLAG M2 beads. The resulting complex was resolved by 4–12% gradient SDS-PAGE and silver staining and analyzed by mass spectrometry. The identified proteins are indicated. M, molecular mass markers; TAP, tandem affinity purification. B, U2OS cell extracts were immunoprecipitated (IP) with anti-hVps34 or anti-Rubicon antibody, and the resulting immunoprecipitates were analyzed by Western blotting (immunoblotting (IB)).
Rubicon Binds Directly to UVRAG and hVps34
Although Rubicon has been shown to interact with the PI3KC3 complex (Fig. 1) (9, 10), the component through which it binds to PI3KC3 directly has not been characterized. To test which component(s) of PI3KC3 directly interact with Rubicon, we purified recombinant full-length Rubicon, hVps34, UVRAG, Beclin 1, and Barkor/Atg14(L) from baculovirus-infected insect cells or mammalian cells (Fig. 2A) and tested their interactions in an in vitro pulldown assay. All of these recombinant proteins were purified to near homogeneity (Fig. 2A). Both Rubicon and Barkor/Atg14(L) possessed an additional His6 tag. These two proteins were bound to a nickel-nitrilotriacetic acid (Ni-NTA) column first, followed by incubation of individual PI3KC3 subunits. In this in vitro pulldown assay, Rubicon directly bound to hVps34 and UVRAG (Fig. 2B). Interestingly, there was no direct interaction between Rubicon and Beclin 1, although Rubicon was identified in the Beclin 1 complex (9, 10). As a control, we found that Barkor/Atg14(L) interacted with Beclin 1, but neither hVps34 nor UVRAG bound directly (Fig. 2B).
FIGURE 2.
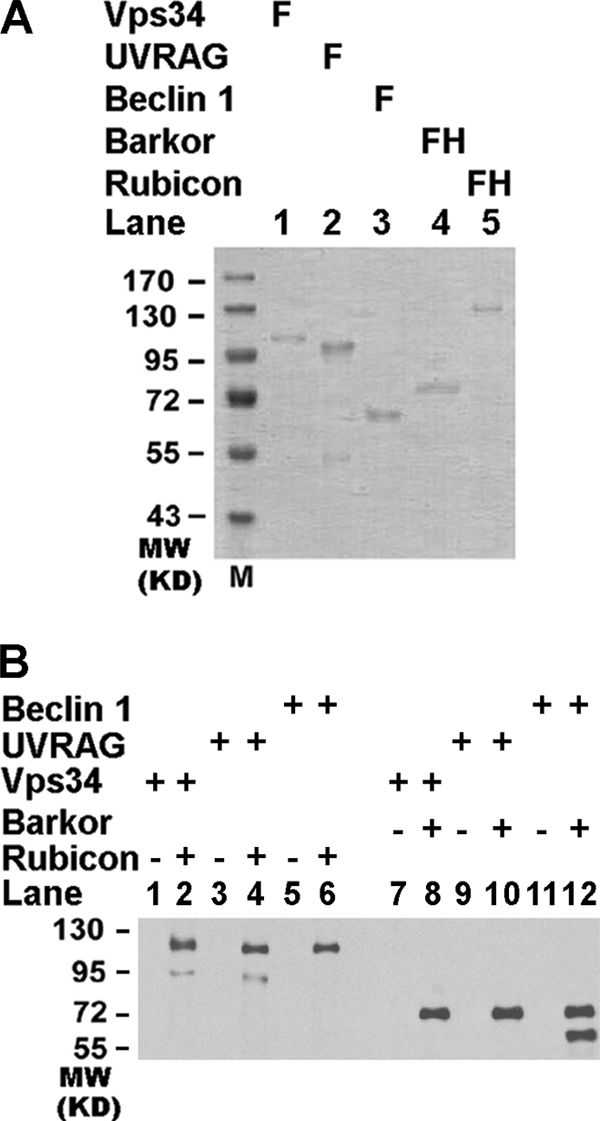
Rubicon physically interacts with UVRAG and hVps34. A, full-length FLAG-tagged (F) or FLAG-His-double-tagged (FH) recombinant hVps34, Beclin 1, and Barkor/Atg14(L). Rubicon and UVRAG were purified from baculovirus-infected insect cells or transfected HEK293T cells. B, Rubicon interacts directly with UVRAG and hVps34 in the purified system. Purified Rubicon-FLAG-His6 or Barkor/Atg14(L)-FLAG-His6 was first bound to Ni-NTA beads. Purified recombinant proteins for hVps34, UVRAG, and Beclin 1 were then allowed to pass through the Ni-NTA beads (even-numbered lanes). Bound proteins were detected by Western blotting using anti-FLAG antibody. The empty Ni-NTA beads were used as controls (odd-numbered lanes).
The RUN Domain of Rubicon Interacts with hVps34
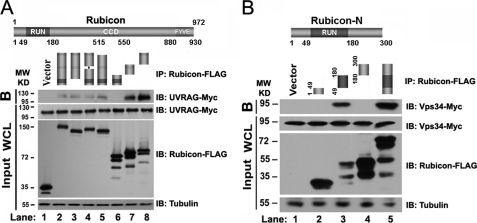
We mapped the interaction domain of Rubicon for UVRAG and hVps34 binding. A series of deletion mutants were generated, including deletions of the RUN domain, coiled-coil domain (CCD), and FYVE-like domain of full-length Rubicon, as well as three large polypeptide fragments containing the RUN domain, CCD, or FYVE domain (Fig. 3A). The interaction of these mutants with UVRAG was evaluated by an immunoprecipitation assay. Deletion of the RUN domain, CCD, or FYVE-like domain was not enough to abolish the interaction between Rubicon and UVRAG (Fig. 3A, lanes 3–5). Two fragments of Rubicon encompassing amino acids (aa) 300–600 and 600–972, but not the N-terminal fragment 1–300, interacted with UVRAG efficiently (Fig. 3A), which is consistent with a previous report that a large region (aa 393–849) of Rubicon binds to UVRAG (9).
FIGURE 3.
Rubicon binds to hVps34 via the RUN domain. A, in Rubicon, the three highlighted domains are the RUN domain, CCD, and FYVE-like domain. FLAG-tagged wild-type Rubicon and deletion mutants, including deletions of the RUN domain, CCD, or FYVE-like domain from full-length Rubicon, as well as three large polypeptide fragments containing the RUN domain, CCD, or FYVE-like domain were coexpressed with Myc-UVRAG in HEK293T cells. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibody. The bound proteins (B) or the 5% input extract was detected by immunoblotting (IB) using the antibodies indicated. WCL, whole cell lysate. B, the N terminus of Rubicon (aa 1–300) was further divided into three FLAG-tagged fragments containing aa 1–48, 49–180 (RUN domain), and 181–300, respectively. These deletion constructs were coexpressed with Myc-hVps34 in HEK293T cells. The cell extracts were immunoprecipitated with FLAG M2 beads, followed by Western blotting for Myc-hVps34 and FLAG-tagged Rubicon fragments.
Because we detected a direct interaction between Rubicon and hVps34, we further investigated the interaction domain of Rubicon with hVps34. The C terminus of Rubicon (aa 300–972) could pull down hVps34 probably through UVRAG (supplemental Fig. S1) (9, 10). Unexpectedly, the N-terminal region of Rubicon (aa 1–300), which does not interact with UVRAG (9, 10), strongly bound to hVps34 in the co-immunoprecipitation assay (Fig. 3B). We further divided the first 300 amino acids into three fragments, including the RUN domain alone (aa 49–180). Although the expression of the RUN domain alone was much lower compared with the other two fragments, the weakly expressed RUN domain bound to hVps34 strongly (Fig. 3B).
The RUN Domain Contributes to Rubicon Inhibition of hVps34 Lipid Kinase Activity
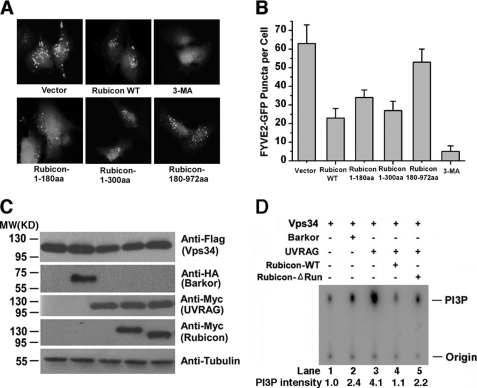
Binding of the Rubicon RUN domain to hVps34 led to the speculation that the RUN domain might be involved in PI3KC3 regulation. We measured PI3KC3 lipid phosphorylation activity in HEK293T cells expressing wild-type or mutant Rubicon. PI3KC3 phosphorylates the 3′-hydroxyl position of the phosphatidylinositol ring to produce PI(3)P (19, 20). The production of PI(3)P by PI3KC3 could be visualized and quantified by fluorescence of the GFP-tagged double FYVE finger of the Hrs protein (15). Because the FYVE probe specifically binds to PI(3)P, we could measure PI3KC3 activity by detecting FYVE fluorescence. PI(3)P production was compromised in Rubicon-expressing cells compared with that in wild-type cells (Fig. 4, A and B), consistent with the inhibitory role of Rubicon in PI3KC3 lipid phosphorylation (10). We then expressed different fragments of Rubicon to test their effect on PI3KC3 activity. Because the expression of the RUN domain alone was low probably because its expression requires the surrounding regions (Fig. 3B), we expressed the N terminus of Rubicon (aa 1–300) and a large C terminus (aa 180–972) to score their effect on PI(3)P production. The RUN domain-containing fragment was efficient in suppressing PI(3)P production (Fig. 4, A and B), suggesting that Rubicon binding of hVps34 via the RUN domain plays an important role in hVps34 inhibition. As a positive control, PI(3)P production could be efficiently blocked by treatment with the PI3K inhibitor 3-methyladenine (Fig. 4, A and B). In addition, the N terminus of Rubicon also reduced the number of LC3 puncta (supplemental Fig. S2), suggesting that autophagy is also blocked by this fragment.
FIGURE 4.
The RUN domain of Rubicon contributes to efficient inhibition of hVps34 lipid kinase activity. A, vector alone, the N-terminal region (aa 1–300), and the C-terminal region (aa 181–972) of Rubicon were coexpressed with GFP-2×FYVE in HEK293T cells. GFP-2×FYVE puncta were observed under a fluorescence microscope. 3-MA, 3-methyladenine. B, quantitative analysis (summarized from 100 cells) of GFP-2×FYVE puncta in the cells described in A. C, FLAG-tagged hVps34 was coexpressed in HEK293T cells with one of the following Myc-tagged proteins: Beclin 1, Barkor/Atg14(L), UVRAG, Rubicon, or Rubicon-ΔRUN. D, cell lysates from the cells described in C were immunoprecipitated with M2 beads and assayed for PI3K kinase activity in a TLC assay. The phosphorylation product [32P]PI(3)P was separated by TLC and further visualized using a Fuji phosphorimaging scanner.
To consolidate the function of the Rubicon RUN domain in PI3KC3 regulation, we measured the lipid phosphorylation activity in an in vitro TLC lipid kinase assay. In this assay, the production of PI(3)P by PI3KC3 could be detected and quantified by radiolabeling the phosphate group of phosphoinositol (19, 20). hVps34 was coexpressed with Barkor/Atg14(L), UVRAG, and Rubicon (Fig. 4C), and its lipid kinase activity was tested accordingly. Both Barkor/Atg14(L) and UVRAG stimulated hVps34 lipid kinase activity (Fig. 4D), supporting their positive roles in hVps34 activation. Coexpression of wild-type Rubicon efficiently blocked UVRAG-mediated hVps34 activity (Fig. 4D, lane 4), indicating that Rubicon can antagonize UVRAG-mediated hVps34 stimulation. A Rubicon mutant with a RUN domain deletion significantly reduced the inhibitory activity of Rubicon in PI(3)P production (∼40%) (Fig. 4D, lane 5). This suggests that the RUN domain of Rubicon contributes to its efficient suppression of PI3KC3 lipid kinase activity.
The RUN Domain Is Important for Rubicon Function in Autophagy
Two recent studies suggested that Rubicon plays a negative role in autophagosome maturation (9, 10). We investigated the function of the RUN domain in autophagy regulation. We employed two of the most representative approaches for autophagy regulation, electron microscopy analysis and p62 degradation, to study the structural requirement of the RUN domain in autophagy.
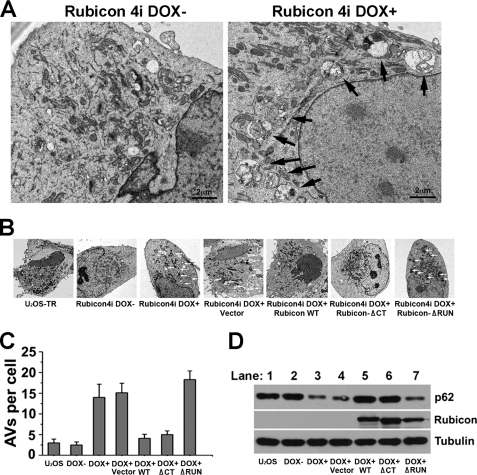
In Rubicon-depleted cells, autophagosome maturation was promoted, which led to the accumulation of late autophagic vacuoles, probably due to the inability of lysosomes to digest increased autophagosomes in time. Autophagic vacuoles could be detected under the electron microscope. We performed the electron microscopy analysis with wild-type and Rubicon RNAi-depleted cells. Rubicon was depleted by an shRNA against Rubicon that was inducibly expressed upon the addition of doxycycline in U2OS cells. In Rubicon-depleted cells, we observed abundant mature autophagic vacuoles that were highly enriched in cytosolic content and organelles (Fig. 5A, arrows). Autophagic vacuoles were rarely observed in parental U2OS or shRNA-non-induced cells (Fig. 5A). This observation is consistent with previously published results (9, 10).
FIGURE 5.
The Run domain is important for autophagy suppression by Rubicon. A, Rubicon shRNA-non-induced U2OS cells (4i DOX−) and doxycycline-induced RNAi-depleted cells (4i DOX+) were observed under a transmission electron microscope. Autophagic vacuoles are indicated by arrows. Clone 4 expressing shRNA against Rubicon (Rubicon 4i) was used in this study. Scale bars = 2 μm. B, Parental U2OS cells (U2OS cells expressing the tet repressor (U2OS-TR)), Rubicon shRNA-non-induced U2OS cells (4i DOX−), RNAi-depleted cells (4i DOX+), and Rubicon-depleted cells complemented with vector alone, FLAG-tagged wild-type Rubicon, the Rubicon C-terminal deletion (ΔCT; aa 800–972), and the Rubicon RUN domain deletion (ΔRUN; aa 1–180) were observed under a transmission electron microscope. Autophagic vacuoles are marked by arrows. C, quantitative results (summarized from 50 cells/cell line) of the autophagic vacuoles (AVs) described in B. D, Rubicon was detected by anti-FLAG M2 antibody; p62 and tubulin were detected with the respective antibodies by immunoblotting of the cells described in B.
Rubicon depletion also led to the accumulation of LC3 cytosolic puncta representing autophagic vacuoles (supplemental Fig. S3). A significant portion of LC3 cytosolic puncta colocalized with Rab5 (an early endosome marker), Rab7 (a late endosome marker), and LAMP1 (a lysosome marker) (supplemental Fig. S1), suggesting that late autophagic vacuoles that fuse with endosomes and lysosomes accumulate in Rubicon-depleted cells. These data confirmed that autophagosome maturation is accelerated when Rubicon is depleted, consistent with previously published results (9, 10).
We further tested whether ectopically expressed Rubicon could slow autophagosome maturation and therefore relieve the accumulation of mature autophagic vacuoles. We established Rubicon RNAi-depleted cells stably complemented with RNAi-resistant wild-type Rubicon (supplemental Fig. S4). Rubicon RUN domain deletion (aa 1–180) and C-terminal deletion (aa 800–972) mutants were also stably expressed in Rubicon-depleted U2OS cells. Autophagic vacuoles were observed and counted in these cells under an electron microscope. Expression of wild-type Rubicon or the Rubicon mutant with the C-terminal deletion was sufficient to ablate the autophagic vacuole accumulation in Rubicon-depleted cells, but expression of the Rubicon mutant with the RUN domain deletion failed to complement this phenotype (Fig. 5, B and C). Hence, the RUN domain is crucial for the negative function of Rubicon in autophagosome maturation.
Degradation of the selective autophagy substrate p62 (21, 22) was also examined in Rubicon-depleted and Rubicon-complemented cells. p62 autophagic degradation was accelerated in Rubicon-depleted cells, which was rescued by complementation with wild-type Rubicon and the C-terminal deletion mutant, but not with the RUN domain deletion mutant (Fig. 5D). This result is consistent with the EM results, indicating that the RUN domain of Rubicon is important for the negative regulator role of Rubicon in autophagic degradation.
DISCUSSION
Rubicon is a recently identified PI3KC3 subunit that is required for suppression of autophagosome maturation. In this study, we demonstrated that Rubicon preferentially interacts with UVRAG in the endosomal PI3KC3 subcomplex. In addition to a physical interaction with UVRAG, Rubicon also binds to the catalytic subunit hVps34 via its RUN domain. This binding contributes to its suppression of hVps34 lipid kinase activity. Finally, the RUN domain is critical for Rubicon function in autophagosome maturation and autophagic degradation. Although the RUN domain is proposed to interact with small GTPases, we have revealed here a unique role of the RUN domain in hVps34 interaction and autophagy regulation.
Several proteins have been reported to interact with hVps34, including Rab5 (23–27). Recent biochemical purifications from several laboratories have disclosed six to seven subunits of PI3KC3, including hVps34, p150/Vps15, Beclin 1/Atg6, UVRAG, Barkor/Atg14(L), and Rubicon (7, 9–11). All of them except Rubicon have a functional counterpart in yeast. Also, similar to the yeast PI3K complex, mammalian PI3KC3 forms two mutually exclusive subcomplexes (7, 9–11). One subcomplex contains the core complex and Barkor/Atg14(L). Because of its localization on early autophagosomes and functionality in autophagosome formation (7), we refer to this complex as the autophagosomal PI3KC3 complex. The other subcomplex contains the core complex, UVRAG, and Rubicon. This complex is localized mainly to endosomes; we therefore refer to it as the endosomal PI3KC3 complex. Emerging evidence suggests that this subcomplex is involved in both autophagosome and endosome maturation (9, 10, 12).
However, the architecture of the mammalian PI3KC3 complex is slightly different from its counterpart in yeast. For example, Barkor/Atg14(L) and UVRAG interact with hVps34 via Beclin 1 in mammalian cells (7, 13), whereas two regulatory proteins, Atg14 (Barkor-like) and Vps38 (UVRAG-like) mediate the interaction between hVps34 and Beclin 1/Atg6 (3). Furthermore, because Rubicon and its autophagy-inhibiting functions through the PI3KC3 complex have no homologous function in yeast, the autophagy inhibition activity of Rubicon could be unique to multicell organisms. However, the mechanism of PI3KC3 and autophagy inhibition by Rubicon is largely unknown. Interestingly, we found no direct interaction between Beclin 1 and Rubicon, although Rubicon was named as a Beclin 1-interacting protein (9, 10). Instead, Rubicon interacts with UVRAG and hVps34 directly (Fig. 2). The physical interaction between Rubicon and hVps34 might help to explain its inhibitory effect on PI3K activity. The data presented in this study support a critical role of the RUN domain in the inhibitory function of Rubicon. However, the interaction between the central region of Rubicon and UVRAG (9, 10) might also contribute to the negative regulation of Rubicon, which needs further investigation. We also cannot exclude the possibility that Rubicon could interact with hVps34 through region(s) other than the RUN domain. Nevertheless, our data demonstrate that the RUN domain is not only sufficient for hVps34 binding but also critical for regulating PI3KC3 lipid kinase activity and autophagy.
The RUN domain was originally found in a group of proteins that interact with small GTPases (15–17). It was therefore proposed as a binding surface for small GTPases. Our results indicate that the RUN domain can also bind hVps34. This may provide a possible link between many RUN domain-containing proteins and PI3K regulation, the detailed mechanism of which is still largely unknown and needs to be further explored through more structural and functional analyses.
In this study, we have identified a previously unknown function of the RUN domain of Rubicon in PI3KC3 and autophagy regulation. Dissecting the molecular mechanism of this molecule will provide exciting insight into the mechanism of the autophagic pathways. Such information should be critical for the development of novel therapeutic tools for multiple human pathological conditions with dysfunctions of autophagy.
Supplementary Material
Acknowledgments
We thank Cell Signaling and Abgent for anti-Rubicon antibodies and Livy Wilz for critical reading of the manuscript.
The work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA133228 (to Q. Z.). The work was also supported by a New Investigator Award for Aging from the Ellison Medical Foundation and the Hellman Family Fund (to Q. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PI3KC3
- class III phosphatidylinositol 3-kinase
- hVps34
- human Vps34
- PI(3)P
- phosphatidylinositol 3-phosphate
- Ni-NTA
- nickel-nitrilotriacetic acid
- CCD
- coiled-coil domain
- aa
- amino acids.
REFERENCES
- 1. Sagona A. P., Nezis I. P., Pedersen N. M., Liestøl K., Poulton J., Rusten T. E., Skotheim R. I., Raiborg C., Stenmark H. (2010) Nat. Cell Biol. 12, 362–371 [DOI] [PubMed] [Google Scholar]
- 2. Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. (1993) Science 260, 88–91 [DOI] [PubMed] [Google Scholar]
- 3. Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S. (2008) Nat. Cell Biol. 10, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao Y., Klionsky D. J. (2007) Cell Res. 17, 839–849 [DOI] [PubMed] [Google Scholar]
- 6. Obara K., Sekito T., Ohsumi Y. (2006) Mol. Biol. Cell 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Itakura E., Kishi C., Inoue K., Mizushima N. (2008) Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009) Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 10. Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009) Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun Q., Fan W., Zhong Q. (2009) Autophagy 5, 713–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun Q., Westphal W., Wong K. N., Tan I., Zhong Q. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19338–19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. (2006) Nat. Cell Biol. 8, 688–699 [DOI] [PubMed] [Google Scholar]
- 14. Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mari M., Macia E., Le Marchand-Brustel Y., Cormont M. (2001) J. Biol. Chem. 276, 42501–42508 [DOI] [PubMed] [Google Scholar]
- 16. Kukimoto-Niino M., Takagi T., Akasaka R., Murayama K., Uchikubo-Kamo T., Terada T., Inoue M., Watanabe S., Tanaka A., Hayashizaki Y., Kigawa T., Shirouzu M., Yokoyama S. (2006) J. Biol. Chem. 281, 31843–31853 [DOI] [PubMed] [Google Scholar]
- 17. Recacha R., Boulet A., Jollivet F., Monier S., Houdusse A., Goud B., Khan A. R. (2009) Structure 17, 21–30 [DOI] [PubMed] [Google Scholar]
- 18. Fan W., Tang Z., Chen D., Moughon D., Ding X., Chen S., Zhu M., Zhong Q. (2010) Autophagy 6, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Backer J. M. (2008) Biochem. J. 410, 1–17 [DOI] [PubMed] [Google Scholar]
- 20. Lindmo K., Stenmark H. (2006) J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 21. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 23. Stein M. P., Feng Y., Cooper K. L., Welford A. M., Wandinger-Ness A. (2003) Traffic 4, 754–771 [DOI] [PubMed] [Google Scholar]
- 24. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. (2006) Cell 126, 191–203 [DOI] [PubMed] [Google Scholar]
- 25. Simonsen A., Lippé R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. (1998) Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
- 26. Wurmser A. E., Emr S. D. (2002) J. Cell Biol. 158, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. (1999) Nat. Cell Biol. 1, 249–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.